Abstract
Objective
Exemestane is a third-generation aromatase inhibitor used in the treatment of breast cancer in postmenopausal women. Reduction to form 17-dihydroexemestane and subsequent glucuronidation to exemestane-17-O-glucuronide is a major pathway for exemestane metabolism. The goal of this study was to analyze 17-dihydroexemestane anti-aromatase activity, characterize the 17-dihydroexemestane glucuronidation pathway, and determine whether the functional polymorphisms in active UGTs could play a role in altered 17-dihydroexemestane glucuronidation.
Methods
Homogenates from a HEK293 aromatase-overexpressing cell line (HEK293-aro) were used to examine exemestane versus 17-dihydroexemestane anti-aromatase activities. UGT-overexpressing cell lines and a panel (n = 110) of human liver microsome (HLM) were screened for glucuronidation activity against 17-dihydroexemestane. UGT2B17 genotyping and liver mRNA expression were performed by real-time PCR.
Results
The inhibition of estrone formation from androst-4-ene-3,17-dione in HEK293-aro cell homogenates was similar for 17-dihydroexemestane (IC50 = 2.3 ± 0.83 μmol/l) and exemestane (IC50 = 1.4 ± 0.42 μmol/l). UGTs 2B17 and 1A4 were high-expression hepatic UGTs that exhibited activity against 17-dihydroexemestane, with UGT2B17 exhibiting a 17-fold higher Vmax/KM than UGT1A4. The rate of exemestane-17-O-glucuronide formation was shown to be significantly (P < 0.001) decreased (14-fold) in HLMs exhibiting the UGT2B17(*2/*2) deletion genotype versus wild-type UGT2B17(*1/*1) HLMs; a 36-fold lower Vmax/KM (P = 0.023) was observed in UGT2B17(*2/*2) versus UGT2B17(*1/*1) HLMs. A significant (P < 0.0001, R2 = 0.72) correlation was observed between HLM exemestane-17-O-glucuronide formation and liver UGT2B17 expression.
Conclusion
These data suggest that 17-dihydroexemestane is an active metabolite of exemestane and that the UGT2B17 deletion polymorphism could play an important role in determining levels of excretion of 17-dihydroexemestane and overall exemestane metabolism.
Keywords: aromatase inhibitor, breast cancer, exemestane 17-dihydroexemestane, glucuronidation, metabolism, pharmacogenetics, UDP glucuronosyltransferase
Introduction
Breast cancer is the most common malignancy among women, with a lifetime risk for breast cancer in women of 10%. US National Cancer Institute estimated 192 370 new cases and 40 170 death from breast cancer in females in 2009 in the United States [1]. In the past 4 decades, treatment with tamoxifen for 5 years has been a standard of care for estrogen-dependent breast cancer [2–5]. However, limitations of tamoxifen therapy include significant adverse effects like increased risk for endocrine cancer [6,7], have prompted the search for alternative endocrine therapies with increased efficacy and fewer long-term complications.
Aromatase inhibitors (AIs) work by blocking the aromatase enzyme in the final step of estrogen synthesis, thus lowering circulating estrogen levels. Exemestane is a noncompetitive third-generation AI that is structurally related to the natural aromatase substrate, androstene-dione. Evidence from several clinical trials indicates that exemestane may be superior to tamoxifen as first-line therapy for postmenopausal women with metastatic breast cancer [8]. Results from at least eight major clinical trials indicate that AIs are associated with longer disease-free survival than therapy with tamoxifen alone [9]. Exemestane is effective for the treatment of postmenopausal women with early-stage or advanced breast cancer [10]. The major toxicity associated with AIs including exemestane is joint pain and bone loss, the propensity for bone fractures, and increased risk for osteoporosis [11–13], a pattern consistent with the lower hip and lumbar spine bone mineral density observed in patients on AIs [14]. Although not as severe as tamoxifen, other toxicities reported for AIs include a variety of gynecological events including vaginal bleeding or discharge and hot flashes [15,16]. Variability in changes to lipid profiles, manifestation of osteoporosis, and time of recurrence, were observed in patients in many AI clinical trials [9] and the mechanism underlying this variability in response to AIs and to their toxicities remains unclear.
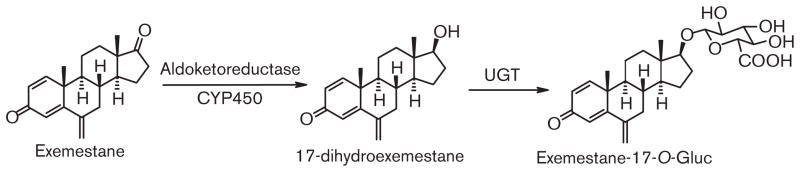
Limited studies have been performed examining exemestane metabolism. Reduction of the 17-keto group by aldoketoreductase to form 17-dihydroexemestane is a major pathway for exemestane metabolism (Fig. 1; [17–21]). Urinary and fecal excretion of exemestane was similar, both around 42%, with the glucuronide of 17-dihydroexemestane (at the 17-O-position) a major metabolite found in urine [19,22]. The glucuronidation pathway of excretion for exemestane is important because 17-dihydroexemestane was reported to exhibit significant anti-aromatase and androgenic activities in vitro [23,24]. The goal of this study was three-fold: (i) to confirm the relative anti-aromatase activity of the major exemestane metabolite, 17-dihydroexemestane; (ii) to identify the UGT enzymes involved in the metabolism of 17-dihydroexemestane; and (iii) to determine whether potentially functional genetic variations in active UGTs play a role in altered 17-dihydroexemestane glucuronidation and overall exemestane metabolism.
Fig. 1.
Schematic of exemestane metabolism.
Materials and methods
Chemicals and materials
Exemestane was purchased from Hangzhou HETD Industry Co. Ltd. (Zhejiang, China). UDPGA, alamethicin, and β-glucuronidase were purchased from Sigma-Aldrich (St Louis, Missouri, USA) and [14C] UDPGA (200 mCi/mmol) was purchased from American Radiolabeled Chemicals (St Louis, Missouri, USA). The NADPH regenerating system including solutions A (26.1 mmol/l NADP, 66 mmol/l glucose-6-phosphate, and 66 mmol/l MgCl2) and B (40 U/ml glucose-6-phosphate dehydrogenenase, 5 mmol/l sodium citrate) was purchased from BD Biosciences (Woburn, Massachusetts, USA). Androst-[4-14C]-ene-3,17-dione was purchased from Perkin Elmer (Boston, Massachusetts, USA). The high-performance liquid chromatography (HPLC) scintillation solution, Ecoscint Flow, was purchased from National Diagnostics (Atlanta, Georgia, USA) whereas Dulbecco’s phosphate-buffered saline (minus calcium-chloride and magnesium-chloride), fetal bovine serum, penicillin-streptomycin and geneticin (G418) were all purchased from Gibco (Grand Island, New York, USA). The Platinum Pfx DNA polymerase and the pcDNA3.1/V5-His-TOPO mammalian expression vector were obtained from Invitrogen (Carlsbad, California, USA) whereas the QIAEX II gel extraction kit was purchased from Qiagen (Valencia, California, USA). The BCA protein assay kit was purchased from Pierce (Rockford, Illinois, USA). Aromatase transfection-ready cDNA was purchased from Origene (Rockville, Maryland, USA) and the anti-aromatase antibody was purchased from Abcam (Cambridge, Massachusetts, USA). The human UGT1A western blotting kit that includes an anti-UGT1A polyclonal antibody and UGT2B7 protein standard, were purchased from Gentest (Woburn, Massachusetts, USA). The anti-β-actin monoclonal antibody was purchased from Sigma-Aldrich. All other chemicals used in these studies were purchased from Fisher Scientific (Pittsburgh, Pennsylvania, USA) unless otherwise specified.
Tissues and cell lines
A description of the normal human liver tissue specimens used for these studies was described earlier [25]. In brief, tissues were quick-frozen at −70°C within 2 h postsurgery. Liver microsomes were prepared through differential centrifugation as described earlier [26] and were stored (10–20 mg protein/ml) at −80°C. Genomic DNA was extracted from each of the same liver specimens from nuclei isolated during the microsome purification process. Microsomal protein concentrations were measured using the BCA assay. All protocols involving the analysis of tissue specimens were approved by the institutional review board at the Penn State College of Medicine and in accordance with assurances filed with and approved by the United States Department of Health and Human Services. UGT2B17 genotyping for all liver genomic DNA specimens and UGT2B17 mRNA expression in the same human liver were performed by real-time PCR as described earlier [27,28].
An aromatase-overexpressing HEK293 cell line (termed HEK293-aro) was generated for the aromatase activity experiments outlined in this study. Using the aromatase cDNA-containing pCMV6-XL4 vector from Origene as template (100 ng), aromatase cDNA was PCR-amplified using Pfx DNA polymerase (1.25 units; Invitrogen), 0.3 mmol/l of each dNTPs, and 20 pmol each of sense (5′-CCAGACGTCGCGACTCTAAATTG-3′) and antisense (5′-CTGTGAGGATGACACTATTGGC-3′) primers, corresponding to nucleotides from −49 to −26 and +1580 to +1601, respectively, relative to the aromatase translation start site. PCR amplification of aromatase cDNA was performed in a GeneAmp 9700 thermocycler (Applied Biosystems, Foster City, California, USA) using as follows: 1 cycle of 94°C for 2min, 35 cycles of 94°C for 15s, 58°C for 30 s, and 68°C for 2min, followed by a final cycle of 68°C for 10min. After a subsequent post-amplification step using Taq DNA polymerase (10min at 72°C; Denville Scientific, South Plainfield, New Jersey, USA) to ensure the addition of 3′-overhanging adenines, the PCR product (1793 bp) was purified after electrophoresis in 1.5% agarose using the QIAEX II gel extraction kit (Qiagen) and sequenced in full using the same aromatase PCR primers. The PCR product was subsequently sub-cloned into the pcDNA3.1/V5-His-TOPO mammalian expression vector using standard methodologies. The cloned vector was sequenced in full (using the same PCR primers described above) and transfected into HEK293 cells (purchased from ATCC; Rockville, Maryland, USA) by electroporation (200 V and 1000 μF) using 10 μg of pcDNA3.1/V5-His-TOPO/aromatase plasmid DNA with 5×106 HEK293 cells in 0.5 ml serum-free media in a GenePulser Xcell (Bio-Rad, Hercules, California, USA). Cells stably overexpressing the individual aromatase were selected with G418 (Invitrogen). Aromatase expression in HEK293 cells was monitored by western blot analysis, by loading 40 μg of total protein lysate of HEK293 (negative control) and HEK293-aro cells, and using the anti-aromatase antibody (#ab18995; Abcam; used in a 1: 200 dilution). Beta-actin was used as a loading control and was monitored using a 1: 5000 dilution of the monoclonal anti-β-actin antibody (Sigma-Aldrich).
Cells over-expressing individual UGT or aromatase were harvested to prepare homogenates as described previously [29]. Total homogenate protein concentrations were measured using the BCA protein assay.
Western blot analysis of UGT proteins
Levels of UGT1A and 2B proteins were determined by western blot analysis as described previously using a 1: 5000 dilution of antibody [29,30]. UGT1A4 protein was detected using the UGT1A antibody from Gentest whereas UGT2B17 expression was measured using a newly-synthesized affinity-purified chicken anti-UGT2B antibody generated against the peptide CKWDQFYSEVLGRPTTL, which is common to all human UGT2B family members (Pocono Rabbit Farm, Canadensis, Pennsylvania, USA). Proteins were detected by chemiluminescence using the SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology, Inc., Rockford, Illinois, USA). Secondary antibodies supplied with the Dura ECL kit (anti-rabbit and anti-mouse) were used at 1: 3000. UGTs 1A4 and 2B17 were quantified against 100–250 ng of human UGT1A and UGT2B7 protein (Gentest), respectively, by densitometric analysis of X-ray film exposures (1 s–2min) of western blots using a GS-800 densitometer with Quantity One software (Bio-Rad). All cell homogenate protein levels were normalized to the levels of calnexin or β-actin observed in each lane (quantified by densitometric analysis of western blots as described above). Determinations of exemestane-17-O-glucuronide formation in UGT1A4 and UGT2B17-over-expressing cell lines were calculated relative to the levels of UGT expression in the respective cell lines. X-ray film bands were always below densitometer saturation levels as indicated by the densitometer software. Densitometric results were always consistent irrespective of the exposure time. Western blot and subsequent densitometric analysis was performed in triplicate on three separate occasions, using the same UGT-containing cell homogenates used for activity assays, with relative UGT protein levels expressed as the mean of these experiments.
Synthesis of 17-dihydroexemestane
17-dihydroexemestane was prepared from exemestane as described previously [19] with minor modifications. Briefly, exemestane and NaBH4 were individually dissolved in methanol/water (4:1, v/v). For reduction of the 17-keto functional group of exemestane, NaBH4 was slowly added to the suspended exemestane in a 1: 2 molar ratio. The mixture was left at ambient temperature for at least 1 h and then assayed by thin layer chromatography to ensure reaction completeness before solvent removal by vacuum. After dissolving the dry residue in 1.5 mol/l HCl, 1 mol/l KOH (500 μl) was added. The mixture was extracted by ethyl ether, dried by vacuum, and re-dissolved in 30 ml ethyl ether:hexane (1: 1) at 50°C. After 3 days, the formed crystals were washed with hexane and dried by vacuum. The synthesized 17-dihydroexemestane structure was characterized by a Bruker 500MHz nuclear magnetic resonance (NMR) spectrometer and HPLC/MS/MS, with the MS spectrum resulting in a clear peak at m/z 299 [M]+ and a daughter product at m/z 135.
Both exemestane and 17-dihydroexemestane were dissolved in 100% ethanol and the stock solutions were kept at −20°C.
Aromatase-inhibiting activity assays
Aromatase activity was determined by measuring the levels of estrone formation using androst-[4-14C]-ene-3,17-dione (5 μmol/l) as substrate and HEK293-aro cell homogenates as the source of aromatase. Exemestane and 17-dihydroexemestane (3.3 nmol/l–10μmol/l) were used for the determination of their aromatase inhibiting activities. Incubations were performed in 10mmol/l potassium phosphate (pH 7.4) buffer containing 100mmol/l KCl, 1mmol/l EDTA, 1mmol/l DTT, a NADPH generating system (with final concentrations of 0.26mmol/l NADP+, 0.66mmol/l G6P, 0.66mmol/l MgCl2 and 0.4 μ/ml G6P dehydrogenenase), 125 μg HEK293-aro cell homogenate, 5 μmol/l androst-[4-14C]-ene-3,17-dione and various concentrations of exemestane or 17-dihydroexemestane, in a final reaction volume of 50 μl at 37°C for 2h. Reactions were terminated by the addition of 50 μl cold acetonitrile on ice. Mixtures were centrifuged for 10 min at 4°C at 16 100 g and the supernatants were collected.
Estrone formation was analyzed by radioflow-HPLC using a Gold 126 Solvent Module HPLC system (Beckman Coulter, Fullerton, California, USA) equipped with an automatic sampler (Model 508), a UV detector (Model 166) and a radioactive flow detector with a 1000-μl flow cell (INUS System, Tampa, Florida, USA). HPLC was performed using a 5 μ Luna C18 analytical column (4.6mm×250 mm, Phenomenex, Torrance, California, USA) combined with a 5 μ Gemini C18 analytical column (4.6mm×250 mm, Phenomenex) in series with a Luna C18 guard column (4mm L×3.0mm ID, Phenomenex). The gradient elution conditions consisted of a flow rate of 0.5 ml/min as follows: starting with 55% acetonitrile and 45% buffer A (5 mmol/l ammonium acetate, pH 5.0) for 5 min, a subsequent linear gradient to 75% acetonitrile/25% buffer A over 15 min was performed and then maintained at 75% acetonitrile for 10 min. The formed [14C] estrone peak was determined by HPLC-radioactive flow detection and was confirmed by retention time comparisons with an estrone standard (Sigma-Aldrich) detected by UV. Total aromatase activity of HEK293-aro cell homogenates was determined as the ratio of the levels of estrone versus androst-[4-14C]-ene-3,17-dione. The concentration of each compound required to reduce the total aromatase activity by 50% (IC50) was calculated using GraphPad Prism 5 software (Graph-Pad Software, San Diego, California, USA). HEK293 cell homogenate without overexpressed aromatase was used as a negative control. All experiments were performed in triplicate in independent assays.
Glucuronidation assays
For all glucuronidation assays, cell line homogenate (100 μg–1mg protein) or human liver microsomes (HLM; 2.5–10 μg protein) were pre-incubated with alamethicin (50 μg/mg protein) for 15min in an ice bath. Glucuronidation reactions were performed in a final reaction volume of 50 μl at 37°C for 60min in 50mmol/l Tris–HCl buffer (pH 7.4), 10mmol/l MgCl2, and 4mmol/l UDPGA with 1 μl (2% ethanol: v/v) 17-dihydroexemestane or exemestane. For screening of UGT-over-expressing cell lines for glucuronidating activity against exemestane and 17-dihydroexemestane, incubations contained up to 3.5mmol/l of exemestane or up to 600 μmol/l 17-dihydroexemestane. For kinetic assays, 4–300 μmol/l 17-dihydroexemestane was utilized. Reactions were terminated by the addition of 50 μl cold acetonitrile on ice. Mixtures were centrifuged for 10 min at 4°C at 16 100 g and the supernatants were collected. Experiments were always performed in triplicate in independent assays.
17-dihydroexemestane glucuronidation was analyzed using a Waters ACQUITY ultra-pressure liquid chromatography (UPLC) System (Milford, Massachusetts, USA) with a 1.7 μ ACQUITY UPLC BEH C18 analytical column (2.1mm×50 mm, Waters, Ireland) in series with a 0.2 μm Waters assay frit filter (2.1 mm, Waters, USA). The gradient elution conditions, using a flow rate of 0.3 ml/min, were as follows: starting with 19% acetonitrile and 81% buffer A (5 mmol/l ammonium acetate, pH 5.0) for 1min, a subsequent linear gradient to 75% acetonitrile/25% buffer over 2 min was performed and then maintained at 75% acetonitrile for 2 min.
Exemestane-17-O-glucuronide was confirmed by its stability in 1mol/l NaOH but sensitivity to the treatment of β-glucuronidase. In addition, incubation products (up to 5 μl) were loaded onto an UPLC/MS/MS for confirmation of exemestane-17-O-glucuronide formation. An UPLC identical to that described above was used in tandem with a Waters TQD triple quadrupole MS system. By using a positive mode, the parent compound [M+H]+ peak and their glucuronide [M-Gluc.+H]+ peak were characterized.
Ten HLMs were randomly chosen from the 110 HLM panel for kinetic analysis – five from UGT2B17 (*1/*1) homozygous wild-type individuals and five from UGT2B17 (*2/*2) homozygous deletion individuals. As controls, glucuronidation assays were regularly performed using HLM (as a positive control for glucuronidation activity) and untransfected HEK293 cell homogenate protein (as a negative control for glucuronidation activity) as described previously [30,31].
Statistical analysis
The Student’s t-test (2-sided) was used for comparing rates and kinetic values of glucuronide formation for individual UGT1A and UGT2B enzymes and variants against 17-dihydroexemestane. Kinetic data were always calculated using Vmax’s that were normalized based on UGTexpression in the different active UGT-overexpressing cell homogenates, with UGTexpression determined by western blot analysis [29,32]. Aromatase inhibition activity and kinetic constants were determined using Graphpad Prism 5 software. The rate of exemestane-17-O-glucuronide formation in HLM was compared by UGT2B17 gene deletion genotype (homozygotes, heterozygotes and wild type) by trend test and Student’s t-test using SPSS statistical software (version 15.0, SPSS Inc., Chicago, Illinois, USA).
Results
Synthesis of 17-dihydroexemestane
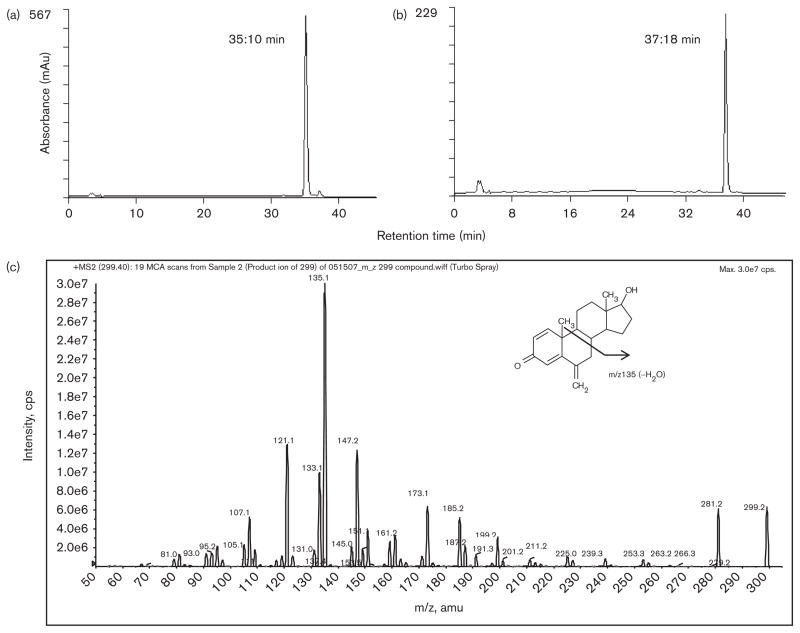
As described in the Materials and methods section, the synthesis and purification of 17-dihydroexemestane was performed using a previously published protocol with modifications [19]. As shown in Fig. 2 (panel a), the HPLC chromatogram revealed a 99% pure product whose retention time (35.1 min) was about 2 min shorter than the parent compound, exemestane (Fig. 2, panel b). The product shown in panel a was characterized by UPLC/MS/MS (Fig. 2, panel c), with the MS spectrum showing a clear [M+H]+ peak at m/z 299.2, which is identical to the predicted molecular weight of 17-dihydroexemestane, and a second major fragment at m/z 135.1. The structure was further confirmed to be 17-dihydroexemestane by 1H NMR: 1H NMR (CDCl3, 500 MHz) δ 0.83 (s, 3H), 1.04–1.08 (m, 1H), 1.16 (s, 3H), 1.24–1.31 (m, 1H), 1.36–1.40 (m, 1H), 1.48–1.52 (m, 1H), 1.64–1.72 (m, 4H), 1.78–1.83 (m, 3H), 1.90–1.93 (m, 1H), 2.08–2.12 (m, 1H), 2.51 (d, 1H, J = 9.5 Hz), 3.68 (t, 1H, J = 8.0 Hz), 4.96 (s, 1H), 5.03 (s, 1H), 6.17 (s, 1H), 6.26 (d, 1H, J = 10.0 Hz), 7.11 (d, 1H, J = 10.0 Hz).
Fig. 2.
Characterization of newly-synthesized 17-dihydroexemestane by HPLC/MS/MS. Panel (a), chromatogram of 17-dihydroexemestane; panel (b), chromatogram of exemestane; panel (c), mass spectrum of 17-dihydroexemestane.
Determination of aromatase-inhibiting activity of 17-dihydroexemestane
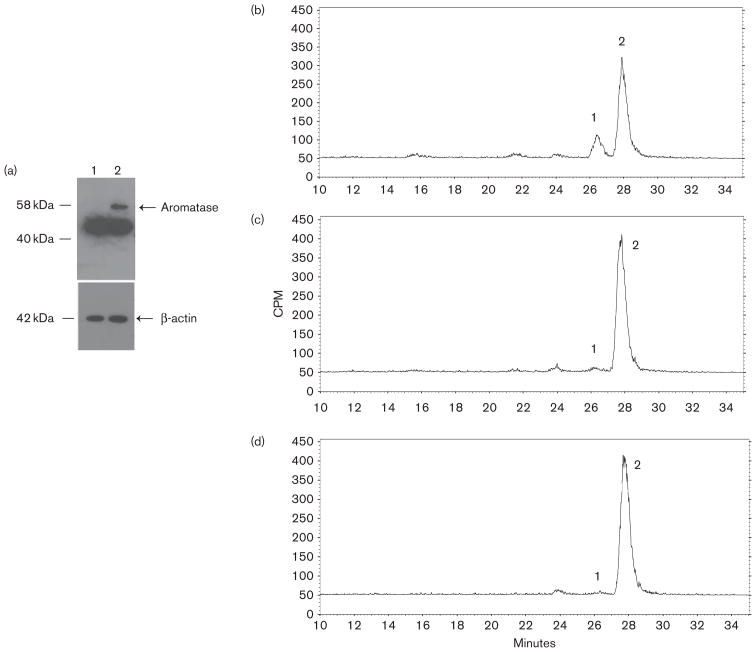
In order to assess the aromatase inhibition by 17-dihydroexemestane in vitro, HEK293-aro cells were created by stable transfection of full-length human aromatase cDNA into HEK293 cells. The expression of aromatase protein in the HEK293-aro cells was characterized by the western blot analysis using an anti-aromatase antibody as described in the Materials and methods section. Although a non-specific band of approximately 42KDa was observed in both HEK293-aro and parental HEK293 cell lysates, a clear band at 58KDa was recognized in HEK293-aro cell homogenates which matched the expected molecular weight of aromatase; no 58KDa band was observed in lysate of untransfected HEK293 cells (Fig. 3, panel a).
Fig. 3.
Aromatase expression in HEK293-aro cells and aromatase-inhibiting activity of exemestane versus 17-dihydroexemestane in HEK293-aro cell homogenates. Panel (a), Western blot analysis of aromatase protein expression was performed using lysates from the aromatase-over-expressing HEK293 cell line used for the aromatase activity assays described in this study. Lane 1, parent HEK293 cell lysate (40 μg total protein); lane 2, HEK293-aro cell lysate (40 μg total protein). Panel (b), Estrone formation in HEK293-aro cells+5 μmol/l androst-[4-14C]-ene-3,17-dione. Panel (c), Inhibition of estrone formation in HEK293-aro cells+5 μmol/l androst-[4-14C]-ene-3,17-dione by 3 μmol/l 17-dihydroexemestane. Panel (d), Inhibition of estrone formation in HEK293-aro cells+5 μmol/l androst-[4-14C]-ene-3,17-dione by 3 μmol/l exemestane. For panels b–d, aromatase activity was determined using HEK293-aro cell homogenates (125 μg) and quantified by radioflow-HPLC as described in the Materials and methods section. Aromatase activity was determined by measuring the formation of 14C-labeled estrone from androst-[4-14C]-ene-3,17-dione, the major endogenous substrate for aromatase. Peak 1, estrone; peak 2, androst-[4-14C]-ene-3,17-dione. No estrone formation was observed using (– minus aromatase) HEK293 cell homogenates (data not shown).
The enzymatic activity of HEK293-aro cells was determined using HPLC by measuring the formation of estrone from androst-4-ene-3,17-dione in incubations with HEK293-aro cell homogenate ± exemestane or 17-dihydroexemestane. As shown in Fig. 3, an obvious estrone peak was observed when no exemestane or 17-dihydroexemestane added (panel b). The aromatase enzymatic activity was inhibited by both 17-dihydroexemestane (panel c) and the parent compound, exemestane (panel d). The IC50 values for exemestane (1.4 ± 0.42) μmol/l and 17-dihydroexemestane (2.3 ± 0.83) μmol/l were comparable and not statistically different (P = 0.15).
Characterization of 17-dihydroexemestane glucuronidation
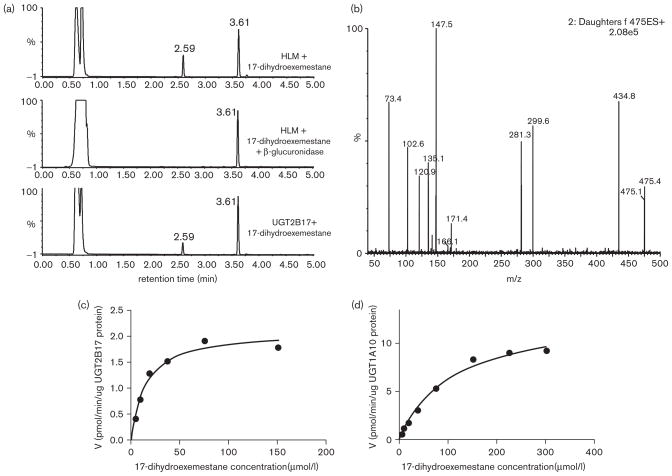
Previous studies have reported that as little as 1% of exemestane is excreted as parent compound [22]. After β-glucuronidase treatment, the level of 17-dihydroexemestane in the urine from subject taking exemestane was significantly increased and was the major exemestane metabolite observed, suggesting that the major mode of exemestane metabolism in the urine was as a 17-dihydroexemestane glucuronide conjugate [19]. To better characterize the glucuronide conjugate of 17-dihydroexemestane, incubations with HLM were examined by UPLC. In addition to a peak corresponding to the parent 17-dihydroexemestane (retention time = 3.61 min), a potential glucuronide peak at a retention time of 2.59 min was observed (Fig. 4, panel a, top trace). This peak was not sensitive to treatment with 1 mol/l NaOH (results not shown) but was sensitive to treatment with β-glucuronidase (Fig. 4, panel a, middle trace), suggesting that this glucuronide peak was an O-glucuronide conjugate. The mass spectrum of this peak as determined by UPLC/MS/MS demonstrated a [M+] peak at m/z 475.4 for exemestane-17-O-glucuronide (the glucuronide conjugate of 17-dihydroexemestane), a [M+H]+ peak at m/z 299.6 for 17-dihydroexemestane after loss of the glucuronide acid moiety (molecular weight = 176 g/mol), and a m/z 281.3 fragment after loss of a H2O molecule from 17-dihydroexemestane (Fig. 4, panel b).
Fig. 4.
Characterization of exemestane-17-O-glucuronide by UPLC/MS/MS and kinetic analysis of the glucuronidation of 17-dihydroexemestane by UGToverexpressing cells. Panel (a), Representative UPLC trace using 250 μg human liver microsome (HLM) and 300 μmol/l 17-dihydroexemestane incubated for 2 h (top trace); representative UPLC trace using 250 μg HLM and 300 μmol/l 17-dihydroexemestane incubated with β-glucuronidase for 16 h (middle trace) and representative UPLC trace using 250 μg UGT2B17 cell homogenates and 300 μmol/l 17-dihydroexemestane incubated for 2 h (bottom trace). Panel (b), Mass spectrum of peak at retention time of 2.59 min. Peak at retention time of 3.61 min is 17-dihydroexemestane. Panel (c), Representative concentration curve for exemestane-17-O-glucuronide formation from UGT2B17-overexpressing cell homogenates. The concentrations of 17-dihydroexemestane used were: 4.7, 9.4, 18.8, 37.5, 75, and 150 μmol/l. Panel (d), Representative concentration curve for exemestane-17-O-glucuronide formation from UGT1A10-overexpressing cell homogenates. The concentrations of 17-dihydroexemestane used were: 4.7, 9.4, 18.8, 37.5, 75, 150, and 300 μmol/l. All other conditions are described in the Materials and methods section.
To identify the UGTs responsible for the glucuronidation of 17-dihydroexemestane, homogenates from HEK293 cells individually over-expressing UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B10, 2B11, 2B15 and 2B17 were screened for glucuronidation activity. Four UGTs exhibited detectable activity against 17-dihydroexemestane: the hepatic UGTs 1A4 and 2B17, and UGTs 1A8 and 1A10 whose expression in liver is low or undetectable [33,34]. None of the other UGTs screened in our assays exhibited any glucuronidation activity using up to 1 mg of UGT-overexpressing cell homogenate. As exemplified for incubations with UGT2B17-overexpressing cell homogenates (Fig. 4, panel a, bottom trace), the peak retention time by UPLC for all of the active UGTs was identical to that observed for exemestane-17-O-glucuronide in HLM (Fig. 4, panel a, top trace). Representative plots of glucuronidarion rate versus substrate concentration are shown in Fig. 4 for the UGTs exhibiting the highest activity against 17-dihydroexemestane (UGT2B17, panel c; UGT1A10, panel d). After normalizing for UGT expression by western blot analysis [29,32], the relative glucuronidation activity against 17-dihydroexemestane based on Vmax/KM was UGT2B17 > UGT1A10 > UGT1A8 > UGT1A4 (Table 1; reported in a recent review [35]). The hepatic UGT2B17 exhibited a similar binding affinity (KM = 15 μmol/l) against 17-dihydroexemestane to UGT1A8 and a significantly (P < 0.001) lower KM than that observed for UGTs 1A4 and 1A10. The relative overall activity of UGT2B17 as determined by Vmax/KM was 1.4-fold and 6.2-fold higher than that observed for UGTs 1A10 and 1A8, respectively, and 17-fold that observed for UGT1A4, the other active hepatic glucuronidating enzyme. While UGT1A8 exhibited a lower KM than UGT1A10, the overall activity of UGT1A10 was 4.5-fold that of UGT1A8 as determined by Vmax/KM.
Table 1.
Kinetic analysis of UGTs active against 17-dihydroexemestane
| UGT | Vmax (pmol/min/μg UGT protein)a | KM (μmol/l) | Vmax/KM (nl/min/μg UGT protein)a |
|---|---|---|---|
| UGT1A8 | 0.30 ± 0.06 | 14 ± 3.9 | 22 ± 2.1 |
| UGT1A10 | 12 ± 1.8 | 124 ± 15 | 100 ± 7.9 |
| UGT2B17 | 2.0 ± 0.25 | 15 ± 2.7 | 137 ± 17 |
| UGT1A424Pro/48Leu | 0.27 ± 0.01 | 34 ± 3.9 | 8.1 ± 0.9 |
| UGT1A424Thr/48Leu | 0.26 ± 0.04 | 28 ± 7.8 | 9.8 ± 1.9 |
| UGT1A424Pro/48Val | 0.40 ± 0.03* | 36 ± 5.4 | 12 ± 2.4 |
Analysis of 17-dihydroexemestane glucuronidation by hepatic UGT variants and HLM
UGTs 1A4 and 2B17 are hepatic enzymes that exhibit prevalent polymorphisms that were shown to be functional in earlier studies [30,36]. HEK293 cell lines over-expressing the UGT1A424Pro/48Leu, UGT1A424Thr/48Leu, and UGT1A4 24Pro/48Val variants were described earlier [30,37,38]. Although a significant 1.5-fold increase in Vmax was observed for the UGT1A424Pro/48Val variant against 17-dihydroexemestane as compared to either of the other UGT1A4 variants, no significant difference in overall 17-dihydroexemestane glucuronidation as determined by Vmax/KM were observed for the three variant UGT1A4 isoforms (Table 1).
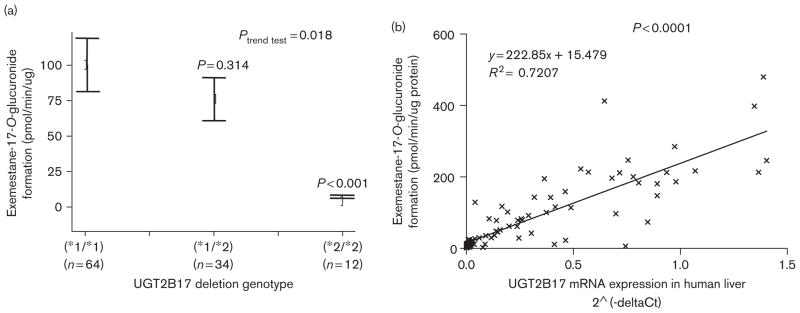
The prevalence of the polymorphic UGT2B17 whole-gene deletion is approximately 30% in Caucasians [27,36,39]. To explore a possible relationship between 17-dihydroexemestane glucuronidation and the UGT2B17 deletion, a series of 110 HLM were examined. The rate of exemestane-17-O-glucuronide formation was determined by UPLC using 9.4 μmol/l 17-dihydroexemestane, a concentration that lies within the linear range of kinetic analysis for HLM against 17-dihydroexemestane. As shown in Fig. 5 (reported in a recent review [35]), there was a significant (P<0.001) 14-fold decrease in glucuronidation activity against 17-dihydroexemestane in HLM from individuals exhibiting the homozygous UGT2B17 deletion genotype (*2/*2) as compared with HLM from individuals wild type for UGT2B17 (*1/*1) (panel a). There was a significant (P=0.018) trend of decreasing HLM exemestane-17-O-glucuronide formation and increasing numbers of the UGT2B17 deletion allele in the specimens examined. A significant (P<0.0001, R2=0.72) correlation was observed between exemestane-17-O-glucuronide formation in HLM and UGT2B17 expression in the same HLM (Fig. 5, panel b). In addition, HLM from individuals exhibiting the UGT2B17 (*2/*2) genotype exhibited a KM that was on average 1.7-fold higher (P=0.012), a Vmax that was on average 22-fold lower (P=0.025), and a Vmax/KM that was on average 36-fold lower (P=0.023) than HLM from individuals with the UGT2B17 (*1/*1) genotype (Table 2).
Fig. 5.
Human liver microsome (HLM) glucuronidating activity against 17-dihydroexemestane versus UGT2B17 genotype and UGT2B17 mRNA expression in human liver. Panel (a), exemestane-17-O-glucuronide formation versus UGT2B17 genotype in HLM. Glucuronidation activity assays were performed using 9.4 μmol/l 17-dihydroexemestane and the glucuronide of 17-dihydroexemestane was separated by UPLC as described in the Materials and methods section. Real-time PCR was performed using genomic DNA from the same liver specimens for which HLMs were also prepared and used to determine UGT2B17 (*1/*1), (*1/*2), and (*2/*2) genotypes. Comparative analysis was performed using HLM from individuals with the UGT2B17 (*1/*1) genotype as the reference, with the P value shown for HLM from individuals with the UGT2B17 (*1/*2), (*2/*2) genotype. Panel (b), exemestane-17-O-glucuronide formation versus UGT2B17 mRNA expression in human liver. UGT2B17 mRNA expression was determined relative to PPIA as the housekeeping gene by real-time PCR by our group previously as described in the Materials and methods section.
Table 2.
Kinetic analysis of 17-dihydroexemestane glucuronidation by individual HLM specimens from individuals stratified by UGT2B17 genotype
| HLM No. | UGT2B17 genotype | Vmax (pmol/min/mg)a | KM (μmol/l) | Vmax/KM (μl/min/mg)a |
|---|---|---|---|---|
| 972 | (*1/*1) | 51.6 | 10.6 | 4.9 |
| 1183 | (*1/*1) | 322 | 7.2 | 45.0 |
| 1392 | (*1/*1) | 393 | 7.1 | 55.0 |
| 2167 | (*1/*1) | 38.9 | 5.1 | 7.6 |
| 2412 | (*1/*1) | 213 | 6.7 | 32.0 |
| mean ± SD | 204 ± 158 | 7.3 ± 2.0 | 28.8 ± 22.3 | |
| 383 | (*2/*2) | 17.0 | 9.3 | 1.8 |
| 416 | (*2/*2) | 5.6 | 17.0 | 0.3 |
| 2174 | (*2/*2) | 13.0 | 12.4 | 1.0 |
| 2175 | (*2/*2) | 2.3 | 13.0 | 0.2 |
| 3873 | (*2/*2) | 8.1 | 11.0 | 0.8 |
| mean ± SD | 9.2 ± 5.9 | 12.6 ± 3.0 | 0.8 ± 0.6 | |
| P valueb | 0.025 | 0.012 | 0.023 |
HLM, human liver microsome.
Kinetic data expressed per mg HLM protein.
Comparing the UGT2B17 (*1/*1) versus (*2/*2) groups.
Discussion
Limited studies have examined the pathways involved in exemestane metabolism. Results from previous studies suggest that reduction of the 17-keto group by aldoketoreductase to form 17-dihydroexemestane appears to be a major pathway for exemestane metabolism [17–21]. The importance of 17-dihydroexemestane in the exemestane metabolic pathway is reinforced by data from preliminary studies demonstrating that 17-dihydroexemestane comprises 48 and 83% of all exemestane metabolites in 1 h incubations containing a NADPH-regenerating system with HLM or human liver cytosol, respectively (mean from four liver specimens); in addition to 17-dihydroexemestane, four metabolites were observed in HLM and one was observed in liver cytosol (Sun and Lazarus; unpublished data). A single study examining the relative activities of exemestane versus 17-dihydroexemestane using placental tissue as the aromatase protein source suggested that the two compounds exhibited similar anti-aromatase activities [23]. This similarity in anti-aromatase activity was confirmed in experiments performed in the present study using an aromatase-over-expressing cell line. 17-dihydroexemestane exhibited an IC50 (2.3 ± 0.83 μmol/l) that was comparable to that observed for exemestane (1.4 ± 0.42 μmol/l). This suggests that as 17-dihydroexemestane is an important metabolite of exemestane, it may contribute significantly to exemestane’s anti-aromatase activity in vivo.
Clinical studies have demonstrated that the oral clearance of exemestane was reduced in patients with significant hepatic or renal impairment, suggesting that altered CYP450 and/or UGT enzyme function might affect exemestane metabolism [40]. As the glucuronide of 17-dihydroexemestane (at the 17-O-position) is an important metabolite found in urine [19], its glucuronidation likely plays a major role in eliminating exemestane-induced activity. In this study, the UGT enzymes active against 17-dihydroexemestane were identified. Four UGTs exhibited activity against 17-dihydroexemestane in vitro: the hepatic UGTs 1A4 and 2B17 and the extra-hepatic UGTs 1A8 and 1A10, with UGT2B17 exhibiting a 2.3-fold lower KM and a 7.4-fold higher Vmax than the other active hepatic UGT, UGT1A4. These data suggest that UGT2B17 is the most active hepatic UGT against 17-dihydroexemestane in vivo. In addition, the KM observed for UGT2B17 against 17-dihydroexemestane in vitro (15 μmol/l) closely approximates the KM observed for HLM (~7 μmol/l), further suggesting that UGT2B17 is the major active liver enzyme responsible for the glucuronidation of 17-dihydroexemestane. Although UGTs 1A8 and 1A10 are either undetectable or expressed at low levels in liver [33,34], they are expressed abundantly in many extra-hepatic tissues [41,42]. As these UGTs also exhibit high activity against 17-dihydroexemestane in vitro, one or both enzymes could also potentially contribute to 17-dihydroexemestane glucuronidation, particularly in target tissues like breast where both enzymes were shown to be expressed [43,44].
The importance of the UGT2B17 enzyme in the hepatic glucuronidation of 17-dihydroexemestane is also strongly supported by the genotype-phenotype correlation observed between HLM exemestane-17-O-glucuronide formation and the presence of the knock-out UGT2B17 deletion allele, with the near-elimination of exemestane-17-O-glucuronide formation in HLM from individuals with the homozygous UGT2B17 deletion genotype. It is likely that the 17-dihydroexemestane glucuronidation activity observed in HLM from UGT2B17 (*2/*2) individuals are because of the presence of active UGT1A4. This possibility is supported by the fact that the approximately 16-fold difference in overall 17-dihydroexemestane glucuronidation activity (Fig. 4) and 36-fold difference in Vmax/KM (Table 2) for HLM from UGT2B17 (*1/*1) versus UGT2B17 (*2/*2) individuals was similar to the decrease in overall kinetic activity observed for UGT2B17 versus UGT1A4 over-expressing cell homogenates. Interestingly, no significant alteration in the overall activity was observed for UGT1A4 variants for 17-dihydroexemestane glucuronidation as determined by Vmax/KM, suggesting that missense variations in UGT1A4 are unlikely to significantly contribute to variations in 17-dihydroexemestane metabolism.
There was some variation in levels of HLM exemestane-17-O-glucuronide formation even within UGT2B17 genotype-stratified groups. As HLM exemestane-17-O-glucuronide formation was correlated with levels of expression of UGT2B17, this suggests that other factors including those that affect UGT2B17 expression may also potentially play a role in regulating exemestane-17-Oglucuronide formation. Interestingly, a similar UGT2B17 genotype-substrate glucuronidation phenotype correlation was observed for other substrates for which UGT2B17 is highly active, including suberoylanilide hydroxamic acid [28] and tobacco-specific nitrosamines like 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol [25,45]. By contrast, HLM specimens that exhibited lower activity in the present study (including sample numbers 972 and 2167 in Table 2) exhibited relatively high activity against other substrates for which UGT2B17 is inactive, including tamoxifen metabolites and nicotine [29,38,46]. These data further support a UGT2B17-related effect on exemestane-17-O-glucuronide formation in human liver.
Earlier reports have indicated that 17-dihydroexemestane may mediate some of the drug effects of exemestane and may thus have clinical importance [24]. The absolute concentrations of 17-dihydroexemestane accounts for about 10–15% of total exemestane dose in the plasma of a small group of individuals treated with exemestane [18,21]. Interestingly, in one of these studies, the 17-dihydroexemestane plasma level in one of three patients was five-fold that observed in the other two patients relative to the levels of parent compound [18]. In addition, a recent study showed that the level of 17-dihydroexemestane was roughly 35–40% of parent exemestane in the plasma of individuals taking exemestane [47]. These studies show that there is variability in the ratio of 17-dihydroexemestane: exemestane in the plasma of individuals taking exemestane. Our study predicts that higher circulating levels of 17-dihydroexemestane may be associated with the UGT2B17 gene deletion – if an individual was deleted for the UGT2B17 gene, the results of this study would suggest that 17-dihydroexemestane glucuronidation and elimination would significantly decrease, increasing overall circulating levels of 17-dihydroexemestane. This increase in circulating 17-dihydroexemestane could increase the exemestane-induced aromatase inhibiting activity and/or exemestane androgenic properties. Owing to the high frequency of the UGT2B17 gene deletion (~30% prevalence in Caucasians [27,36,39]) and its potential significant importance in the metabolism of 17-dihydroexemestane, clinical studies incorporating UGT2B17 genotype as a determinant of exemestane metabolic profiles and overall patient response should be performed.
Acknowledgments
The authors thank the NMR Core and the Molecular Genetics Core facilities at Penn State University College of Medicine for performing NMR of 17-dihydroexemestane and DNA sequencing of aromatase-over-expressing cell lines, respectively. These studies were supported by Public Health Service R01-DE13158 (National Institute for Dental and Craniofacial Research) from the National Institutes of Health, Department of Health and Human Services to P. Lazarus. Some of this work (cited in the text of the manuscript) was reported earlier in a review article (Lazarus P., Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev 2010; 42:176–88).
References
- 1.US National Institutes of Health. 2009 [cited; Available from: http://www.cancer.gov/cancertopics/types/breast] [PubMed]
- 2.Howell A, Howell SJ, Evans DG. New approaches to the endocrine prevention and treatment of breast cancer. Cancer Chemother Pharmacol. 2003;52 (Suppl 1):S39–S44. doi: 10.1007/s00280-003-0645-5. [DOI] [PubMed] [Google Scholar]
- 3.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 4.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 5.Cuzick J, Powles T, Veronesi U, Forbes J, Edwards R, Ashley S, et al. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- 6.Rutqvist LE, Johansson H, Signomklao T, Johansson U, Fornander T, Wilking N Stockholm Breast Cancer Study Group. . Adjuvant tamoxifen therapy for early stage breast cancer and second primary malignancies. J Natl Cancer Inst. 1995;87:645–651. doi: 10.1093/jnci/87.9.645. [DOI] [PubMed] [Google Scholar]
- 7.Van Leeuwen FE, Benraadt J, Coebergh JW, Kiemeney LA, Gimbrere CH, Otter R, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–452. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- 8.Ferretti G, Bria E, Giannarelli D, Felici A, Papaldo P, Fabi A, et al. Second-and third-generation aromatase inhibitors as first-line endocrine therapy in postmenopausal metastatic breast cancer patients: a pooled analysis of the randomised trials. Br J Cancer. 2006;94:1789–1796. doi: 10.1038/sj.bjc.6603194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen A, Trudeau M, Shelley W, Messersmith H, Pritchard KI. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: a systematic review. Cancer Treat Rev. 2008;34:157–174. doi: 10.1016/j.ctrv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Deeks ED, Scott LJ. Exemestane: a review of its use in postmenopausal women with breast cancer. Drugs. 2009;69:889–918. doi: 10.2165/00003495-200969070-00007. [DOI] [PubMed] [Google Scholar]
- 11.Howell A, Cuzick J, Baum M, Buzdar A, Dowsett M, Forbes JF, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365:60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 12.Coates AS, Keshaviah A, Thurlimann B, Mouridsen H, Mauriac L, Forbes JF, et al. Five years of letrozole compared with tamoxifen as initial adjuvant therapy for postmenopausal women with endocrine-responsive early breast cancer: update of study BIG 1–98. J Clin Oncol. 2007;25:486–492. doi: 10.1200/JCO.2006.08.8617. [DOI] [PubMed] [Google Scholar]
- 13.Coleman RE, Banks LM, Girgis SI, Kilburn LS, Vrdoljak E, Fox J, et al. Skeletal effects of exemestane on bone-mineral density, bone biomarkers, and fracture incidence in postmenopausal women with early breast cancer participating in the Intergroup Exemestane Study (IES): a randomised controlled study. Lancet Oncol. 2007;8:119–127. doi: 10.1016/S1470-2045(07)70003-7. [DOI] [PubMed] [Google Scholar]
- 14.Perez EA, Josse RG, Pritchard KI, Ingle JN, Martino S, Findlay BP, et al. Effect of letrozole vs. placebo on bone mineral density in women with primary breast cancer completing 5 or more years of adjuvant tamoxifen: a companion study to NCIC CTG MA 17. J Clin Oncol. 2006;24:3629–3635. doi: 10.1200/JCO.2005.05.4882. [DOI] [PubMed] [Google Scholar]
- 15.Karaer O, Oruc S, Koyuncu FM. Aromatase inhibitors: possible future applications. Acta Obstet Gynecol Scand. 2004;83:699–706. doi: 10.1111/j.0001-6349.2004.00562.x. [DOI] [PubMed] [Google Scholar]
- 16.Grana G. Adjuvant aromatase inhibitor therapy for early breast cancer: a review of the most recent data. J Surg Oncol. 2006;93:585–592. doi: 10.1002/jso.20564. [DOI] [PubMed] [Google Scholar]
- 17.Lonning PE. Pharmacological profiles of exemestane and formestane, steroidal aromatase inhibitors used for treatment of postmenopausal breast cancer. Breast Cancer Res Treat. 1998;49(Suppl 1):S45–S52. doi: 10.1023/a:1006048722559. discussion S73–S77. [DOI] [PubMed] [Google Scholar]
- 18.Evans TR, Di Salle E, Ornati G, Lassus M, Benedetti MS, Pianezzola E, et al. Phase I and endocrine study of exemestane (FCE 24304), a new aromatase inhibitor, in postmenopausal women. Cancer Res. 1992;52:5933–5939. [PubMed] [Google Scholar]
- 19.Mareck U, Geyer H, Guddat S, Haenelt N, Koch A, Kohler M, et al. Identification of the aromatase inhibitors anastrozole and exemestane in human urine using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:1954–1962. doi: 10.1002/rcm.2545. [DOI] [PubMed] [Google Scholar]
- 20.Di Salle E, Ornati G, Paridaens R, Coombes RC, Lobelle JP, Zurlo MG. Preclinical and clinical pharmacology of the aromatase inhibitor exemestane (FCE 24304) In: Motta M, Serio M, editors. Sex hormones and antihormones in endocrine dependent pathology: basic and clinical aspects. Amsterdam: Elsevier; 1994. pp. 279–286. [Google Scholar]
- 21.Traina TA, Poggesi I, Robson M, Asnis A, Duncan BA, Heerdt A, et al. Pharmacokinetics and tolerability of exemestane in combination with raloxifene in postmenopausal women with a history of breast cancer. Breast Cancer Res Treat. 2008;111:377–388. doi: 10.1007/s10549-007-9787-1. [DOI] [PubMed] [Google Scholar]
- 22.Aromasin Exemestane Tablets. 2007 [cited; Available from: http://www.pfizer.com/files/products/uspi_aromasin.pdf]
- 23.Buzzetti F, Di Salle E, Longo A, Briatico G. Synthesis and aromatase inhibition by potential metabolites of exemestane (6-methylenandrosta-1, 4-diene-3,17-dione) Steroids. 1993;58:527–532. doi: 10.1016/0039-128x(93)90029-m. [DOI] [PubMed] [Google Scholar]
- 24.Ariazi EA, Leitao A, Oprea TI, Chen B, Louis T, Bertucci AM, et al. Exemestane’s 17-hydroxylated metabolite exerts biological effects as an androgen. Mol Cancer Ther. 2007;6:2817–2827. doi: 10.1158/1535-7163.MCT-07-0312. [DOI] [PubMed] [Google Scholar]
- 25.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–1196. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 26.Coughtrie MW, Burchell B, Bend JR. A general assay for UDP-glucuronosyltransferase activity using polar amino-cyano stationary phase HPLC and UDP[U-14C]glucuronic acid. Anal Biochem. 1986;159:198–205. doi: 10.1016/0003-2697(86)90328-3. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sexspecific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:823–828. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- 28.Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res. 2009;69:2981–2989. doi: 10.1158/0008-5472.CAN-08-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, et al. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos. 2007;35:2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 30.Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos. 2004;32:72–79. doi: 10.1124/dmd.32.1.72. [DOI] [PubMed] [Google Scholar]
- 31.Fang JL, Beland FA, Doerge DR, Wiener D, Guillemette C, Marques MM, et al. Characterization of benzo(a)pyrene-trans-7,8-dihydrodiol glucuronidation by human tissue microsomes and overexpressed UDP-glucuronosyltransferase enzymes. Cancer Res. 2002;62:1978–1986. [PubMed] [Google Scholar]
- 32.Dellinger RW, Chen G, Blevins-Primeau AS, Krzeminski J, Amin S, Lazarus P. Glucuronidation of PhIP and N-OH-PhIP by UDP-glucuronosyltransferase 1A10. Carcinogenesis. 2007;28:2412–2418. doi: 10.1093/carcin/bgm164. [DOI] [PubMed] [Google Scholar]
- 33.Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, et al. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos. 2009;37:1759–1768. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- 34.Itaaho K, Court MH, Uutela P, Kostiainen R, Radominska-Pandya A, Finel M. Dopamine is a low-affinity and high-specificity substrate for the human UDP-glucuronosyltransferase 1A10. Drug Metab Dispos. 2009;37:768–775. doi: 10.1124/dmd.108.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarus P, Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev. 42:176–188. doi: 10.3109/03602530903208652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson W, III, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 37.Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of udp-glucuronosyltransferase 1a10 (ugt1a10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the ugt1a10139lys isoform. Drug Metab Dispos. 2006;34:943–949. doi: 10.1124/dmd.105.009100. [DOI] [PubMed] [Google Scholar]
- 38.Blevins-Primeau AS, Sun D, Chen G, Sharma AK, Amin S, Lazarus P, et al. Functional significance of polymorphic variants of UDP-glucuronosyltransferase (UGTs) active against tamoxifen metabolites. Cancer Res. 2009;69:1892–1900. doi: 10.1158/0008-5472.CAN-08-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher CJ, Ahn K, Knipe AL, Dyer AM, Richie JP, Jr, Lazarus P, et al. Association between haplotypes of manganese superoxide dismutase (SOD2), smoking, and lung cancer risk. Free Radic Biol Med. 2009;46:20–24. doi: 10.1016/j.freeradbiomed.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jannuzzo MG, Poggesi I, Spinelli R, Rocchetti M, Cicioni P, Buchan P. The effects of degree of hepatic or renal impairment on the pharmacokinetics of exemestane in postmenopausal women. Cancer Chemother Pharmacol. 2004;53:475–481. doi: 10.1007/s00280-004-0774-5. [DOI] [PubMed] [Google Scholar]
- 41.Strassburg CP, Oldhafer K, Manns MP, Tukey RH. Differential expression of the UGT1A locus in human liver, biliary, and gastric tissue: identification of UGT1A7 and UGT1A10 transcripts in extrahepatic tissue. Mol Pharmacol. 1997;52:212–220. doi: 10.1124/mol.52.2.212. [DOI] [PubMed] [Google Scholar]
- 42.Strassburg CP, Manns MP, Tukey RH. Expression of the UDP-glucuronosyltransferase 1A locus in human colon. Identification and characterization of the novel extrahepatic UGT1A8. J Biol Chem. 1998;273:8719–8726. doi: 10.1074/jbc.273.15.8719. [DOI] [PubMed] [Google Scholar]
- 43.Lehmann L, Wagner J. Gene expression of 17beta-estradiol-metabolizing isozymes: comparison of normal human mammary gland to normal human liver and to cultured human breast adenocarcinoma cells. Adv Exp Med Biol. 2008;617:617–624. doi: 10.1007/978-0-387-69080-3_64. [DOI] [PubMed] [Google Scholar]
- 44.Starlard-Davenport A, Lyn-Cook B, Radominska-Pandya A. Identification of UDP-glucuronosyltransferase 1A10 in non-malignant and malignant human breast tissues. Steroids. 2008;73:611–620. doi: 10.1016/j.steroids.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15:769–778. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 46.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp > Tyr) polymorphism. Cancer Res. 2007;67:9024–9029. doi: 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- 47.Corona G, Elia C, Casetta B, Diana C, Rosalen S, Bari M, et al. A liquid chromatography-tandem mass spectrometry method for the simultaneous determination of exemestane and its metabolite 17-dihydroexemestane in human plasma. J Mass Spectrom. 2009;44:920–928. doi: 10.1002/jms.1566. [DOI] [PubMed] [Google Scholar]