Abstract
The matrix protein VP40 plays a critical role in Ebola virus assembly and budding, a process that utilizes specialized membrane domains known as lipid rafts. Previous studies with purified protein suggest a role for oligomerization of VP40 in this process. Here, we demonstrate VP40 oligomers in lipid rafts of mammalian cells, virus-like particles, and in the authentic Ebola virus. By mutagenesis, we identify several critical C-terminal sequences that regulate oligomerization at the plasma membrane, association with detergent-resistant membranes, and vesicular release of VP40, directly linking these phenomena. Furthermore, we demonstrate the active recruitment of TSG101 into lipid rafts by VP40. We also report the successful application of the biarsenic fluorophore, FlAsH, combined with a tetracysteine tag for imaging of Ebola VP40 in live cells.
Keywords: rafts, FlAsH, TSG101, filovirus
The filoviruses Ebola (EBOV) and Marburg (MARV) are nonsegmented negative-strand RNA viruses, that cause severe hemorrhagic fever with high mortality rates in humans (1, 2). Natural outbreaks, although still limited, have risen in recent years. There is growing concern that these viruses could be used as agents of bioterrorism. Therefore, filoviruses remain a serious threat to global public health.
The matrix protein VP40 is critical for assembly and budding of EBOV, as in other negative-strand RNA viruses (3). When expressed in cells, VP40 is primarily associated with the plasma membrane, which is consistent with its proposed role in EBOV budding (4). Truncated VP4031-326 protein is monomeric in solution (5), consisting of two domains each with a β-sandwich folding loosely attached by a flexible linker (6). Oligomerization has been reported for two truncated forms of VP40, but the significance of oligomerization in vivo and the stoichiometry of full-length VP40 remains unknown (6-8). Scianimanico et al. (7) found that destabilizing domain interactions triggers a conformational change resulting in hexamerization of purified VP4031-326, leading to the speculation that such a conformational switch induces viral assembly and budding. However, it remains unclear if and how this conformational change occurs in vivo.
Previous studies (5, 7) mainly focused on the interaction of purified VP40 with synthetic liposomes. Studies using cellular membranes have not differentiated between plasma and internal membranes or between regions of membrane with distinct physical and functional properties (9). Cholesterol-enriched detergent-resistant membrane (DRM) domains within plasma membrane (lipid rafts) play a critical role in a variety of biological functions from cell signaling (10) to virus trafficking (11). We recently showed that Ebola glycoprotein (GP) is located in lipid rafts and determined a critical role for rafts in the entry and release of EBOV (12). In the present study, we demonstrate that VP40, along with TSG101, localizes to plasma membrane rafts in human cells and show the enrichment of VP40 oligomers in DRMs, virus-like particles (VLPs), and in the authentic virus. We also identify C-terminal residues critical for membrane association and vesicular budding. Finally, we have successfully used a biarsenic compound (FlAsH) and a tetracysteine (TC) tag for visualization of VP40 in live cells.
Materials and Methods
Transfections, Cell Lysates, Western Blotting, and Antibodies. The 293T cells were transfected by using Lipofectamine plus (Invitrogen) according to manufacturer's instructions. After 24 h, cells were lysed and Western blot analyses were performed as described (12). Antibodies used were as follows: anti-VP40 mAb (AE11), anti-GP mAb (6D8), anti-Myc mAb (9E10), anti-HA mAb (Santa Cruz Biotechnology), and horseradish peroxidase (HRP)-labeled goat anti-mouse as secondary antibody. Rabbit anti-VP40 antibody was generated by immunizing rabbits with a peptide corresponding to the C-terminal 16 amino acids of Ebola VP40. Convalescent sera from guinea pigs infected with EBOV were used as anti-EBOV antibody (Tom Geisbert, U.S. Army Medical Research Institute of Infectious Diseases).
cDNA Constructs. For live staining of VP40 with the biarsenic fluorophore FlAsH we engineered a 16-aa TC tag (WEAAAREACCRECCARA) at the N terminus of VP40 cDNA (TC-VP401-326) (13). Truncation and point mutants were generated by a Stratagene QuikChange kit and TC-VP401-326 as template. A Myc epitope was introduced into the C terminus of TSG101 and an HA tag was introduced into the N terminus of VP40 by PCR.
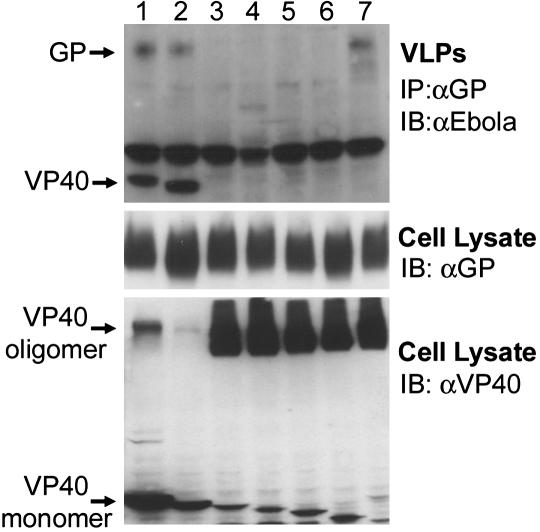
Ebola VLP Production. To generate VLPs, 293T cells were transfected with Ebola VP40 alone or with GP. After 48 h, supernatants cleared from cell debris were overlaid on 20% sucrose and were ultracentrifuged at 100,000 × g at 4°C for 2 h. The pellet was then used directly or further purified on a sucrose step gradient (60%, 40%, and 10%). After ultracentrifugation (100,000 × g for 2 h at 4°C), the VLPs were recovered from the interface of 10% and 40% sucrose and analyzed by immunoblotting. To detect VLPs containing GP+VP40, VLPs were immunoprecipitated from the supernatant by using anti-GP antibody and anti-mouse magnetic beads (Dynal, Great Neck, NY). The coprecipitated VP40 was detected by immunoblotting.
DRM/Lipid Raft Preparation. DRM/lipid raft and nonraft membrane fractions were generated as described (14). Briefly, cells were lysed in hypotonic buffer by Dounce homogenization, the nuclear fraction was pelleted, and supernatants were ultracentrifuged (100,000 × g for 45 min at 4°C). The cytoplasmic supernatants were collected and the pellet containing the membrane and cytoskeletal proteins was solubilized in raft extraction buffer (14) containing 0.5% Triton X-100. Rafts and detergent-soluble membrane (DSM) fractions were then prepared from this lysate as described (14) and were analyzed by immunoblotting.
Confocal Microscopy. The 293T cells grown on chambered cover-slips and transfected with TC-VP40 constructs were rinsed with 400 μM ethane-di-thiol (EDT) in Hanks' balanced salt solution (HBSS)/0.1% glucose/1 mM sodium pyruvate and incubated with 0.5 μM FlAsH (Panvera, Madison, WI) in supplemented HBSS for 1 h at 37°C and washed in EDT-HBSS. For concurrent labeling of rafts, cells were subsequently stained with 40 μg/ml Alexa 594-conjugated cholera toxin subunit B (CTB; Molecular Probes) for 30 min at room temperature, and immunostaining was performed with primary and labeled secondary antibodies (Molecular Probes) as described (12). Fluorescence images were recorded by a Bio-Rad 2000MP confocal system attached to a Nikon TE300 inverted microscope.
Results
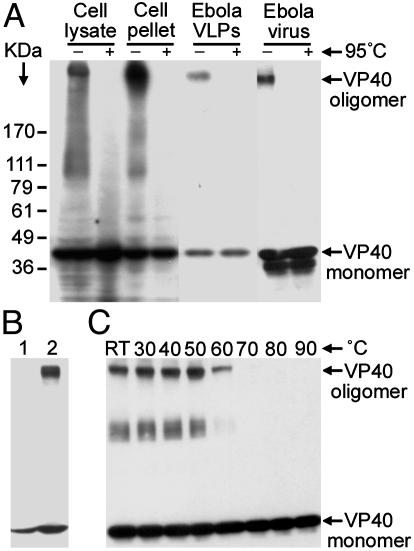
Ebola VP40 Oligomers Associated with Cellular Membranes Are SDS-Resistant but Are Heat-Sensitive. To determine whether VP40 forms oligomers within mammalian cells, we analyzed Western blots of detergent-soluble lysates and insoluble pellets of VP40-expressing cells with and without boiling. An SDS-resistant, but heat-sensitive, VP40 oligomer was detected in both fractions (Fig. 1A, lanes 1-4). VP40 oligomers were also detected in Ebola VLPs and in UV-inactivated EBOV (Fig. 1 A, lanes 5-8). Oligomeric VP40 was exclusively associated with membranes, because no oligomers were detected in the cytosolic fraction obtained by hypotonic lysis (Fig. 1B), thus suggesting that oligomerization of VP40 requires membrane association. To analyze the heat sensitivity of VP40 oligomers, VLPs were lysed in SDS sample buffer and heated at increasing temperatures. VP40 oligomers were resistant to temperatures as high as 50-60°C, suggesting a very stable intermonomer association (Fig. 1C).
Fig. 1.
VP40 oligomers associated with cellular membranes are SDS-resistant but are heat-sensitive. (A) The 293T cells were transfected with VP40 and lysed, and the detergent-soluble lysates and insoluble pellets were subjected to Western blotting (with and without boiling in SDS sample buffer) with anti-VP40 mAb. Ebola VLPs and UV-inactivated EBOV were solubilized in SDS sample buffer and were analyzed similarly. (B) The 293T cells transfected with VP40 were subjected to hypotonic lysis and freeze-thaw cycle, and the cytosolic (lane 1) and membrane (lane 2) fractions were analyzed by Western blotting. (C) Ebola VLPs were solubilized in SDS sample buffer, heated at increasing temperatures, and analyzed by anti-VP40 immunoblotting.
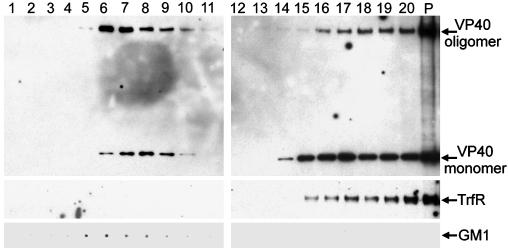
Ebola VP40 Oligomers Are Enriched in Lipid Rafts. To further examine VP40 subcellular localization, DSM and DRM/raft fractions were prepared by sucrose gradient centrifugation from VP40-transfected 293T cells. Fractions (200 μl) from the gradient were analyzed by immunoblotting. A substantial portion of VP40 was found in the raft fractions (fractions 6-10) and appeared to contain more oligomeric VP40 than monomeric VP40 (Fig. 2). In contrast, the soluble fractions (fractions 15-20) contained more monomeric VP40. VP40 was also found in the pellet, which mainly consists of cytoskeleton-associated proteins (Fig. 2, lane P). As a control, we analyzed the fractions for the presence of the raft marker, GM1, and the raft-excluded protein transferrin receptor (TrfR). As expected, we found GM1 only in the low-density detergent-insoluble fractions, whereas TrfR was excluded from these fractions (Fig. 2).
Fig. 2.
Ebola VP40 oligomer is enriched in rafts. Lipid rafts were extracted from membrane preparation of VP40-transfected 293T cells by sucrose gradient ultracentrifugation. Equal aliquots (200 μl) were collected from the gradient starting from the top and were analyzed by Western blotting with anti-VP40 antibody. P, cytoskeletal pellet. The blot was reprobed with anti-TrfR antibody. GM1 was detected by dot blotting using HRP-conjugated cholera toxin B followed by enhanced chemiluminescence.
Characterization of VP40 Truncation and Point Mutants. Prior in vitro studies have revealed that deleting the C-terminal seven residues induces spontaneous multimerization of VP40 without affecting its association with liposomes (7), although removing residues 213-326 triggers oligomerization and simultaneously disrupts the ability of VP40 to associate with liposomes (5). However, those studies were performed by using a purified VP40 mutant lacking the first 30 N-terminal amino acids. Therefore, we sought to examine the role of the C-terminal residues in the context of the full-length protein and in a biologically more relevant system. In addition, it was important to determine which C-terminal sequences are specifically involved in VP40 membrane localization.
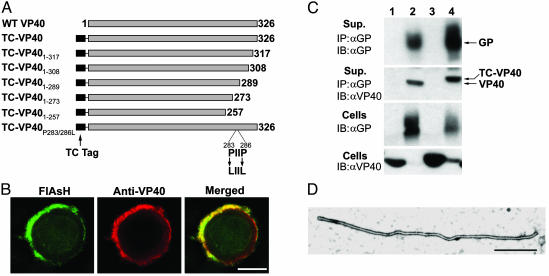
To monitor the subcellular localization of different VP40 mutants in live cells and to correlate this localization with functional data, we used a traceable fluorescent tag. We initially generated a GFP fusion of EBOV VP40 and found that this fusion protein failed to efficiently form VLPs, presumably due to the large size of GFP. To circumvent this problem, we used a tagging approach, recently developed by Griffin and colleagues (15, 16) that exploits the ability of trivalent biarsenic compounds to covalently bind pairs of thiol groups. One such compound, FlAsH, fluoresces green on binding to a TC motif (CCXXCC) engineered into recombinant proteins (13, 16). FlAsH is cell-permeable and is thus useful for live staining. To examine the role of specific C-terminal residues, we engineered a 16-aa TC tag (16) at the N terminus of VP40 (TC-VP40) and in a series of VP40 mutants (Fig. 3A). To verify the specificity of FlAsH staining, TC-VP40 was expressed in 293T cells and was visualized by concurrent staining with FlAsH and anti-VP40 antibody. As shown in Fig. 3B, TC-VP40 showed bright superimposable membrane staining with both FlAsH and anti-VP40 antibody. To evaluate the functionality of TC-VP40, we tested this construct in a VLP release assay. We had previously shown that coexpression of VP40 and GP results in efficient release of filamentous particles (12). To detect the release of VLPs, VP40 was coimmunoprecipitated with anti-GP mAb from culture supernatants. Both wild-type VP40 and TC-VP40 coprecipitated from the supernatant with anti-GP antibody (Fig. 3C). Electron microscopy (EM) analysis of VLPs produced by TC-VP40 and GP revealed a filamentous structure (Fig. 3D) typical for EBOV (12). These data show that the TC tag did not interfere with VP40 function.
Fig. 3.
TC-tagged Ebola VP40 constructs. (A) A schematic representation of the wild-type VP40, TC-VP40, and the different mutant constructs used in this study. (B) FlAsH and Ebola VP40 antibody staining are superimposable. Confocal images of 293T cells transfected with TC-VP40 and simultaneously stained with FlAsH (green) and rabbit anti-VP40 antibody (red). (C) VLPs released into culture supernatant of 293T cells transfected with GP and VP40 or TC-VP40 were immunoprecipitated with anti-GP antibody, and cells were lysed. Immunoprecipitates and lysates were analyzed by Western blotting with anti-GP and VP40 mAbs. (D) EM image of Ebola VLP generated by coexpression of GP and TC-VP40 and purified by sucrose gradient centrifugation. (Scale bar, 1 μm.)
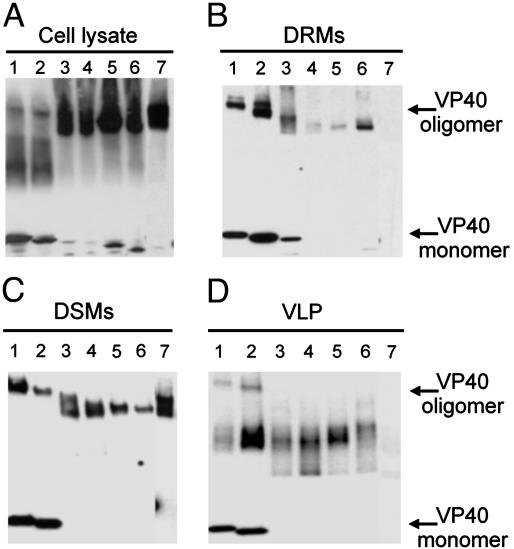
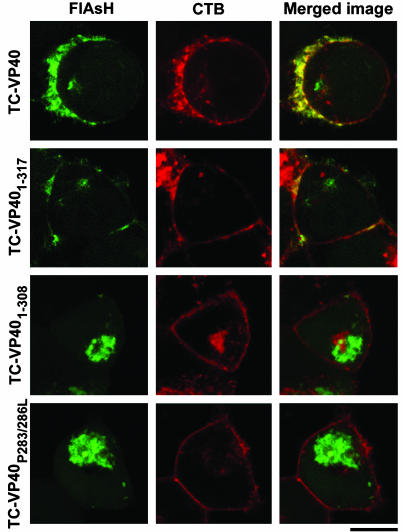
Next, the biochemical and functional properties of the full-length and mutant TC-VP40 constructs were examined. As expected, TC-VP40 (Fig. 4, lane 1) formed DRM-associated oligomers and also produced VLPs, further confirming that the TC tag did not affect the biochemical properties of VP40. TC-VP401-317 also formed oligomers that associated with the DRMs and produced VLPs (Fig. 4, lane 2). The overall oligomer to monomer ratio was not significantly changed by the deletion of the last nine amino acids. In contrast, the truncation of 18 C-terminal residues (TC-VP401-308) resulted in predominantly oligomeric protein that mainly associated with the DSM fraction (Fig. 4 A and B, lane 3). Lack of DRM association of TC-VP401-308 suggests a specific role for residues 309-317 in VP40 interaction with DRMs. Interestingly, this mutant did not produce VLPs (Fig. 4D, lane 3). Other VP40 truncation mutants 1-289, 1-273, and 1-257, were all predominantly oligomeric, and also failed to form VLPs (Fig. 4D, lanes 4-6). The association of mutants 1-289 and 1-273 with DRM was drastically reduced, whereas association with DSMs remained readily detectable (Fig. 4 B and C, lanes 4 and 5). The TC-VP401-257 showed an increased DRM association compared with other truncations (Fig. 4B, lane 6). This result is consistent with a previous report (9) suggesting that deletion of the C-terminal 150 residues enhances the integral membrane character of VP40.
Fig. 4.
Characterization of Ebola VP40 mutant constructs. The 293T cells were transfected with TC-VP401-326 (lane 1), TC-VP401-317 (lane 2), TC-VP401-308 (lane 3), TC-VP401-289 (lane 4), TC-VP401-273 (lane 5), TC-VP401-257 (lane 6), and TC-VP40P283/286L (lane 7). Total cell lysates (A), DRMs (B), DSMs (C), and VLPs (D) were prepared, and, without boiling, were analyzed for the existence of monomeric and oligomeric VP40 by immunoblotting with anti-VP40 mAb.
Destabilization of interdomain interaction has been suggested to be involved in VP40 function (7). Prolines 283 and 286 are part of a hydrophobic zipper in the interdomain interface and a computer analysis of VP40 sequence (http://scansite.mit.edu) revealed that these residues could function as an SH3 domain-binding site. Movements of the two domains could free this motif for interaction with cellular protein, which may be important for VP40 function. Therefore, we mutagenized the paired prolines to leucines. As shown in Fig. 4 (lane 7), TC-VP40P283/286L was almost exclusively in oligomeric form in total cell lysate and DSM fractions but was completely absent from the DRM fraction. Despite high levels of expression, this mutant did not generate VLPs. Taken together, these data strongly suggest that association with DRMs is required for vesicular release of VP40, and that amino acids 309-317, as well as prolines 283 and 286, are crucial for VP40 association with these microdomains as well as vesicular release.
Coexpression of GP greatly enhances the VLP release (12). Therefore, to examine whether the presence of GP has any effect on the behavior of VP40 mutants, wild-type and mutant TC-VP40 were coexpressed with GP, and VLP release was assayed by immunoprecipitation. Similar to results in Fig. 4, only TC-VP40 and 1-317 mutant produced GP/VP40-containing VLPs (Fig. 5). In the presence of VP40P283/286L, some GP was released into the supernatant, but no VP40 was associated with the released GP. In contrast, no GP release was observed in the presence of the truncation mutants. This effect of VP40 mutants on GP release warrants further investigation.
Fig. 5.
The 293T cells were transfected with GP, along with TC-VP401-326 (lane 1), TC-VP401-317 (lane 2), TC-VP401-308 (lane 3), TC-VP401-289 (lane 4), TC-VP401-273 (lane 5), TC-VP401-257 (lane 6), and TC-VP40P283/286L (lane 7). (Top) VLPs were immunoprecipitated and analyzed by a guinea pig anti-Ebola antibody. (Middle) Immunoblotting of cell lysates with anti-GP mAb. (Bottom) Analysis of VP40 expression in cell lysates (samples not boiled).
Confocal Imaging of VP40 Subcellular Localization. In the above biochemical experiments, it was not possible to distinguish between plasma membrane and internal lipid bilayers, which may behave similarly in the extraction process. In fact, lipid rafts have been found in internal membranes (17). On the other hand, EM data indicate that EBOV buds from the plasma membrane (2, 4). Therefore, it was important to verify the raft localization of VP40 and its mutants in intact cells. To address this question, 293T cells transfected with the TC-VP40 constructs were analyzed for VP40 localization in live cells after staining with FlAsH. As expected (18), TC-VP40 was mainly found at the plasma membrane (Figs. 3B and 6). However, we also found a small fraction of VP40 with a globular, and sometimes tubular, appearance inside the cells. When cells were costained with Alexa 594 CTB to visualize rafts, the membrane-bound VP40 was found to colocalize with rafts (Fig. 6, and Movie 1, which is published as supporting information on the PNAS web site). These results confirm the association of VP40 with lipid rafts in intact cells. TC-VP401-317 displayed a different staining pattern. This mutant was associated with the cell membrane at varying degrees in different cells. However, overall membrane association was reduced as compared with TC-VP40 (Fig. 6). This finding suggests that the C-terminal nine amino acids partially contribute to membrane binding. Interestingly, a significant portion of TC-VP401-317 was organized into fibril-like patterns in the cytoplasm (Fig. 6, and Movie 2, which is published as supporting information on the PNAS web site). Similar to full-length VP40, the membrane-associated 1-317 mutant colocalized with lipid rafts (Fig. 6). In contrast, truncation of the C-terminal 18 residues (TC-VP401-308) completely abrogated the binding of VP40 to the plasma membrane and lipid rafts (Fig. 6, and Movie 3, which is published as supporting information on the PNAS web site). This mutant accumulated in the cytoplasm in a large globular form. Mutants 1-289, 1-273, and 1-257 behaved similarly (data not shown, and Fig. 9 and Movie 4, which are published as supporting information on the PNAS web site). The TC-VP40P283/286L mutant also failed to bind to plasma membrane and appeared as large aggregates in the cytoplasm (Fig. 6, and Movie 5, which is published as supporting information on the PNAS web site). Similar results were also obtained when these experiments were performed in the presence of GP (Fig. 10, which is published as supporting information on the PNAS web site), suggesting that GP does not affect the localization of VP40 and its mutants. Whether the aggregates of mutant VP40 are associated with any subcellular organelles remains to be determined.
Fig. 6.
Localization of TC-VP40 and mutant VP40 in 293T cells. Cells were transfected with the indicated plasmids and stained with FlAsH (green) and Alexa 594-conjugated CTB (red) and analyzed by confocal microscopy. Areas of colocalization of VP40 and rafts appear yellow in the overlay. Three-dimensional renderings of the merged images are shown in Movies 1-5. (Scale bar, 10 μm.)
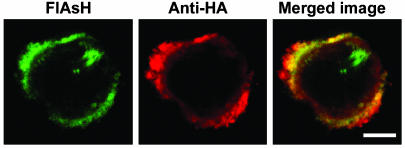
To ascertain that the globular appearance of the VP40 mutants was not a result of misfolding, we examined whether TC-VP401-308 can be redirected to membrane by full-length VP40. When TC-VP401-308 was coexpressed with HA-tagged full-length VP40, most FlAsH staining was observed at the plasma membrane, suggesting that VP40 heterooligomers are capable of localizing to the plasma membrane (Fig. 7). Thus, the globular appearance of the mutants in the cytosol is not due to misfolding, but as a result of loss of C-terminal tail function. Taken together, these data strongly suggest a direct correlation between membrane and possibly raft localization of VP40 and its ability to bud from the cells.
Fig. 7.
Full-length VP40 redirects 1-308 mutant to plasma membrane. The 293T cells were cotransfected with HA-VP40 and TC-VP401-308, stained with FlAsH and anti-HA mAb, and analyzed by confocal microscopy. (Scale bars, 5 μm.)
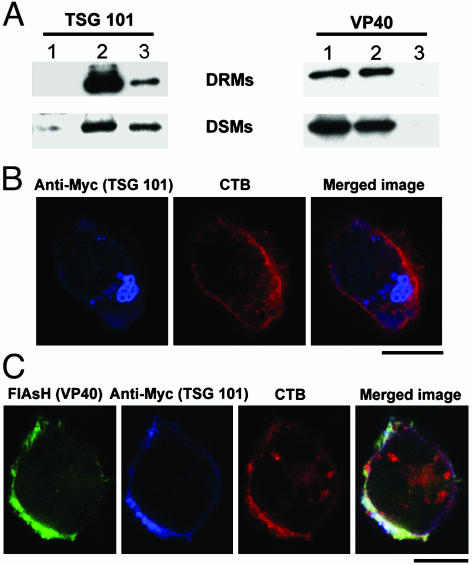
VP40 Redirects TSG101 to Lipid Rafts. Ebola VP40 binds TSG101, a component of the vacuolar protein sorting machinery, through a PTAP motif (18, 19). This interaction appears to be crucial for VP40 vesicular release (20). If raft localization of VP40 oligomers is critical for assembly and budding, then TSG101 should also be recruited into rafts. To test this hypothesis, we expressed VP40, a Myc-tagged TSG101, or a combination of both, in 293T cells, and extracted DRMs and DSMs. As shown in Fig. 8A, TSG101 was equally distributed between the DRM and DSM fractions (Left, lane 3). However, expression of VP40 greatly increased the association of TSG101 with DRMs with little effect on DSM-bound TSG101 (Fig. 8A, lane 2). In the absence of VP40, TSG101 mainly associated with cytosolic vesicular structures and was not detectable at the plasma membrane (Fig. 8B), suggesting that the TSG101 copurified with membrane fractions in Fig. 8A originated from internal membranes. In the presence of TC-VP40, TSG101 was almost entirely found at the plasma membrane in association with VP40 and GM1, showing that VP40 actively redirected TSG101 to cell-surface lipid rafts (Fig. 8C). These findings suggest that the budding complex containing VP40, TSG101, and possibly other components of the ESCRT complex (21), accumulate in lipid rafts to orchestrate a coordinated egress.
Fig. 8.
VP40 recruits TSG101 to lipid rafts. (A) DRM and DSM fractions were prepared from 293T cells transfected with wild-type VP40 alone (lane 1), VP40+TSG101-Myc (lane 2), and TSG101-Myc alone (lane 3) and were analyzed by immunoblotting with anti-VP40 and anti-Myc mAbs. (B) The 293T cells were transfected with TSG101-Myc alone or with TC-VP40, stained with FlAsH (green), anti-Myc antibody (blue), and CTB (red), and analyzed by confocal microscopy. Areas of colocalization of the three markers are white. (Scale bars, 10 μm.)
Discussion
Multimerized viral matrix proteins are believed to form a lattice at the plasma membrane, which triggers membrane bulging, a process that can be driven, in part, by the binding energy released during multimerization (3). This process, in turn, triggers the budding of enveloped viruses (3). Previous studies, using purified protein, showed that Ebola VP40 oligomerizes upon truncation of its C-terminal 8 (7) or 114 amino acids (5). However, the exact mechanism and the biological significance of VP40 oligomerization remain unknown. Here, we demonstrate the existence of SDS-resistant, but heat-sensitive, VP40 oligomers in cells, VLPs, and, importantly, in the authentic virus. The oligomerization of full-length VP40 in cells is completely dependent on membrane binding (Fig. 1B).
Cholesterol-enriched lipid rafts play an important role in several host-pathogen interactions (22-24). Compartmentalization of viral assembly in a specialized microdomain, such as rafts, may increase the efficiency of virus budding and decrease the frequency of defective, noninfectious particles. Our data indicate that VP40 oligomer is selectively enriched in DRMs consisting of lipid rafts, and that all mutants that are defective in plasma membrane and DRM localization also fail to produce VLPs, strongly suggesting a direct relationship between translocation of VP40 to these domains and viral assembly and budding. We previously reported the raft localization of Ebola GP (12). By concentrating GP and oligomerized VP40, rafts may provide a specialized site for the assembly and release of the virus.
Biochemical studies with purified protein have suggested that both the oligomerization and membrane binding of VP40 are regulated by a conformational change that involves movement of the two domains against each other (7). In the crystal structure of the monomeric VP40, the C-terminal tail packs into the interdomain interface and stabilizes interdomain association (6, 7). Our findings indicate that removal of the C-terminal 18 residues of VP40 results in deregulated oligomerization and complete loss of association with the plasma membrane and lipid rafts. This phenotype was rescued by providing the C-terminal tail in trans through coexpression of wild-type VP40 (Fig. 7). Because the two domains in the monomeric VP40 are loosely associated (6), removing the C-terminal tail may expose the interdomain interface. In support of this notion, the recently solved octameric crystal structure of VP4055-194 revealed that approximately the same interdomain surface serves as the interface between individual monomers (8). Therefore, it is possible that the C-terminal 18 amino acids directly interact with components of rafts at the plasma membrane, resulting in their displacement from the interdomain interface and in oligomerization of VP40 at the plasma membrane by exposing the multimerization surface.
Mutation of Pro-283 and Pro-286 also resulted in a phenotype similar to the 1-308 mutant. These two prolines are part of a hydrophobic zipper at the interdomain interface (6). Thus, their removal may have a similar destabilizing effect on interdomain interactions as C-terminal truncations. It was striking that, despite retaining the intact C-terminal tail, the mutation of the prolines completely abolished DRM and plasma membrane association while still allowing association with DSMs (Fig. 6 B and C). These membranes appear to be internal, because confocal analysis showed no association of this mutant with the plasma membrane (Fig. 6). Fusion of the C-terminal 18 residues of VP40 with red fluorescence protein (RFP) did not cause RFP to localize to the plasma membrane (data not shown). These data suggest that the tail alone is not sufficient for membrane targeting, and that the interdomain interface may also play a direct role. The C-terminal tail, once displaced from the interdomain interface, may make the initial contacts with the plasma membrane, resulting in the exposure of a membrane-binding surface in VP40 and simultaneously enabling VP40 to oligomerize. Therefore, this region may functionally couple the membrane association with oligomerization.
SH3 domains are polyproline-binding modules involved in a variety of protein-protein interactions of critical importance for cellular signaling, endosomal sorting, cytoskeletal rearrangements, as well as endocytic vesicle formation; a process that resembles viral budding (25, 26). P283 and P286 are part of a potential SH3 domain-binding motif (27). Therefore, the phenotype of this mutant may relate to loss of binding to SH3-containing cellular proteins that may act as molecular scaffolds during assembly. Binding of the two prolines to SH3 domains may free the oligomerization surface of the N-terminal domain, and, at the same time, lead the molecule to the budding site at the plasma membrane.
Our mutagenesis studies revealed that deleting the last nine residues did not disrupt oligomerization, raft localization, or VLP release, but did partially affect the subcellular distribution of VP40. A larger fraction of 1-317 mutant was found in the cytoplasm partly associated with tubular structures (Fig. 6 and Movie 2). Association with tubular structures was also occasionally observed in cells expressing full-length VP40. The significance of these observations is not clear but may represent a snap shot of cellular trafficking of VP40. The nature of this compartment may be the subject of further investigation. These data suggest that the C-terminal nine amino acids play a partial role in the final translocation to the plasma membrane.
Association of cellular proteins with lipid rafts is often regulated by acylation (28). Whereas no acylation signal is present in VP40, our data indicate that VP40 uses a distinct mechanism for raft localization, which involves a finely regulated conformational change. This mechanism may include a direct interaction between the C-terminal tail and some lipid components of rafts or involve intermediate proteins. It must be noted that, so far, we have not been able to differentiate between raft localization and plasma membrane association by our mutational analysis. This finding suggests that these two processes may be inherently linked. Recently, Licata et al. (20) suggested a role for TSG101, a component of vacuolar protein sorting, in raft localization of VP40. However, in that study, TSG101 and VP40 were shown in postlysis pellet, a fraction that also contains other proteins besides raft proteins. In the absence of VP40, we detected some basal level of TSG101 in DRMs (Fig. 8A), which probably represents association with endosomal compartments with raft-like properties, as was recently shown for recycling endosomes (17). However, TSG101 was detected at the plasma membrane only in presence of VP40, where it colocalized perfectly with VP40 and the raft marker, CTB, suggesting that VP40 mediates TSG101 translocation to rafts. To the best of our knowledge, this is the first report of active recruitment of TSG101 into lipid rafts. Because TSG101 plays a critical role in the budding of late domain-containing viruses (29), this finding further emphasizes a critical role for lipid rafts in the assembly and budding of EBOV.
Throughout our experiments, we consistently observed both oligomeric and monomeric VP40. It is not clear whether this observation truly reflects the coexistence of two different conformations or whether it results from a postlysis equilibrium between the two species. However, we did observe different oligomer:monomer ratios in rafts versus nonraft regions of the membrane, suggesting that the monomer may not be a mere product of postlysis dissociation. In addition, several mutants were almost entirely oligomeric, suggesting that the oligomer, once formed, is largely resistant to SDS. If indeed two conformational pools of VP40 exist in virions and cell membranes, it would raise the question of whether they are carrying out distinct functions.
An important aspect of this study is the visualization of VP40 trafficking in live cells by using the biarsenic compound, FlAsH (13). Whereas our biochemical data indicate that several mutants associate with cellular membranes, live staining shows that some of them do not bind to plasma membrane, but are trapped in the cytoplasm, possibly associated with internal membranes. Thus, it is the combination of biochemical analysis and live staining that enables us to conclude that the ability of VP40 to bind to the plasma membranes and DRMs, and to oligomerize at the plasma membrane directly correlates with its budding capability.
In summary, we have demonstrated that (i) VP40 forms oligomers in cells, VLPs, and the authentic EBOV, (ii) oligomeric full-length VP40 is formed only in association with cellular membranes, (iii) oligomeric VP40 is enriched in lipid rafts, (iv) membrane association and oligomerization of VP40 at the plasma membrane are dependent on the C-terminal tail, and (v) membrane association of VP40 directly correlates with its budding capability. These findings greatly enhance our understanding of the molecular mechanisms of EBOV pathogenesis and suggest that the C-terminal 18 amino acids, as well as the prolines 283 and 286 of VP40, may prove to be important targets for developing therapeutics against Ebola infection.
Supplementary Material
Acknowledgments
We thank Drs. Roger Tsien, Guido M. Gaietta, and Mark Ellisman, [National Center for Microscopy and Imaging Research at the University of California at San Diego, La Jolla, CA, which is supported by National Institutes of Health National Center for Research Resources Grant P41-04050 (to Mark Ellisman)], for valuable advice regarding the live imaging; Melody P. Javid (Montgomery Blair High School, Silver Spring, MD) for excellent technical assistance; Drs. Catherine Bosio and Kelly Warfield for preparation of EBOV; Drs. Alan Schmaljohn, Brad Stiles (U.S. Army Medical Research Institute of Infectious Diseases); and James Burnett (Science Applications International Corporation) for critical review of the manuscript. This work was supported by U.S. Army Medical Research and Material Command Research Plan 03-4-4J-012.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EBOV, Ebola virus; GP, glycoprotein; VLP, virus-like particle; DRM, detergent-resistant membrane; DSM, detergent-soluble membrane; TC, tetracysteine; CTB, cholera toxin subunit B.
References
- 1.Feldmann, H. & Klenk, H. D. (1996) Adv. Virus Res. 47, 1-52. [DOI] [PubMed] [Google Scholar]
- 2.Feldmann, H. & Kiley, M. P. (1999) Curr. Top. Microbiol. Immunol. 235, 1-21. [DOI] [PubMed] [Google Scholar]
- 3.Garoff, H., Hewson, R. & Opstelten, D. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisbert, T. W. & Jahrling, P. B. (1995) Virus Res. 39, 129-150. [DOI] [PubMed] [Google Scholar]
- 5.Ruigrok, R. W., Schoehn, G., Dessen, A., Forest, E., Volchkov, V., Dolnik, O., Klenk, H. D. & Weissenhorn, W. (2000) J. Mol. Biol. 300, 103-112. [DOI] [PubMed] [Google Scholar]
- 6.Dessen, A., Volchkov, V., Dolnik, O., Klenk, H. D. & Weissenhorn, W. (2000) EMBO J. 19, 4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scianimanico, S., Schoehn, G., Timmins, J., Ruigrok, R. H., Klenk, H. D. & Weissenhorn, W. (2000) EMBO J. 19, 6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomis-Ruth, F. X., Dessen, A., Timmins, J., Bracher, A., Kolesnikowa, L., Becker, S., Klenk, H. D. & Weissenhorn, W. (2003) Structure (London) 11, 423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasenosky, L. D., Neumann, G., Lukashevich, I. & Kawaoka, Y. (2001) J. Virol. 75, 5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. A. & London, E. (2000) J. Biol. Chem. 275, 17221-17224. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, S. M., Crowe, S. M. & Mak, J. (2001) J. Clin. Virol. 22, 217-227. [DOI] [PubMed] [Google Scholar]
- 12.Bavari, S., Bosio, C. M., Wiegand, E., Ruthel, G., Will, A. B., Geisbert, T. W., Hevey, M., Schmaljohn, C., Schmaljohn, A. & Aman, M. J. (2002) J. Exp. Med. 195, 593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaietta, G., Deerinck, T. J., Adams, S. R., Bouwer, J., Tour, O., Laird, D. W., Sosinsky, G. E., Tsien, R. Y. & Ellisman, M. H. (2002) Science 296, 503-507. [DOI] [PubMed] [Google Scholar]
- 14.Aman, M. J., Tosello-Trampont, A. C. & Ravichandran, K. (2001) J. Biol. Chem. 276, 46371-46378. [DOI] [PubMed] [Google Scholar]
- 15.Griffin, B. A., Adams, S. R. & Tsien, R. Y. (1998) Science 281, 269-272. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, B. A., Adams, S. R., Jones, J. & Tsien, R. Y. (2000) Methods Enzymol. 327, 565-578. [DOI] [PubMed] [Google Scholar]
- 17.Gagescu, R., Demaurex, N., Parton, R. G., Hunziker, W., Huber, L. A. & Gruenberg, J. (2000) Mol. Biol. Cell 11, 2775-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2001) Nat. Med. 7, 1313-1319. [DOI] [PubMed] [Google Scholar]
- 19.Li, L. & Cohen, S. N. (1996) Cell 85, 319-329. [DOI] [PubMed] [Google Scholar]
- 20.Licata, J. M., Simpson-Holley, M., Wright, N. T., Han, Z., Paragas, J. & Harty, R. N. (2003) J. Virol. 77, 1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pornillos, O., Garrus, J. E. & Sundquist, W. I. (2002) Trends Cell Biol. 12, 569-579. [DOI] [PubMed] [Google Scholar]
- 22.Gatfield, J. & Pieters, J. (2000) Science 288, 1647-1650. [DOI] [PubMed] [Google Scholar]
- 23.Scheiffele, P., Rietveld, A., Wilk, T. & Simons, K. (1999) J. Biol. Chem. 274, 2038-2044. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, D. H. & Hildreth, J. E. (2000) J. Virol. 74, 3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scaife, R. M. & Margolis, R. L. (1997) Cell Signalling 9, 395-401. [DOI] [PubMed] [Google Scholar]
- 26.Simpson, F., Hussain, N. K., Qualmann, B., Kelly, R. B., Kay, B. K., McPherson, P. S. & Schmid, S. L. (1999) Nat. Cell Biol. 1, 119-124. [DOI] [PubMed] [Google Scholar]
- 27.Aman, M. J., Bosio, C. M., Panchal, R. G., Burnett, J. C., Schmaljohn, A. & Bavari, S. (2003) Microbes Infect. 5, 639-649. [DOI] [PubMed] [Google Scholar]
- 28.Simons, K. & Toomre, D. (2000) Nat. Rev. 1, 31-39. [DOI] [PubMed] [Google Scholar]
- 29.Freed, E. O. (2002) J. Virol. 76, 4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.