Abstract
Induction of the Arf tumor suppressor gene by elevated thresholds of mitogenic signals activates a p53-dependent transcriptional response that triggers either growth arrest or apoptosis, thereby countering abnormal cell proliferation. Conversely, Arf inactivation is associated with tumor development. Expression of Arf in tissues of adult mice is difficult to detect, possibly because its induction leads to the arrest or elimination of incipient tumor cells. We replaced coding sequences of exon 1β of the mouse cellular Arf gene with a cDNA encoding GFP, thereby producing Arf-null animals in which GFP expression is driven by the intact Arf promoter. The Arf promoter was induced in several biologic settings previously shown to elicit mouse p19Arf expression. Inactivation of Arf in this manner led to the outgrowth of tumor cells expressing GFP, thereby providing direct evidence that the Arf promoter monitors latent oncogenic signals in vivo.
The Ink4a-Arf locus encodes two tumor suppressor proteins, p16Ink4a and p19Arf, that up-regulate the activities of the retinoblastoma protein (Rb) and the p53 transcription factor, respectively (1). The p16Ink4a protein inhibits the activity of cyclin D-dependent kinases, thereby maintaining Rb in its hypophosphorylated, growth-suppressive state, whereas p19Arf antagonizes Mdm2 activity to induce a p53 transcriptional response that leads to cell cycle arrest or apoptosis, depending on the biologic setting. Targeted disruption of Ink4a, Arf, or both genes in the mouse strongly predisposes them to tumor development (2-5); similarly, their inactivation by mutation, deletion, or epigenetic silencing is observed in many human cancers (6, 7).
The Ink4a-Arf locus is unusual because the transcripts of two alternative first exons are spliced to that of a shared second exon, whose sequences are translated in two different reading frames (8). However, despite this economical gene structure, the separate Ink4a and Arf promoters can differentially respond to input signals and be independently silenced in tumors. Arf is not usually expressed in normal tissues but is induced by sustained and elevated mitogenic signals that may stem from oncogene activation (1). For example, whereas physiologic thresholds of Myc and Ras signaling do not activate Arf gene expression, overexpression of Myc (9) and sustained signaling by oncogenic Ras (10) induce Arf to trigger p53 activity. This process counters aberrant mitogenic signaling by inducing growth arrest or apoptosis in cells that might otherwise give rise to tumors. Metaphorically, then, Arf acts as a fuse that monitors mitogenic current and is activated when signaling circuits are overloaded. Understanding how the Arf promoter distinguishes between normal and abnormal signaling thresholds remains a challenging problem (1).
Although much of p19Arf's activity depends on p53, inactivation of Arf in mice lacking both Mdm2 and p53 leads to a much more rapid appearance of tumors than that observed in animals lacking any one or two of these genes (11). Tumors spontaneously appear even before these triple knockout animals reach reproductive age, and multiple tumors routinely arise from different tissue types in individual animals. Moreover, introduction of Arf into cells lacking p53, or both Mdm2 and p53, can arrest proliferation, albeit much less efficiently than in cells that retain Mdm2 and p53 function (11, 12). Therefore, p19Arf, Mdm2, and p53 cannot function in a strictly linear signaling pathway, and p19Arf has activities that do not depend on Mdm2 and p53. At least some of the p53-independent effects of p19Arf might be mediated by its ability to inhibit ribosomal RNA processing (13) and to indirectly induce genes regulating other antiproliferative and apoptotic programs (14, 15).
It is unlikely that the normal physiologic role of Arf is to guard against tumorigenesis, particularly in short-lived species, such as mice, that rarely develop cancers spontaneously. To date, the only context in which Arf loss has been found to affect normal development is in the eyes of newborn mice, where excess retrolental tissue and persistence of the hyaloid vasculature in the vitreous result in destruction of both the lens and neuroretina (16). To ultimately discern how the Arf gene is regulated in vivo, we replaced Arf exon 1β coding sequences with a cassette specifying GFP, thereby disabling Arf function and placing GFP under the control of the cellular Arf promoter. Although most normal tissues of Arf GFP/GFP mice expressed negligible GFP levels, tumors and ocular masses exhibited vivid green fluorescence, indicating that the Arf promoter responds to aberrant signals in these pathologic settings.
Materials and Methods
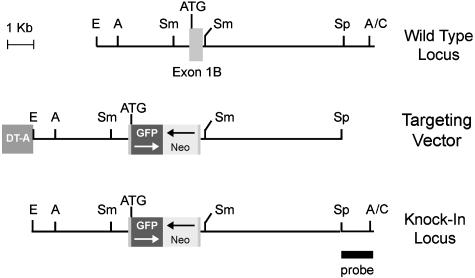
Targeting Vector. A 1-kb SmaI fragment containing exon 1β (Fig. 1) was altered by using PCR-based primers to create a site of restriction for NcoI at the Arf ATG codon and an XhoI site 16 nucleotides 5′ to the 3′ splice donor site. An NcoI-HindIII restriction fragment containing cDNA specifying enhanced GFP and a 3′ polyadenylation signal (Clontech) was ligated in orientation opposite to a neomycin resistance (neo) gene flanked by its own 5′ promoter and 3′ polyadenylation signal (Stratagene) to create an NcoI-XhoI fragment that was substituted for coding sequences in Arf exon 1β. This substitution left intact the Arf promoter and 5′ untranslated sequences and recreated the ATG codon to enable GFP translation. Sequences encoding the exon 1β splice donor site were retained 3′ of the neo gene but are prevented from being transcribed by the GFP polyadenylation signal. A cassette encoding the diphtheria toxin A-chain (DT-A) was appended to the 5′ end of the vector to allow selection against nonhomologous recombination elsewhere in the mouse genome (17).
Fig. 1.
Targeting of the Arf locus surrounding exon 1β. A schematic map of the region flanking exon 1β (Top), relevant sequences in the targeting vector (Middle), and the knock-in allele (Bottom) are illustrated. Arf coding sequences were replaced by a cassette encoding enhanced GFP and the neomycin-resistance gene (neo) in opposite orientations (arrows). The neo gene includes its own 5′ promoter, whereas GFP expression is driven by the Arf promoter; both neo and GFP terminate with 3′ polyadenylation signals. The targeting vector contains a gene encoding the diphtheria toxin A chain (DT-A), which is toxic unless eliminated and therefore selects against nonhomologous recombination of the targeting vector elsewhere in the mouse genome. The probe used to score the different alleles is illustrated at the bottom right. ATG refers to the position of the GFP initiation codon. Restriction sites for EcoR1 (E), AflII (A), SmaI (Sm), SpeI (Sp), and ClaI (C) are indicated.
Homologous Recombination and Generation of Knock-in Mice. W9.5 129/SvJ embryonic stem (ES) cells were electroporated with the linearized targeting vector and selected with G418 (Gibco Invitrogen) as described (3). DNAs from 490 drug-resistant colonies were screened by Southern blotting after restriction with AflII and hybridization with a unique sequence SpeI-ClaI probe (Fig. 1 Bottom). Three ES clones exhibiting correct homologous recombination and normal karyotypes were injected into C57BL/6 blastocysts, which were implanted into the uteri of pseudopregnant B6CBA F1/J foster mothers and allowed to develop to term. Male chimeras derived from the three ES clones were mated to C57BL/6 females that transmitted the knock-in allele through the germ line. As anticipated (3), heterozygous offspring gave rise to normally developing animals that segregated the knock-in allele at the expected Mendelian frequency. F1 animals were interbred to generate F2 littermates used in subsequent studies. The three Arf-GFP strains were phenotypically identical.
Mouse Maintenance, Interbreeding, and Imaging. Where indicated, cohorts of Arf GFP/GFP animals received single 4-Gy exposures of ionizing radiation 5 days after birth or were left untreated and monitored for tumor development. ArfGFP/GFP mice were bred to Eμ-Myc C57BL/6 transgenic mice to generate Arf+/GFP F1 offspring expressing the Myc transgene. Animals were examined at least twice weekly for signs of disease. Mice were maintained and humanely killed when moribund in accordance with Institutional Animal Care and Use Committee guidelines. Tumors were harvested immediately and were snap-frozen in liquid nitrogen. Survival curves were calculated as described (18).
Animals were examined by total-body fluorescence imaging (19, 20) during the course of tumor development. Selective excitation of GFP was produced by using an Illumatool TLS system with an excitation band-pass filter at 470 nm, and emitted fluorescence was collected through a long-pass filter at 515 nm (Lightools Research, Encinitas, CA) on a three-chip cooled color charge-coupled device camera (C5810, Hamamatsu Photonics Systems, Hamamatsu City, Japan). IMAGE PRO PLUS 4.5.1 software (Media Cybernetics, Silver Spring, MD) was used to analyze digital images of 1,024 × 724 pixels captured directly on a computer equipped with a high-resolution light-emitting diode screen.
Flow Cytometry. Bone marrow, spleen, and lymph node cells were harvested from precancerous and lymphomatous mice and red blood cells were lysed in hypotonic buffer (18). Cells collected by low-speed centrifugation were suspended at 1 × 106 per 0.1 ml and incubated for 30 min on ice in 2% FBS in PBS containing a 1:200 dilution of photofluor-conjugated antibody to B220 (RA3-6B2, BD Biosciences Pharmingen). Cells were washed, resuspended in staining buffer, and analyzed by flow cytometry for GFP (excitation at 488 nm; emission at 519 nm) and for B220 binding (excitation at 633 nm; emission at 660 nm).
Mouse Embryo Fibroblast (MEF) Cultures and Viral Transduction. MEFs from midgestation embryos were explanted and propagated as described (3). pBABE retroviral vectors expressing either a puromycin-resistance gene (puro) alone or both puro and oncogenic Harvey Ras (21) (a gift from S. Lowe, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) were packaged by using ecotropic helper virus in human kidney 293T cells and used to infect MEFs (9). Primary MEFs at passages 2-5 were plated under sparse conditions (2 × 105 cells per 100-mm-diameter culture dish) on day -1, infected on day 0 with retroviruses, and selected for 2 days after infection with 2 μg/ml puromycin. Cultures were harvested on days 3 and 4 after infection. Cells diluted to the original concentration and replated on day 4 were harvested 3 days later (day 7). Cells were assayed by flow cytometry for GFP fluorescence and by immunoblotting for GFP protein (Ab7.1 and 13.1, 0.4 μg/ml, Roche Diagnostics) and p19Arf expression (0.5 μg/ml Ab80, AbCam, Cambridge, MA) (9).
Tumor Pathology and GFP Staining. Formalin-fixed, paraffinembedded tumor specimens were sectioned at 5 μm, stained with hematoxylin and eosin, and examined by light microscopy. Immunocytochemistry for various discriminating markers was performed as described in detail (22, 23). Partially dissected eyes from newborn mice were studied by light and fluorescence microscopy. Detection of GFP was performed with 12-μm frozen sections of testis fixed in 4% paraformaldehyde in PBS. Sections were incubated for 30 min at room temperature in PBS containing 0.1% Triton X-100 and 10% goat serum followed by overnight incubation at 4°C with rabbit polyclonal anti-GFP (1:1,000; Molecular Probes) and 1-h incubation at room temperature with cy-3-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch). Antibodies were diluted in PBS containing 0.1% Triton X-100 and 2% goat serum. Stained tissues were mounted by using vectashield (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole and imaged with a Zeiss 510 NLO multiphoton/confocal laser-scanning microscope. The multiphoton infrared laser was used to excite 4′,6-diamidino-2-phenylindole (blue), and the confocal HeNe laser was used to excite Cy3 (red).
Results
Arf GFP/GFP MEFs Express GFP in Response to “Culture Shock” and Oncogenic Ras. With the exception of the yolk sac (L. Nilsson and F.Z., unpublished data) and postnatal eye (16), Arf is not detectably expressed during normal mouse development (24). However, MEFs derived from midgestation embryos and serially passaged in culture are induced to synthesize p19Arf, which accumulates as the cells lose their proliferative capacity and eventually senesce (9). Conversely, MEFs lacking Ink4a-Arf (2) or Arf alone (3) do not senesce and readily emerge as immortalized cell lines sensitive to transformation by oncogenic Ras. Although Arf induction in primary MEFs normally accompanies the stress of ex vivo culture, the process is accelerated by oncogenic Ras, resulting in earlier senescence (25).
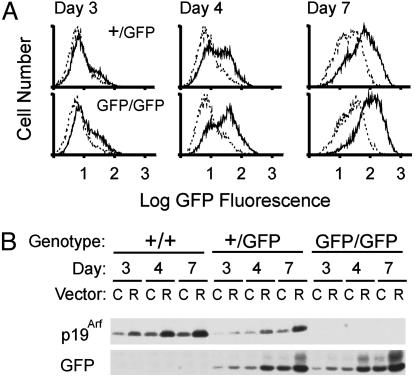
MEFs prepared from Arf+/+, Arf+/GFP, or ArfGFP/GFP embryos were infected with a control retroviral vector or one encoding oncogenic Ras. Cells plated in sparse cultures and allowed to proliferate began to express GFP, which accumulated during the first week of culture (Fig. 2 A and B Bottom). MEFs expressing oncogenic Ras exhibited faster induction of GFP, whether they retained a copy of the wild-type Arf allele or not. Arf+/+ and Arf+/GFP MEFs also expressed increasing levels of p19Arf protein that accumulated more rapidly in cells expressing Ras (Fig. 2B Top). MEFs from Arf+/GFP mice expressed approximately half of the p19Arf protein detected in matched Arf+/+ controls and less GFP protein than ArfGFP/GFP MEFs. Thus, in cells from heterozygotes, the GFP knock-in allele had no discernable effect on expression of the wild-type Arf gene or vice versa. ArfGFP/GFP MEFs did not senesce and were maintained for as many as 20 passages as cell lines; these cells were sensitive to morphologic transformation by oncogenic Ras (data not shown), consistent with the idea that they are functionally Arf-null. As expected (3), p16Ink4a was continuously expressed, as documented by immunoblotting (data not shown). Therefore, despite the lack of p19Arf protein production and the failure of ArfGFP/GFP MEFs to undergo senescence, the Arf promoter appeared to properly regulate GFP expression.
Fig. 2.
Analysis of GFP and p19Arf expression in MEFs. (A) Arf+/GFP (Upper) and ArfGFP/GFP (Lower) MEFs infected with a control retroviral vector (dotted lines) or a vector encoding oncogenic Ras (solid lines) were scored for green fluorescence by flow cytometry at the indicated times after plating in sparse culture. (B) Equal quantities of protein from detergent lysates of wild-type (+/+) MEFs and from the cells of the indicated genotypes analyzed in A were separated on denaturing gels, transferred to a membrane, and blotted with antibodies to p19Arf or GFP as indicated at the bottom. Blotting with antibodies to actin served as an internal loading control (data not shown). Cells infected with the control vector (C) or the one encoding Ras (R) were analyzed at the same times after plating as those in A.
ArfGFP/GFP Mice Are Prone to Tumor Development. The majority of Arf-null mice develop tumors in their first year of life, and virtually all succumb to cancers in their second (3, 26). The most prevalent tumors are sarcomas and lymphomas, although carcinomas and gliomas are detected less frequently. Although Arf is not induced by ionizing radiation (3, 27), tumor development in Arf-null strains is accelerated in animals exposed neonatally to x-rays or chemical carcinogens (26).
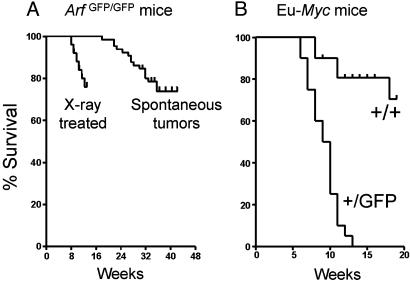
Untreated ArfGFP/GFP mice also developed tumors, which arose more rapidly in animals that received a single 4-Gy dose of total body irradiation 5 days after birth (Fig. 3A). Histopathologic analysis of 26 mice that spontaneously developed tumors revealed a broad disease spectrum. Most (65% in this series) were highly invasive sarcomas of various types, including those with pleomorphic morphology (seven cases), gastrointestinal stromal tumors (three cases), hemangiosarcomas (three cases), malignant peripheral nerve sheath tumors (two cases), and histiocytic sarcomas (two cases). Lymphomas (33%) comprised the remainder. Although we analyzed a relatively small cohort of animals, the 2:1 frequency of sarcomas versus lymphomas mimics that seen in Arf-null mice (26).
Fig. 3.
Mouse survival curves. (A) Rates of spontaneous tumor development in Arf GFP/GFP mice observed for up to 44 weeks after birth, as compared with those arising in animals that received a single sublethal dose of ionizing irradiation at 5 days of age. (B) Rates of lymphoma development in Eμ-Myc transgenic mice contrasted on an Arf+/+ or Arf+/GFP genetic background.
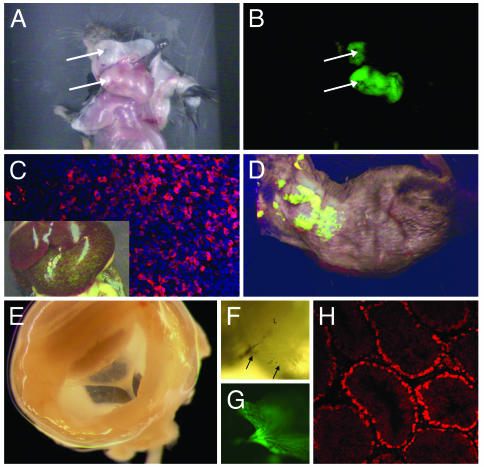
Depending on tumor type, size, and anatomic location, many solid tumors exhibited vivid green fluorescence (Fig. 4 A and B). Spontaneously arising lymphomas that metastasized to liver sometimes appeared macroscopically as green fluorescent nodules (Fig. 4C Inset). Even when metastatic foci were not visible by eye, immunohistochemical analysis and the use of a more sensitive two-photon confocal laser microscope demonstrated invasion of the liver parenchyma by GFP-positive lymphoma cells (Fig. 4C, GFP in red, 4′,6-diamidino-2-phenylindole in blue). Half of the mice had enlarged spleens with extensive foci of extramedullary hematopoiesis, a finding observed in animals lacking Ink4a-Arf or Arf alone (2, 3). Together, these results argue that although ArfGFP/GFP mice have lost Arf tumor suppressor activity, the retained Arf promoter continues to actively drive GFP expression in tumor cells.
Fig. 4.
GFP expression in mouse tissues. (A and B) Arf GFP/GFP mouse with a green fluorescent sarcoma (arrows) in the neck region. (C) Illustration of macroscopic foci of GFP-positive lymphoma cells that metastasized to liver (Inset) and microscopic foci visualized by immunofluorescence (red) and counterstained with 4′,6-diamidino-2-phenylindole (blue). (D) Whole-body imaging of a shaved Arf +/GFP,Eμ-Myc mouse with lymphoma. A whole mount of a dissected eye from an ArfGFP/GFP mouse (E) illustrates a funnel-shaped mass stretching from the lens (top left) toward the optic cup at the rear. A closer view (F) illustrates elements of the hyaloid vasculature (arrows) within the green fluorescent mass (G). (H) Immunohistochemical staining of GFP (red) in the testis of an 8-month-old mouse. The position of stained cells within the tubules closely corresponds to regions containing spermatogonia and immature (leptotene) spermatocytes in meiosis I.
Myc Activates the Arf-GFP Allele in Vivo. When overexpressed, c-Myc is a potent Arf inducer, and its proapoptotic activities are mediated in part by its ability to trigger p19Arf-dependent expression of p53 (9). Conversely, loss of either p19Arf or p53 function greatly dampens Myc's ability to kill cells and thereby allows overexpressed Myc to act as a more potent promoter of cell growth and proliferation. These effects can be readily appreciated in the Eμ-Myc mouse model of Burkitt's lymphoma, in which a Myc transgene driven by the Ig heavy-chain promoterenhancer (Eμ) induces B cell tumors (28). Mice expressing the transgene routinely develop B cell lymphomas with a mean latency of ≈26 weeks, and all die of disease by 1 year after birth (Fig. 3B). When crossed with Arf+/- heterozygotes, disease latency is shortened to ≈12 weeks, and >80% of the lymphomas that arise lose the wild-type Arf allele, consistent with the idea that Arf functions as a prototypic “two-hit” tumor suppressor (18, 29, 30). Similar results were obtained in Arf+/GFP mice (Fig. 3B), except that the resulting lymphomas exhibited green fluorescence. Indeed, we have been able to observe disease development in real time in Arf+/GFP animals (Fig. 4D), as have others who used transplanted lymphoma cells that were engineered to express GFP (20, 31).
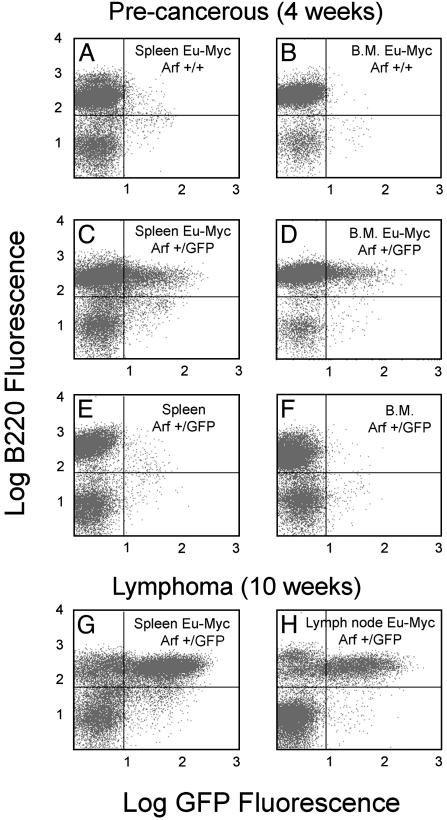
Lymphoid organs from precancerous Eμ-Myc transgenic mice exhibit a high proliferative index that is offset by apoptosis. Only later in the course of disease does inactivation of p53 or Arf cancel apoptosis and result in the rapid expansion of Myc-expressing lymphoma cells (18). To determine whether we might be able to detect Myc-induced Arf promoter activity before the emergence of frank disease, we harvested splenocytes and bone marrow cells from young Arf+/GFP mice and simultaneously assayed B220-positive B cells for GFP expression. Neither spleen nor bone marrow cells from Arf +/+, Eμ-Myc transgenic mice expressed GFP (Fig. 5 A and B), whereas a substantial fraction of B220-positive B cells from Arf+/GFP, Eμ-Myc transgenic mice exhibited green fluorescence (Fig. 5 C and D). A much smaller fraction of B220-negative cells in the spleen expressed GFP (Fig. 5C); these cells are likely to be more mature B cells that are not present in the bone marrow (Fig. 5D) (18, 28). Spleen and bone marrow from nontransgenic Arf+/GFP animals did not express GFP (Fig. 5 E and F), in agreement with previous findings that Eμ-Myc is required to drive Arf expression in B cells. Splenocytes (Fig. 5G) and lymph node cells (Fig. 5H) taken from mice with overt lymphomas exhibited even greater degrees of GFP expression, and involved lymph nodes could be detected in living mice by using whole-body fluorescence imaging (Fig. 4D).
Fig. 5.
Flow cytometric analysis of lymphoid cells expressing GFP. (A-F) Illustration of results obtained from 4-week-old mice that exhibited no overt signs of lymphoma development (precancerous). (G and H) Documentation of GFP expression in older lymphomatous animals. Results with splenocytes are shown in A, C, E, and G, results with bone marrow (B.M.) are shown in B, D, and F, and results with lymph node cells are shown in H. The genotypes of the mice are indicated in the upper right of each panel. Cells were costained for the B cell marker B220 (ordinate) or for GFP (abscissa) and analyzed by dual-color flow cytometry.
Arf-GFP Expression in the Vitreous of the Mouse Eye. During the first week of postnatal development, Arf is induced in the vitreous of the eye where its expression is required for regression of the hyaloid vascular system that nourishes the developing lens and vitreous. In newborn Arf-null mice, the hyaloid vasculature persists and is associated with an abnormal proliferation of perivascular cells that form a funnel-shaped retrolental mass that ultimately disrupts the architecture of the posterior lens and the neuroretina (16).
Eye development in Arf+/GFP mice proceeds normally, although examination of the hyaloid vasculature at the time that Arf is normally expressed allowed us to visualize a few green fluorescent cells (data not shown). As expected, ArfGFP/GFP mice developed the characteristic pathologic findings previously observed in the Arf-null strain, except that the funnel-shaped retrolental mass in the vitreous included green fluorescent cells. Fig. 4E shows a whole mount of a dissected eye from a mouse at postnatal day 4 in which the vitreal mass was clearly visualized. A closer view illustrates vascular elements within the mass (Fig. 4F) surrounded by green fluorescent cells (Fig. 4G). These findings provide further compelling evidence that ArfGFP/GFP animals lack a functional Arf gene and that the intact Arf promoter responds appropriately to physiologic signals that normally promote vascular regression in the eye.
Arf Promoter Expression in the Testis. A previous analysis of newborn mice indicated that the first detectable site of Arf expression in otherwise normal tissue is in the testis (24). Because immunohistochemical identification of p19Arf protein has not so far been achieved, the cells that express p19Arf have not been identified. We prepared sections from the testis of male mice of various ages but were unable to detect GFP expression by fluorescence microscopy. However, immunohistochemical staining allowed ready detection of GFP expression in testicular tubules from adult ArfGFP/GFP mice (Fig. 4H). In sections taken from animals from 2 to 8 months of age, staining was limited to cells lining the tubules, which, based on their location and morphology, most likely represent either spermatogonia or leptotene spermatocytes at the earliest stages of meiosis I (32). Thus, the low levels of Arf expressed within the normal testis appear to be limited to cells that switch from a mitotic to a meiotic division cycle.
Using the same technique, we have visualized small pockets of green fluorescent cells in other ostensibly normal tissues, including the thymic medulla, lung alveoli, and regions of the brain and gastrointestinal tract (data not shown). We are now attempting to use other lineage-specific markers to identify the nature of the GFP-positive cells in young Arf +/GFP and Arf GFP/GFP mice. As yet, we have no clear idea of the identity of these cell populations, but our preliminary analysis indicates that such cells are rare and express only low levels of GFP, in agreement with earlier studies that Arf expression is difficult to detect in most normal tissues (24).
Discussion
The ArfGFP/GFP mouse strain lacks sequences encoding the p19Arf tumor suppressor protein and therefore exhibits phenotypic characteristics seemingly identical with those of Arf-null strains described (3-5). First, MEFs derived from these animals did not undergo senescence and were susceptible to transformation by oncogenic Ras. Second, ArfGFP/GFP mice spontaneously developed tumors, and these arose more rapidly when neonatal mice were exposed to a single sublethal dose of ionizing radiation. Third, lymphomagenesis induced by an Eμ-Myc transgene was greatly accelerated in the Arf+/GFP genetic background, in which the latency of disease was shortened from ≈26 to ≈12 weeks. As expected, the wild type Arf allele was lost from the vast majority of the tumors that arose (ref. 18 and data not shown), consistent with Arf's behavior as a prototypic “two-hit” tumor suppressor in this model system. Finally, newborn Arf GFP/GFP mice exhibited ocular pathology due to the presence of bilateral retrolental masses associated with failure of hyaloid vasculature regression in the vitreous and destruction of the retina and the lens.
We found that GFP was expressed in all these biologic settings. Specifically, cultured MEFs, spontaneously arising tumors, Eμ- Myc-induced lymphomas, and retrolental masses in the vitreous of the eye each exhibited green fluorescence. Therefore, even in the complete absence of functional Arf protein, the Arf-GFP promoter-reporter responds in vivo to signals that would otherwise induce p19Arf expression. We reason that Arf-inductive signals arising in wild-type mice would normally lead to a p53 response that counters cell proliferation by cell cycle arrest or apoptosis, ultimately limiting the number of Arf-expressing cells. In contrast, Arf inactivation permits persistent signaling and aberrant proliferation and, in the Arf-GFP mouse, facilitates the accumulation of GFP-positive cells.
Some solid tumors that arose in Arf GFP/GFP mice and most of the lymphomas in Arf+/GFP, Eμ-Myc transgenic animals produced sufficiently strong signals to allow their visualization by whole-body fluorescence imaging of live mice. Not all tumors exhibited such strong fluorescence at a macroscopic level, but when these were examined microscopically, we were invariably able to detect GFP-positive cells by immunohistochemistry. Many such tumors showed heterogeneous staining due to the presence of mixtures of normal and tumor cells. However, we cannot formally exclude the possibility that some tumor cells might segregate Arf-GFP alleles in the course of tumor progression. The Arf gene is closely flanked by two other tumor suppressors, Ink4a and Ink4b, within a segment of <100 kb in both the mouse and human genomes, and it is conceivable that selection against expression of either of the Ink4 loci could result in deletions or epigenetic silencing of the Arf locus. Indeed, several examples exist in which Arf inactivation is followed by Ink4a loss of function, or vice versa, either during establishment of certain cultured cell lines (33) or in tumor development (4, 34-37).
Northern and immunoblotting assays have not documented Arf gene expression during mouse embryonic development (24). We recently used more sensitive RT-PCR analyses to repeat studies on early- and mid-gestation embryos and found that Arf transcripts are expressed in the yolk sac but are not readily detected in the embryo proper (L. Nilsson and F.Z., data not shown). After birth, low levels of Arf transcription were detected by RT-PCR in some organs of 4-month-old mice, and these remained essentially unchanged when reassayed in 15-month-old animals (24). In both young and old mice, the most prominent site of Arf expression was the testis; much lower levels were detected in spleen and lung, with barely detectable expression in kidney and brain. Although GFP expression could not be detected by using fluorescence microscopy of tissue sections taken from testes of adult ArfGFP/GFP mice, immunohistochemistry performed with antibodies to GFP identified positively staining cells in the testicular tubules that appeared to correspond to spermatogonia and/or very immature spermatocytes. We also observed foci of GFP-positive cells in thymus, lung, intestine, and brain and are currently attempting to characterize these cells in greater detail.
Although the roles that Arf may play in normal tissues largely remain a mystery, the Arf-GFP mouse should facilitate investigations of Arf temporal and spatial expression in various normal tissues. For example, Arf expression in the mouse eye is remarkably restricted. Normally, at birth, the retrolental vitreous contains delicate hyaloid vascular elements composed of endothelial and sparse perivascular cells. These vessels arise from the hyaloid artery at the optic disk, splay through the vitreous, and nourish the developing lens and pupillary membrane. Between postnatal days 6 and 10, this component of the hyaloid vasculature normally regresses, a process that does not occur in Arf-null mice in which excess perivascular cells accumulate (16). In normal animals, RT-PCR specifically identified Arf transcripts in the vitreous, but not in the optic cup or lens and only between postnatal days 1 and 5. A preliminary analysis of eyes from newborn Arf+/GFP mice has identified a few GFP-positive cells within the vitreous (data not shown), and we are now attempting to determine their lineage and how p19Arf prevents their excess accumulation. Regardless of their identity, the presence of fluorescent cells in the retrolental masses arising in ArfGFP/GFP mice now indicates that Arf plays a cell-autonomous role in their development.
Lymphoid cells from the spleen and bone marrow of Arf+/GFP mice bearing an Eμ-Myc transgene expressed GFP before any overt evidence of lymphoma existed. This corresponds to a time in the life history of the disease in which greatly increased lymphoid cell proliferation is offset by Arf- and p53-dependent apoptosis (18, 20, 29). Most GFP expression in lymphoid tissues of Arf+/GFP, Eμ-Myc mice was confined to B220-positive B cells (28). In contrast, lymphoid tissues taken from control animals lacking the Myc transgene did not express GFP, underscoring the role of Myc as a potent Arf inducer and indicating, as well, that Arf-GFP expression in normal splenocytes or bone marrow cells (weakly positive by RT-PCR) was below the limit of detectability of the flow cytometric assay used. Despite the fact that B220positive cells in the spleen and bone marrow of Arf+/GFP,Eμ-Myc transgenic mice expressed a significant number of green fluorescent cells, we could not detect splenic involvement in precancerous mice by whole-body imaging of live animals. However, lymph nodes from lymphomatous mice contained a sufficient concentration of GFP-positive cells to be imaged. Because Arf responds to many different oncogenic signals, including Ras, Abl, β-catenin, and others (38), and is inappropriately activated in animals lacking Arf-repressors such as Bmi-1 (39), breeding the Arf-GFP reporter strain to mice expressing these oncogenes or lacking such repressors, particularly in targeted somatic tissues, should similarly help to identify precancerous cells in other biologic settings.
Acknowledgments
We thank P. McKinnon and H. Russell for help with ES cells; S. Ragsdale for karyotyping; C. Nagy and G. Grosveld for ES injection and blastocyst implantation; E. Van de Kamp, W. den Besten, and J. Doherty for help constructing the targeting vector; G. Murti and K. Barnes for confocal microscopy; R. Ashmun and N. Carpino for flow cytometry; D. Bush and S. Portillo for immunohistochemical analyses; A. Martin for studies of the mouse eye; C. Hornsby and G. Assem for genotyping; and E. Vasquez for animal husbandry. This work was supported by National Institutes of Health Grants CA71907 (to M.F.R.), DK44158 and CA76379 (to J.L.C.), and T32-CA70089 (to R.T.W), Cancer Center Support Grant CA21765, National Research Service Award Grant F32-DK10154 (to T.A.B.), and American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. C.J.S. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: MEF, mouse embryo fibroblast; ES, embryonic stem.
References
- 1.Lowe, S. W. & Sherr, C. J. (2003) Curr. Opin. Genet. Dev. 13, 77-83. [DOI] [PubMed] [Google Scholar]
- 2.Serrano, M., Lee, H.-W., Chin, L., Cordon-Cardo, C., Beach, D. & DePinho, R. A. (1996) Cell 85, 27-37. [DOI] [PubMed] [Google Scholar]
- 3.Kamijo, T., Zindy, F., Roussel, M. F., Quelle, D. E., Downing, J. R., Ashmun, R. A., Grosveld, G. & Sherr, C. J. (1997) Cell 91, 649-659. [DOI] [PubMed] [Google Scholar]
- 4.Krimpenfort, P., Quon, K. C., Mooi, W. J., Loonstra, A. & Berns, A. (2001) Nature 413, 83-86. [DOI] [PubMed] [Google Scholar]
- 5.Sharpless, N. E., Bardeesy, N., Lee, K.-H., Carrasco, D., Castrillon, D. H., Aguirre, A. J., Wu, E. A., Horner, J. W. & DePinho, R. A. (2001) Nature 413, 86-91. [DOI] [PubMed] [Google Scholar]
- 6.Ruas, M. & Peters, G. (1998) Biochim. Biophys. Acta 1378, F115-F177. [DOI] [PubMed] [Google Scholar]
- 7.Esteller, M., Corn, P. G., Baylin, S. B. & Herman, J. G. (2001) Cancer Res. 61, 3225-3229. [PubMed] [Google Scholar]
- 8.Quelle, D. E., Zindy, F., Ashmun, R. A. & Sherr, C. J. (1995) Cell 83, 993-1000. [DOI] [PubMed] [Google Scholar]
- 9.Zindy, F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmero, I., Pantoja, C. & Serrano, M. (1998) Nature 395, 125-126. [DOI] [PubMed] [Google Scholar]
- 11.Weber, J. D., Jeffers, J. R., Rehg, J. E., Randle, D. H., Lozano, G., Roussel, M. F., Sherr, C. J. & Zambetti, G. P. (2000) Genes Dev. 14, 2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carnero, A., Hudson, J. D., Price, C. M. & Beach, D. H. (2000) Nat. Cell Biol. 2, 148-155. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto, M., Kuo, M.-L., Roussel, M. F. & Sherr, C. J. (2003) Mol. Cell 11, 415-424. [DOI] [PubMed] [Google Scholar]
- 14.Kuo, M.-L., Duncavage, E. J., Mathew, R., den Besten, W., Pie, D., Naeve, D., Yamamoto, T., Cheng, C., Sherr, C. J. & Roussel, M. F. (2002) Cancer Res. 63, 1046-1053. [PubMed] [Google Scholar]
- 15.Rocha, S., Campbell, K. J. & Perkins, N. D. (2003) Mol. Cell 12, 15-25. [DOI] [PubMed] [Google Scholar]
- 16.McKeller, R. N., Fowler, J. L., Cunningham, J. J., Warner, N., Smeyne, R. J., Zindy, F. & Skapek, S. X. (2002) Proc. Natl. Acad. Sci. USA 99, 3848-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarrick, J. W., Parnes, J. R., Seong, R. H., Solter, D. & Knowles, B. B. (1993) Transgenic Res. 2, 183-190. [DOI] [PubMed] [Google Scholar]
- 18.Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. (1999) Genes Dev. 13, 2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, M., Baranov, E., Jiang, P., Sun, F. X., Li, X. M., Li, L., Hasegawa, S., Bouvet, M., Al-Tuwaijri, M., Chrishima, T., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 1206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt, C. A., Fridman, J. S., Yang, M., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cancer Cell 1, 289-298. [DOI] [PubMed] [Google Scholar]
- 21.Morgenstern, J. P. & Land, H. (1990) Nucleic Acids Res. 18, 1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue, K., Wren, R., Rehg, J. E., Adachi, M., Cleveland, J. L., Roussel, M. F. & Sherr, C. J. (2000) Genes Dev. 14, 1797-1809. [PMC free article] [PubMed] [Google Scholar]
- 23.Zindy, F., Nilsson, L. M., Nguyen, L., Meunier, C., Smeyne, R. J., Rehg, J. E., Eberhart, C., Sherr, C. J. & Roussel, M. F. (2003) Cancer Res. 63, 5420-5427. [PubMed] [Google Scholar]
- 24.Zindy, F., Quelle, D. E., Roussel, M. F. & Sherr, C. J. (1997) Oncogene 15, 203-211. [DOI] [PubMed] [Google Scholar]
- 25.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. (1997) Cell 88, 593-602. [DOI] [PubMed] [Google Scholar]
- 26.Kamijo, T., Bodner, S., van de Kamp, E., Randle, D. H. & Sherr, C. J. (1999) Cancer Res. 59, 2217-2222. [PubMed] [Google Scholar]
- 27.Kamijo, T., van de Kamp, E., Chong, M. J., Zindy, F., Diehl, A. J., Sherr, C. J. & McKinnon, P. (1999) Cancer Res. 59, 2464-2469. [PubMed] [Google Scholar]
- 28.Adams, J. M., Harris, A. W., Pinkert, C. A., Corcoran, L. M., Alexander, W. S., Cory, S., Palmiter, R. D. & Brinster, R. L. (1985) Nature 318, 533-538. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt, C. A., McCurrach, M. E., De Stanchina, E. & Lowe, S. W. (1999) Genes Dev. 13, 2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs, J. J. L., Scheijen, B., Vonchen, J.-W., Kieboom, K., Berns, A. & van Lohuizen, M. (1999) Genes Dev. 13, 2678-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt, C. A., Fridman, J. S., Yang, M., Lee, S., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cell 109, 335-346. [DOI] [PubMed] [Google Scholar]
- 32.Zindy, F., den Besten, W., Chen, B., Rehg, J. E., Latres, E., Barbacid, M., Pollard, J. W., Sherr, C. J., Cohen, P. E. & Roussel, M. F. (2001) Mol. Cell. Biol. 21, 3244-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randle, D. H., Zindy, F., Sherr, C. J. & Roussel, M. F. (2001) Proc. Natl. Acad. Sci. USA 98, 9654-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachoo, R. M., Maher, E. A., Ligon, K. L., Sharpless, N. E., Chan, S. S., You, M. J., Tang, Y., DeFrances, J., Stover, E., Weissleder, R., et al. (2002) Cancer Cell 1, 269-277. [DOI] [PubMed] [Google Scholar]
- 35.Bardeesy, N., Bastian, B. C., Hezel, A., Pinkel, D., DePinho, R. A. & Chin, L. (2001) Mol. Cell. Biol. 21, 2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You, M. J., Castrillon, D. H., Bastian, B. C., O'Hagan, R. C., Bosenberg, M. W., Parsons, R., Chin, L. & DePinho, R. A. (2002) Proc. Natl. Acad. Sci. USA 99, 1455-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin, L. (2003) Nat. Rev. Cancer 3, 559-570. [DOI] [PubMed] [Google Scholar]
- 38.Sherr, C. J. (2001) Nat. Rev. Mol. Cell. Biol. 2, 731-737. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs, J. J. L., Kieboom, K., Marino, S., DePinho, R. A. & van Lohuizen, M. (1999) Nature 397, 164-168. [DOI] [PubMed] [Google Scholar]