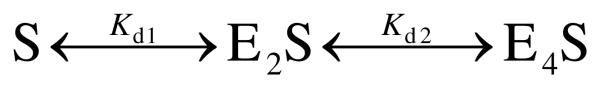Abstract
Benzo[a]pyrene (B[a]P) is a well-characterized environmental polycyclic aromatic hydrocarbon pollutant. In living organisms, B[a]P is metabolized to the genotoxic anti-benzo[a]pyrene diol epoxide that reacts with cellular DNA to form stereoisomeric anti-B[a]PDE-N2-dG adducts. In this study, we explored the effects of adduct stereochemistry and position in double-stranded DNA substrates on the functional characteristics of the catalytic domain of murine de novo DNA methyltransferase Dnmt3a (Dnmt3a-CD). A number of 18-mer duplexes containing site-specifically incorporated (+)- and (−)-trans-anti-B[a]PDE-N2-dG lesions located 3′- and 5′-adjacent to and opposite the target cytosine residue were prepared. The Dnmt3a-CD binds cooperatively to the DNA duplexes with an up to 5-fold greater affinity as compared to the undamaged DNA duplexes. Methylation assays showed a 1.7–6.3 fold decrease of the methylation reaction rates for the damaged duplexes. B[a]PDE modifications stimulated a non-productive binding and markedly favoured substrate inhibition of Dnmt3a-CD independently of DNA methylation status. The latter effect was sensitive to the position and stereochemistry of the B[a]PDE-N2-dG adducts. The overall effect of trans-anti-B[a]PDE-N2-dG adducts on Dnmt3a-CD was less detrimental than in the case of the prokaryotic methyltransferases we previously investigated.
Benzo[a]pyrene (B[a]P) is an ubiquitous and harmful pollutant that is abundant in car exhaust and tobacco smoke (1), and is metabolically activated to the biologically active benzo[a]pyrene-7,8-diol-9,10-epoxides with predominant formation of benzo[a]pyrene-7R,8S-dihydrodiol-9S,10R-epoxide ((+)-anti-B[a]PDE). In the anti-orientation of B[a]PDE, the 7-OH and 9,10-epoxide groups are on opposite sites of the planar polycyclic aromatic ring system (2, 3). Both the (+)- and (−)-anti-enantiomers of B[a]PDE bind covalently to the exocyclic amino group of guanine, with trans or cis opening of the epoxide ring. The most abundant adduct is (+)-trans-anti-B[a]PDE-N2-dG, both in vivo (4) and in vitro (about 90%) (5), with lower amounts of the (+)-cis- and (−)-trans-anti-B[a]PDE-N2-dG adduct being formed when native DNA is reacted with B[a]PDE in vitro (5). The B[a]PDEDNA adducts affect the functional properties of a variety of enzymes that include, for example, bacteriophage T7 RNA polymerase (6), replicative (7) and Y-family DNA polymerases (8), HIV-1 integrase (9), vaccinia DNA topoisomerase and Escherichia coli exonuclease III (10), human topoisomerase I (11), and human DNA polymerase gamma (12). Furthermore, B[a]PDE-DNA adducts exert significant effects on DNA methylation. The treatment of mouse BALB/3T3 CL1-13 cells with B[a]P resulted in a 12% decrease in the overall 5-methylcytosine content of cellular DNA (13). Another study showed that B[a]P exposure of breast cancer cells led to dynamic sequence-specific hypo- and hypermethylation events identified in a number of genomic repeats (14). The B[a]PDE-N2-dG adducts strongly affect DNA methylation catalyzed by the prokaryotic DNA methyltransferases (MTases), M.SssI and M.HhaI (15, 16), and fully abolish methylation when located 5′-adjacent to the target cytosine (15). High levels of B[a]PDE-DNA adducts inhibit DNA methylation by the eukaryotic MTase Dnmt1 (17).
The methylation of DNA is an epigenetic modification that plays an important role in the control of gene expression in mammalian cells (18). Alterations of the DNA methylation status may constitute a key mechanism in tumorigenesis, since global genomic hypomethylation as well as gene-specific hypermethylation are observed in nearly all types of tumors (19, 20). DNA methylation in mammals is carried out by three active MTases, Dnmt1, Dnmt3a, and Dnmt3b, using S-adenosyl-L-methionine as the methyl group donor. Methylation occurs predominantly at the cytosine residues in CpG sequence contexts, yielding 5-methylcytosines (21). Mammalian MTase Dnmt3a is particularly active during embryogenesis, and is commonly thought to be responsible for de novo methylation and establishing methylation patterns (22). Recently, the role of Dnmt3a in maintenance DNA methylation has been proposed (23). It was suggested that the bulk of methylation copying is carried out by the maintenance MTase Dnmt1 when DNA is replicated, while Dnmt3a and Dnmt3b complete the methylation process soon after replication and correct errors that are left behind by Dnmt1. Downregulation of Dnmt3a dramatically inhibits the growth and metastasis of malignant melanoma (24) and the replication of colorectal cancer cells (25). Finally, there are indications that direct Dnmt3a–p53 interactions suppress the p53-mediated transcription of tumor suppressor genes (26).
The main objective of this study was to investigate the effects of B[a]PDE-induced DNA damage on methylation carried out by the catalytic domain of murine Dnmt3a. We have uncovered that the enzyme can cooperatively bind to B[a]PDE-DNA adducts with the formation of non-productive complexes, in particular, at high adduct concentrations (substrate inhibition). However, there is little effect of lesions on the catalytic constant at low adduct concentration. Accordingly, we conclude that the overall effects of trans-anti-B[a]PDE-N2-dG adducts on Dnmt3a-CD activity are less severe than those associated with prokaryotic methyltransferases.
EXPERIMENTAL PROCEDURES
Chemicals and Buffers
S-Adenosyl-L-homocysteine was purchased from Sigma (St. Louis, MO). [CH3-3H]AdoMet (77 Ci/mmol, 13 μM) was obtained from Amersham Biosciences (Little Chalf-ont, U.K.), and (γ-32P)-ATP (1 Ci/μmol) from ≪Isotop≫ (Obninsk, Russia). B[a]PDE-modified oligodeoxynucleotides (Table 1) were synthesized as described previously (15). Other oligodeoxynucleotides were purchased from Syntol (Moscow, Russia) and IDT (Coralville, IA). The fluorescein label (FAM) was introduced at the 5′-end of the oligodeoxynucleotides using an aminoalkyl linker containing six methylene groups. The oligodeoxynucleotide concentrations were determined spectrophotometrically (27).
Table 1.
Oligodeoxynucleotide sequencesa
| Designation | Sequence |
|---|---|
| GCGC | 5′GAGCCAAGCGCACTCTGA |
| GMGC | 5′GAGCCAAGMGCACTCTGA |
| B−CGC | 5′GAGCCAAB−CGCACTCTGA |
| B+CGC | 5′GAGCCAAB+CGCACTCTGA |
| GCB−C | 5′GAGCCAAGCB−CACTCTGA |
| GCB+C | 5′GAGCCAAGCB+CACTCTGA |
| GMB−C | 5′GAGCCAAGMB−CACTCTGA |
| GMB+C | 5′GAGCCAAGMB+CACTCTGA |
| CGCG | 5′TCAGAGTGCGCTTGGCTC |
| CGMG | 5′TCAGAGTGMGCTTGGCTC |
M, 5-methylcytosine; B+ or B−, (+)- or (−)-trans-anti-B[a]PDE-N2-dG, respectively.
The following buffers (B1-B7) were used: B1, 50 mM sodium phosphate (pH 6.0), 1 M NaCl, 10 mM mercaptoethanol, 10% (v/v) glycerol, 0.1% Triton X-100; B2, buffer B1 containing 17 μg/ml phenylmethanesulfonyl chloride, 5 μg/ml leupeptin and 1 μg/ml pepstatin A; B3, B4, and B5, buffer B2 containing 10 mM, 20 mM and 150 mM imidazole-HCl, respectively; B6, 20 mM Tris-HCl (pH 7.4), 0.2 mM EDTA, 2 mM dithiothreitol, 5% (v/v) glycerol; B7, 20 mM HEPES-NaOH (pH 7.0), 100 mM KCl, 1 mM EDTA, 0.2 mM dithiothreitol.
Enzyme Expression and Purification
The N-terminal His6 tag fusion catalytic domain of Dnmt3a was expressed in E. coli BL21 (DE3) cells (Novagen) using the pET28a plasmid containing Dnmt3a-CD as a vector, as described previously (28). Cells were grown in LB medium at 32°C with intensive aeration until A600 ~ 0.7 was attained. Protein expression was initiated by the addition of 1 mM isopropyl β-D-thiogalactopyranoside. After 3 h, cells were harvested by centrifugation at 3000 g for 15 min and disrupted with an MSE Sonifier (Crawley, U.K.) in buffer B2. The cell debris was removed by centrifugation. The supernatant was applied to a Ni-NTA agarose column (BD Biosciences Clontech) equilibrated with B1. After washing with buffers B2, B3 and B4, Dnmt3a-CD was eluted with buffer B5. The eluate was dialyzed overnight against buffer B6, glycerol was added to 50% (v/v), and BSA to a concentration of 200 μg/ml. All protein expression and purification steps were performed at 4°C. The final preparation of Dnmt3a-CD was >90% pure, as assessed with 12.5% Laemmli polyacrylamide gel electrophoresis. The Dnmt3a-CD concentration was determined by the Bradford assay, and expressed in terms of monomer residues.
Fluorescence Polarization Measurements
Fluorescence polarization was measured using FAM-labeled oligodeoxynucleotide duplexes at 25°C in 500 μL of buffer B7 employing a Beacon 2000 Fluorescence Polarization System (PanVera, USA), with excitation at 488 nm and emission at 535 nm. Oligodeoxynucleotide duplexes (5 nM) containing the FAM label at the 5′-end of one DNA strand (5′-FAM-TCAGAGTGCGCTTGGCTC or 5′-FAM-TCAGAGTGMGCTTGGCTC) were preincubated with 0.1 mM AdoHcy, and the fluorescence polarization of free DNA (P0) was measured. Dnmt3a-CD was added in 1–5 μL aliquots of a 8.5 μM stock solution to a final concentration of 325–480 nM, and the polarization value (P) was recorded 2 min after each addition. The titration was continued until the polarization values reached a constant value (Pmax) corresponding to fully bound duplexes. In all cases, the titration curves were determined in at least two independent trials. The binding data were analyzed in terms of Scheme 1 (29): where E2 and E4 represent the homodimeric and homotetrameric Dnmt3a-CD molecules, respectively, and S denotes FAM-labeled DNA. The dissociation constants, Kd1 and Kd2, were derived from the binding curves by simultaneous fitting equations 1-5 with SCIENTIST from MacroMath (St. Louis, MO) to the fluorescence polarization values P at different total enzyme concentrations [E2]0:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Scheme 1.
[E2] and [S] represent the free enzyme and substrate concentrations, respectively, while [S]0 is a total FAM-DNA concentration. The fitting procedure took into account the mass balance and the decrease in concentrations of both free enzyme and DNA upon complex formation.
Methylation Assays
DNA methylation by Dnmt3a-CD was monitored by measuring the amount of tritium incorporated into the oligodeoxynucleotide duplexes in buffer B7 at 37°C (30). The reaction mixtures contained either 0.1–0.3 μM oligodeoxynucleotide duplex, 1.7–7.6 μM Dnmt3a-CD, and 1.2 μM [CH3-3H]AdoMet (i.e., the enzyme was in an at least 10-fold excess over substrate), or, 2–2.5 μM oligodeoxynucleotide duplex, 0.69 μM Dnmt3a-CD and 2 μM [CH3-3H]AdoMet, representing a 2.9–3.6-fold molar excess of a substrate over the enzyme. The reaction was initiated by the addition of [CH3-3H]AdoMet. After 4–90 min of incubation, 4.9 μl aliquots of the reaction mixtures were pipetted onto DE-81 paper disks (Whatman, Brentford, U.K.). The filters were washed four times with 50 mM KH2PO4 and once with water, dried and placed into 5 ml of ZhS-106 scintillation fluid (Reakhim, Russia). The filter-bound radioactivity was measured using a Tracor Analytic Delta 300 scintillation counter (ThermoQuest/CE Instruments, Piscataway, USA), and the amount of methylated DNA was determined according to a previously described protocol (30). The time-course of each of the methylation reactions were fit to Eq 6, where [P] and [P]lim represent the concentrations of methylated DNA at time t, and infinity, respectively, and k is the apparent rate constant of methylation:
| (6) |
The initial rate of the methylation reaction (v) was calculated from the relationship v = k[P]lim, and the final yield of the methylated product (R) was calculated as [P]lim/[S]0, where [S]0 represents the initial substrate concentration. In experiments measuring only the initial linear parts of the product formation curves, the initial rates of methylation were directly determined from the slopes of these curves, as eq 6 can be approximated by [P] = kt[P]lim in this case.
RESULTS
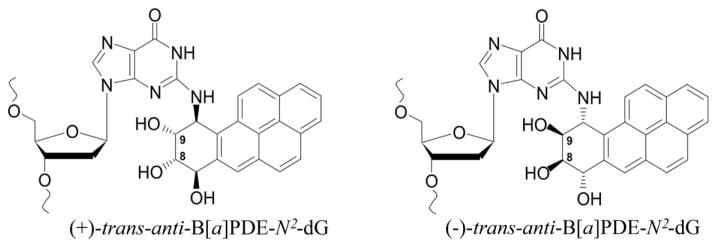
The aim of the present study was to determine whether (+)- and (−)-trans-anti-B[a]PDE-N2-dG lesions (hereafter designated B+ and B−, respectively) (Fig. 1) affect the function of Dnmt3a-CD. We employed the catalytic domain of Dnmt3a, which has a catalytic activity similar to that of the wild-type enzyme (31, 32). Ten different 18-mer oligodeoxynucleotide duplexes with B+ or B− incorporated in one strand of the unmethylated or hemimethylated DNA duplexes within or adjacent to the 5′-side of the Dnmt3a-CD recognition sequence, were prepared (Table 1). Three families of B[a]PDE-damaged duplexes derived from 18-mer undamaged control duplexes GCGC/CGCG, GCGC/CGMG, and GMGC/CGCG were employed in our studies (Table 2). Hemimethylated substrates contained m5dC instead of target dC either in the strand with damaged guanine (GMGC/CGCG family) or in the complementary strand (GCGC/CGMG family). The methylation rates and catalytic constants obtained under conditions of enzyme or DNA excess, as well as the dissociation constants for these three families of DNA duplexes are summarized in Table 2.
Fig. 1.
Structure of (+) and (−)-trans-anti-B[a]PDE-N2-dG adducts.
Table 2.
Binding and kinetic parameters for parental and B[a]PDE-damaged DNA duplexes as substrates of Dnmt3a-CD
| DNA duplexa | Kd1 (nM) | Kd2 (nM) | Kd1Kd2 (nM2) | Rb (%) | v1c (nM/min) | v2 (nM/min) | kcat,1d (h−1) | kcat,2e (h−1) |
|---|---|---|---|---|---|---|---|---|
| GCGC CGCG |
104 ± 26 | 34 ± 10 | 3540 | 95 ± 4 | 25 ± 4 | 3.0 ± 0.3 | 5.2 | 2.7 |
| B−CGC CGCG |
187 ± 54 | 9 ± 3 | 1680 | 94 ± 4 | 34 ± 6 | 1.16 ± 0.03 | 6.9 | 0.59 |
| B+CGC CGCG |
49 ± 12 | 26 ± 6 | 1270 | 100 ± 3 | 17 ± 1 | 3.1 ± 0.3 | 3.4 | 3.7 |
| GCB−C CGCG |
70 ± 25 | 18 ± 7 | 1260 | 70 ± 6 | 27 ± 6 | 1.1 ± 0.1 | 5.5 | 0.88 |
| GCB+C CGCG |
190 ± 59 | 21 ± 7 | 3930 | 74 ± 5 | 30 ± 7 | 0.85 ± 0.03 | 6.2 | 0.54 |
|
| ||||||||
| GMGC CGCG |
170 ± 90 | 22 ± 6 | 3720 | 64 ± 4 | 12 ± 2 | 2.6 ± 0.2 | 2.5 | 1.8 |
| GMB−C CGCG |
73 ± 23 | 30 ± 11 | 2190 | n.d.f | n.d. | 1.6 ± 0.2 | n.d. | 1.7 |
| GMB+C CGCG |
80 ± 27 | 28 ± 13 | 2130 | n.d. | n.d. | 0.89 ± 0.05 | n.d. | 0.92 |
|
| ||||||||
| GCGC CGMG |
Kd1 >> Kd2 | 4400 ± 630 | 28 ± 4 | 7 ± 4 | 2.3 ± 1.3 | 1.4 | 0.94 | |
| B−CGC CGMG |
Kd1 >> Kd2 | 890 ± 60 | 13.7 ± 0.3 | 3.6 ± 0.3 | 0.36 ± 0.01 | 0.73 | 0.14 | |
| B+CGC CGMG |
Kd1 >> Kd2 | 1200 ± 190 | 17 ± 12 | 3.6 ± 0.8 | 0.87 ± 0.03 | 0.73 | 0.45 | |
| GCB−C CGMG |
Kd1 >> Kd2 | 870 ± 120 | 18 ± 1 | 5.0 ± 0.6 | 0.39 ± 0.01 | 1.0 | 0.15 | |
| GCB+C CGMG |
Kd1 >> Kd2 | 990 ± 80 | 17 ± 4 | 3.2 ± 0.6 | 0.36 ± 0.01 | 0.65 | 0.15 | |
The recognition site of Dnmt3a-CD is marked in bold, while the cytosine to be methylated is underlined.
Proportion of DNA converted at the end of the reaction in assays conducted at [E2]0>[S]0.
The initial rate of methylation measured with 100 to 300 nM substrate and normalized to 300 nM substrate.
Apparent catalytic constant obtained by dividing v1 by the concentration of the E4S complex in the corresponding assay. The E4S concentration was calculated using Scheme 1 with the Kd1 and Kd2 values specified in the table.
Apparent catalytic constant obtained by dividing v2 by the concentration of the E4S complex in the corresponding assay.
Not determined.
Effects of B[a]PDE-derived lesions on Dnmt3a-CD Binding
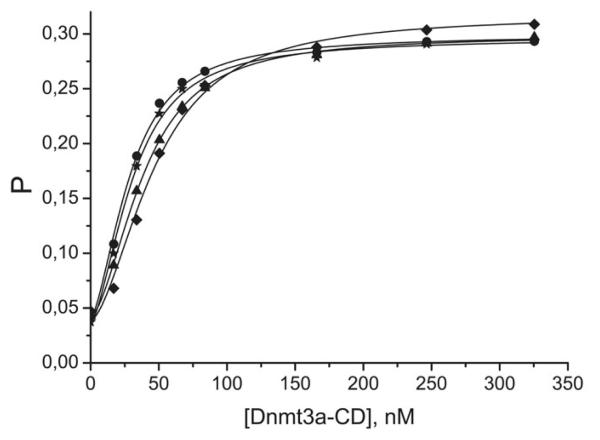
Typical binding curves obtained upon titration of the FAM-labeled 18-mer DNA duplexes with Dnmt3a-CD using a fluorescence polarization technique are presented in Fig. 2. The binding curves of duplexes not shown in this Figure exhibited similar behavior. The co-factor analog, AdoHcy, was added to mimic the experimental conditions of the methylation assays. The binding curves are clearly sigmoidal in shape for the damaged as well as the undamaged DNA duplexes. These results are consistent with the earlier findings that the interactions of Dnmt3a-CD with undamaged and 8-oxoG-containing 30-mer DNA duplexes are characterized by a positively cooperative mode of binding (29).
Fig. 2.
Binding curves obtained upon titration of B[a]PDE-damaged fluorescein-labeled DNA duplexes (10 nM) with increasing amounts of Dnmt3a-CD in the presence of AdoHcy (0.1 mM). P represents the fluorescence polarization value. Total monomeric enzyme concentration is presented. Designations are: (●) B−CGC/CGMG, (▲) B+CGC/CGMG, (✩) GCB−C/CGMG, (◆) GCB+C/CGMG.
The curves can be interpreted in terms of Scheme 1, yielding two macroscopic binding constants, Kd1 and Kd2 (Table 2), for binding of the two Dnmt3a-CD homodimers to the DNA duplex. As shown in Table 2, Kd1 exceeded Kd2 in all cases, whereas Kd1/Kd2 = 0.25 is expected for non-cooperative binding (33). The difference between Kd1 and Kd2 was particularly large in the case of the duplex GCGC/CGMG and its B[a]PDE-derivatives, thus making the fraction of the E2S complex insignificant over the whole range of enzyme concentrations and preventing an evaluation of the magnitudes of the individual binding constants. For these duplexes, only the product Kd1Kd2 could be estimated for comparison with other duplexes.
The binding affinities for B[a]PDE-damaged duplexes, characterized by the values of the dissociation constants product Kd1Kd2, were higher than for the corresponding undamaged duplexes in all families (Table 2), except for GCB+C/CGCG. Notably, the Kd1 values were more sensitive to the B[a]PDE-DNA adducts than the Kd2 values. This finding is consistent with the mode of binding of Dnmt3a, when the second Dnmt3a dimer is attracted to the DNA molecule by interactions with the first, already bound Dnmt3a dimer, a mechanism that should be less sensitive to the presence of the B[a]PDE residues.
Effects of B[a]PDE-derived Lesions on Dnmt3a-CD Catalysis
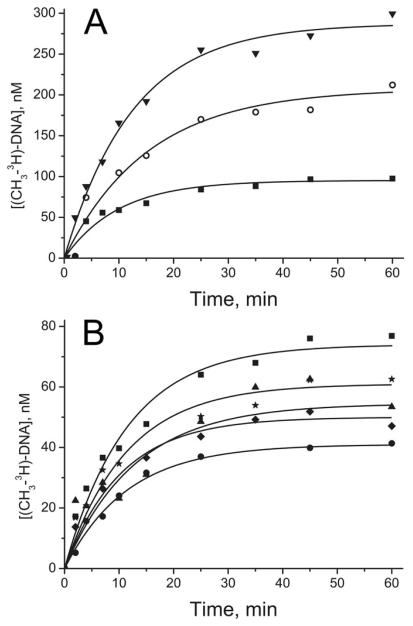
The rates of methylation v1 and v2 were measured at two different sets of enzyme and DNA concentrations: enzyme in excess over substrate or substrate in excess over enzyme (Table 2). In the first case, the measurements spanned the complete time course of the reactions (Fig. 3), and the time course of the reaction was well described by an exponential function (Eq. 6). As the substrate conversion was relatively low in the second set of measurements, the product formation curves were linear (Fig. 4), allowing for the direct determination of the initial rates of methylation (v2) from the slopes. The amounts of methylated products accumulating at the end of the incubation period did not exceed the enzyme concentration in either case, which is indicative of single-turnover conditions. It is worth noting that introduction of B[a]PDE residue into DNA had virtually no effect on v1 values, which were changed by only 1.4-2-fold compared to those for the corresponding undamaged substrates. In contrast, the v2 values were in most cases decreased by the B[a]PDE modifications, by a factor of 1.7–6.3.
Fig. 3.
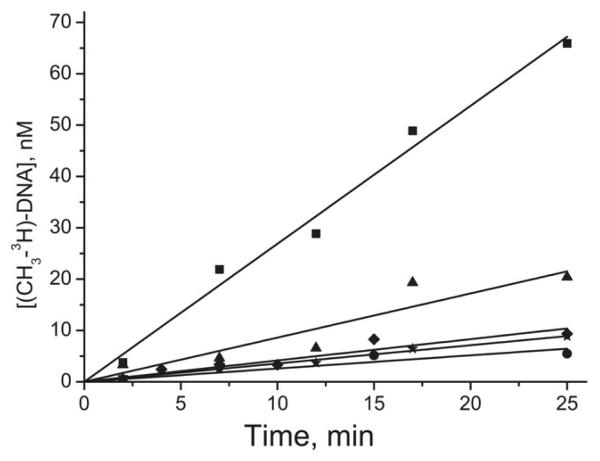
Time-courses of methylation of selected B[a]PDE-damaged (A) and non-damaged (B) DNA duplexes by Dnmt3a-CD under conditions of enzyme excess over DNA. The reaction mixtures contained 300 nM DNA, 5.2-7.6 μM Dnmt3a-CD and 2 μM [CH3-3H]AdoMet. Designations in (A) are: (▼) GCGC/CGCG, (○) GMGC/CGCG, (■) GCGC/CGMG, while those in (B) are as specified for Fig. 2.
Fig. 4.
Time-courses of methylation of selected B[a]PDE-damaged duplexes by Dnmt3a-CD under conditions of DNA excess over enzyme. The reaction mixtures contained 2–2.5 μM DNA, 0.69 μM Dnmt3a-CD, and 2 μM [CH3-3H]AdoMet. Designations are as specified for Fig. 3B.
The initial rates estimated in the two types of assays (v1 and v2 in Table 2) are not directly comparable because the experiments were conducted at different enzyme-substrate concentrations. Therefore, v1 and v2 were converted to apparent catalytic rate constants (kcat,1 and kcat,2, respectively) by dividing these values by the corresponding initial E4S concentrations. The latter was calculated in each case from the total enzyme and substrate concentrations using the Kd1 and Kd2 values determined from the fluorescence titration experiments (Table 2), which was justified by the fact that the FAM label does not affect the DNA-MTase interactions (27). In experiments measuring v1, nearly all of added substrate was present in the form of the E4S complex, whereas 32–88% of the added enzyme formed E4S complexes in the experiments designed to determine v2. The kcat,1 and kcat,2 estimations were based on the earlier finding that E4S is the only catalytically competent complex in this system. The E4S stoichiometry of the substrate complex is consistent with the crystal structure of Dnmt3a-CD complexed with its orthologous protein Dnmt3L, gel-filtration data and the results of mutational analysis (32, 34). Furthermore, this stoichiometry is supported by comparison of Pmax − P0 values for the complexes of GCGC/CGCG with Dnmt3a-CD (0.25–0.26) and M.SssI (0.08-0.09) (data not shown), if one takes into account that the subunit masses of these enzymes are 36 and 44 kDa, respectively, and that M.SssI binds as monomer. Similar stoichiometries were reported for complexes with 30-mer duplexes (29).
The kcat,1 values, derived from enzymatic activities measured at low free substrate concentrations, were several fold lower for the hemimethylated DNA duplexes derived from GMGC/CGCG and GCGC/CGMG than for unmethylated duplexes derived from GCGC/CGCG, but were not significantly affected by the B[a]PDE-N2-dG-adducts (Table 2). In contrast, the kcat,2 values obtained from the activities measured at high free substrate concentrations displayed more marked variations within each of the three different families of DNA duplexes. In general, B[a]PDE-N2-dG lesions led to a decrease in kcat,2 by a factor of 2–7. Furthermore, in most cases, the kcat,2 values were lower than the kcat,1 values, indicative of substrate inhibition, apparently resulting from non-productive binding of additional substrate molecule(s) at high substrate concentration (33, 35, 36). A difference in the catalytic constants for the damaged substrates varied in the three families of duplexes. The largest difference between kcat,1 and kcat,2 (up to 12-fold) values was observed with B[a]PDE-damaged unmethylated substrates, but no inhibition was evident with one such substrate (B+CGC/CGCG). Undamaged substrates showed little substrate inhibition, both in the unmethylated and hemimethylated forms.
Notably, substrate inhibition was negligible at free substrate concentration below 10 nM that were used in the kcat,1 measurements. Therefore, the kcat,1 values refer to the true catalytic constant of Dnmt3a-CD. The differences in kcat,2 values mainly reflect the effects of the structure of the substrate on its ability to form non-productive complex with Dnmt3a-CD at the higher DNA substrate concentrations.
Formation of non-productive complexes
The final yields (R) of methylated products were estimated in assays performed under conditions of enzyme excess. In the case of hemimethylated duplexes, the concentrations of methylated DNA at the end of the reaction were lower than the initial substrate concentrations (0.3 μM) in all cases (Fig. 3). Based on results of experiments with other substrate analogs (29), these phenomenon can be explained in terms of the formation of non-productive enzyme-substrate complexes. The reaction virtually stops after all of the productive complexes are converted, leaving the substrate intact in the non-productive complexes, which dissociates very slowly. The R value may thus vary from 100% (non-productive binding does not occur) to 0% (all of the complexes are non-productive).
For the undamaged, unmethylated substrate (GCGC/CGCG), the R value was nearly 100%, but decreased significantly in the case of the hemimethylated DNA duplexes (GMGC/CGCG and GCGC/CGMG) (Table 2 and Fig. 3A), indicating considerable non-productive substrate binding. The introduction of B[a]PDE-N2-dG-adducts further decreased the R value by approximately 1.5–2-fold (Table 2 and Fig. 3B), the effect being somewhat larger in the case of hemimethylated duplexes in which damaged dG was located in the unmethylated strand.
DISCUSSION
Interactions of Dnmt3a-CD with Non-damaged Substrates
As shown previously, two dimeric Dnmt3a-CD molecules bind cooperatively to 30-mer DNA duplexes to form a catalytically competent E4S complex (29). The present results suggest that the 18-mer DNA duplexes behave similarly to 30-mer duplexes. Specifically, the sigmoidicity of the binding curves in Fig. 2 indicates that two dimeric Dnmt3a-CD molecules bind per DNA duplex and the binding curves exhibit positive cooperativity. The binding affinity is slightly lower, i.e., the Kd1Kd2 value is 4400 nM2 for the 18-mer GCGC/CGMG duplex (Table 2) versus 830 nM2 for a 30-mer GCGC/CGMG duplex (29). This difference is entirely attributable to the Kd1 value since Kd2 values are similar for both duplexes (Table 2 and (29)).
Based on the R values, some inferences can be made on the mode of Dnmt3a-CD binding to DNA. The yields of the methylated products (R) obtained with GCGC/CGCG, GMGC/CGCG and GCGC/CGMG duplexes as substrates were 95:64:28 (Table 2). The R values less than 100% for the hemimethylated duplexes suggest that the active site is oriented towards the already methylated cytosine in the non-productive complex of Dnmt3a-CD. This hypothesis is in agreement with the previous data showing both productive and non-productive binding of 30-mer hemimethylated duplexes (29). The ability of Dnmt3a-CD to bind to methylated strand stems from the observation that the enzyme can interact with fully methylated DNA (29). That the R values are higher for the GMGC/CGCG duplex than for the GCGC/CGMG duplex may indicate preferential Dnmt3a-CD binding to DNA from the side of the bottom strand (TCAGAGTGCGCTTGGCTC and TCAGAGTGMGCTTGGCTC, respectively) in both duplexes, consistent with the preferential methylation of Py−2Pu−1CGPy+1 sequence by Dnmt3a (Py and Pu represent pyrimidine and purine nucleotide, respectively) (37, 38). Based on this, one can further speculate that flanking sequence has a stronger effect on Dnmt3a-CD binding to DNA than the methylation status of the target cytosine.
Interestingly, both kcat,1 and R values for the duplex GMGC/CGCG (2.5 h−1 and 64%, respectively) are higher than those for the duplex GCGC/CGMG (1.4 h−1 and 28%, respectively). This correlation between the kcat,1 and R values may mean that the decreased kcat,1 value for the latter duplex results solely from increased non-productive binding. However, more extensive data are needed to further test this hypothesis.
Interactions of Dnmt3a-CD with B[a]PDE-damaged DNA
The introduction of one B+ or B− lesion stimulated the binding of 18-mer DNA duplexes to Dnmt3a-CD (Table 2), as characterized by the Kd1 and Kd2 values. The accompanying decrease in R suggests that this effect results, at least partly, from the formation of non-productive binding. The increased stabilities of the complexes may result from the contribution of hydrophobic interactions between the B[a]PDE residues and the enzyme. The benzo[a]pyrenyl moieties are positioned in the minor groove of DNA, oriented either towards the 5′- or the 3′-end of the modified strand in the case of the stereoisomeric adducts in the B+ and Bduplexes, respectively (39, 40). However, the catalytic constant estimated at low substrate concentrations (kcat,1) was only slightly affected by the B[a]PDE-N2-dG adducts.
The major effect of the B[a]PDE-N2-dG lesions is to suppress Dnmt3a-CD activity at high substrate concentrations (kcat,2), i.e., to induce strong substrate inhibition. Purdy and co-workers (41) recently reported that there was no inhibition of Dnmt3a-CD activity by unmodified DNA substrates. Our data consistently show that substrate inhibition of Dnmt3a-CD by undamaged 18-mer unmethylated and hemimethylated DNA duplexes is quite small (within the experimental error of kcat,1 and kcat,2) (Table 2). In contrast, DNA duplexes containing the B[a]PDE-N2-dG adducts, except B+CGC/CGCG and, possibly, B+CGC/CGMG, demonstrate strong substrate inhibition of Dnmt3aCD (kcat,1/kcat,2 ~ 4–12). In order for kcat to drop by a factor of 4–12 at the free substrate concentration of approximately 2 μM, as used in kcat,2 measurements, the binding constant describing binding of the inhibiting substrate molecule(s) should be well below 1 μM. The inhibiting substrate molecule(s) may either decrease the true catalytic constant of the productive complex or further decrease R value (i.e., further stimulate non-productive binding). Notably, the R values in Table 2 were derived from assays performed in the absence of substrate inhibition, and may not be the same as those of the substrate-inhibited complexes. We cannot distinguish between these alternatives at present because both effects are expected to decrease the observed kcat value. Another corollary is that Scheme 1 is valid only at low substrate concentrations, such as those employed in fluorescence titration experiments and v1 measurements, and should be supplemented with an E4S2 complex at high substrate concentrations.
The stereochemistry of the B[a]PDE-N2-dG adduct does not appear to be important when located within the CpG site, since the R, kcat,1 and kcat,2 values are similar for the (+)- and (−)-adduct forms (Table 2). However, the stereochemistry is clearly important when the adduct is located on the 5′-side and adjacent to the target cytosine.
Recent studies have shown that the B[a]PDE-N2-dG-adducts strongly affect DNA methylation catalyzed by the prokaryotic MTases, M.EcoRII, M.HhaI, and M.SssI (15, 27). In fact, methylation by M.HhaI and M.SssI was completely abolished when B+ or B− was located on the 5′-side and adjacent to the target cytosine (15). Dnmt3a-CD is therefore considerably less sensitive to these adducts since it displays significant methylation activity although at a several-fold smaller rate. Moreover, the B[a]PDE residues decrease the DNA affinity of M.HhaI up to 100-fold, whereas the same DNA lesions enhance the binding of Dnmt3a-CD to the DNA 4-5 fold. Therefore, we conclude that the methylation activity of Dnmt3a-CD is less affected by the B[a]PDE-N2-dG adducts compared with the prokaryotic MTases. While the evolutionary advantage of such “desensitization” is evident, the elucidation of the underlying mechanism should await determination of three-dimensional structures of the complexes of these enzymes with DNA.
The effects of the B[a]PDE-derived lesions on Dnmt3a-CD differ from those of 7,8-dihydro-8-oxoguanine (8-oxoG), another frequently occurring DNA lesion associated with oxidative stress and also with normal cellular metabolism (42). Guanine oxidation to 8-oxoG generates a strong hydrogen bond acceptor at C8 and converts N7 from a proton acceptor to donor, thus altering the hydrogen bond network between the Dnmt3a-CD and the DNA (29). As a result, the binding of hemimethylated DNA duplexes containing 8-oxoG 5′-adjacent to the target cytosine by Dnmt3a-CD is weakened by a factor of 17, and the rate of methylation determined under conditions of DNA excess is decreased by a factor of 4 (29). The minor groove B[a]PDE-N2-dG adduct produces similar changes in DNA methylation, and the opposite effect on Dnmt3a-CD binding.
In summary, the B[a]PDE-DNA adducts stimulate the binding of Dnmt3a-CD to DNA, enhance the probability of formation of non-productive complexes, and markedly promote substrate inhibition of Dnmt3a-CD. The latter effect is sensitive to the position and the stereochemistry of the adducts. However, the overall effect of B[a]PDE damage on Dnmt3a-CD is less detrimental than it is in the case of prokaryotic methyltransferases.
ACKNOWLEDGEMENTS
We thank Prof. A. Jeltsch for the Dnmt3a-CD plasmid. We are grateful to Prof. N.E. Geacintov for critically reading the manuscript and helpful discussion.
This research was supported by RFBR Grants 09-04-00869, 10-04-00809, 08-04-91109/CRDF RUB1-2919-MO-07, and NIH/NCI grant CA099194.
Abbreviations
- AdoHcy
S-adenosyl-L-homocysteine
- AdoMet
S-adenosyl-L-methionine
- B[a]P
benzo[a]pyrene
- anti-B[a]PDE
r7,t8-dihydroxy-t9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- B+ and B−
(+)- or (−)-trans-anti-B[a]PDE-N2-dG, respectively
- Dnmt3a
DNA methyltransferase Dnmt3a
- Dnmt3a-CD
catalytic domain of Dnmt3a
- EDTA
disodium ethylenediaminetetraacetate
- FAM
6(5)-carboxyfluorescein
- M
5-methylcytosine
- MTase
DNA-(cytosine C5)-methyltransferase
REFERENCES
- 1.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–472. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 2.Conney AH, Chang RL, Jerina DM, Wei SJ. Studies on the metabolism of benzo[a]pyrene and dose-dependent differences in the mutagenic profile of its ultimate carcinogenic metabolite. Drug Metab Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- 3.Boysen G, Hecht SS. Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure-specific methods. Mutat Res. 2003;543:17–30. doi: 10.1016/s1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein IB, Jeffrey AM, Jennette KW, Blobstein SH, Harvey RG, Harris C, Autrup H, Kasai H, Nakanishi K. Benzo(a)pyrene diol epoxides as intermediates in nucleic acid binding in vitro and in vivo. Science. 1976;193:592–595. doi: 10.1126/science.959820. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 6.Choi DJ, Marino-Alessandri DJ, Geacintov NE, Scicchitano DA. Site-specific benzo[a]pyrene diol epoxide-DNA adducts inhibit transcription elongation by bacteriophage T7 RNA polymerase. Biochemistry. 1994;33:780–787. doi: 10.1021/bi00169a020. [DOI] [PubMed] [Google Scholar]
- 7.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 8.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 9.Johnson AA, Sayer JM, Yagi H, Patil SS, Debart F, Maier MA, Corey DR, Vasseur JJ, Burke TR, Jr., Marquez VE, Jerina DM, Pommier Y. Effect of DNA modifications on DNA processing by HIV-1 integrase and inhibitor binding: role of DNA backbone flexibility and an open catalytic site. J Biol Chem. 2006;281:32428–32438. doi: 10.1074/jbc.M605101200. [DOI] [PubMed] [Google Scholar]
- 10.Yakovleva L, Handy CJ, Yagi H, Sayer JM, Jerina DM, Shuman S. Intercalating polycyclic aromatic hydrocarbon-DNA adducts poison DNA religation by Vaccinia topoisomerase and act as roadblocks to digestion by exonuclease III. Biochemistry. 2006;45:7644–7653. doi: 10.1021/bi060158h. [DOI] [PubMed] [Google Scholar]
- 11.Pommier Y, Kohlhagen G, Laco GS, Kroth H, Sayer JM, Jerina DM. Different effects on human topoisomerase I by minor groove and intercalated deoxyguanosine adducts derived from two polycyclic aromatic hydrocarbon diol epoxides at or near a normal cleavage site. J Biol Chem. 2002;277:13666–13672. doi: 10.1074/jbc.M200209200. [DOI] [PubMed] [Google Scholar]
- 12.Graziewicz MA, Sayer JM, Jerina DM, Copeland WC. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32:397–405. doi: 10.1093/nar/gkh213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson VL, Jones PA. Inhibition of DNA methylation by chemical carcinogens in vitro. Cell. 1983;32:239–246. doi: 10.1016/0092-8674(83)90514-7. [DOI] [PubMed] [Google Scholar]
- 14.Sadikovic B, Andrews J, Rodenhiser DI. DNA methylation analysis using CpG microarrays is impaired in benzopyrene exposed cells. Toxicol Appl Pharmacol. 2007;225:300–309. doi: 10.1016/j.taap.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Subach OM, Baskunov VB, Darii MV, Maltseva DV, Alexandrov DA, Kirsanova OV, Kolbanovskiy A, Kolbanovskiy M, Johnson F, Bonala R, Geacintov NE, Gromova ES. Impact of benzo[a]pyrene-2′-deoxyguanosine lesions on methylation of DNA by SssI and HhaI DNA methyltransferases. Biochemistry. 2006;45:6142–6159. doi: 10.1021/bi0511639. [DOI] [PubMed] [Google Scholar]
- 16.Subach OM, Maltseva DV, Shastry A, Kolbanovskiy A, Klimasauskas S, Geacintov NE, Gromova ES. The stereochemistry of benzo[a]pyrene-2′-deoxyguanosine adducts affects DNA methylation by SssI and HhaI DNA methyltransferases. FEBS J. 2007;274:2121–2134. doi: 10.1111/j.1742-4658.2007.05754.x. [DOI] [PubMed] [Google Scholar]
- 17.Ruchirawat M, Becker FF, Lapeyre JN. Mechanism of rat liver DNA methyltransferase interaction with anti-benzo[a]pyrenediol epoxide modified DNA templates. Biochemistry. 1984;23:5426–5432. doi: 10.1021/bi00318a008. [DOI] [PubMed] [Google Scholar]
- 18.Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- 19.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 20.Kanai Y. Alterations of DNA methylation and clinicopathological diversity of human cancers. Pathol Int. 2008;58:544–558. doi: 10.1111/j.1440-1827.2008.02270.x. [DOI] [PubMed] [Google Scholar]
- 21.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 22.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 23.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng T, Kuang Y, Wang L, Li J, Wang Z, Fei J. An essential role for DNA methyltransferase 3a in melanoma tumorigenesis. Biochem Biophys Res Commun. 2009;387:611–616. doi: 10.1016/j.bbrc.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 25.Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, Rocken C, Ebert MP, Kwok TT, Sung JJ. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101:699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YA, Kamarova Y, Shen KC, Jiang Z, Hahn MJ, Wang Y, Brooks SC. DNA methyltransferase-3a interacts with p53 and represses p53-mediated gene expression. Cancer Biol Ther. 2005;4:1138–1143. doi: 10.4161/cbt.4.10.2073. [DOI] [PubMed] [Google Scholar]
- 27.Baskunov VB, Subach FV, Kolbanovskiy A, Kolbanovskiy M, Eremin SA, Johnson F, Bonala R, Geacintov NE, Gromova ES. Effects of benzo[a]pyrenedeoxyguanosine lesions on DNA methylation catalyzed by EcoRII DNA methyltransferase and on DNA cleavage effected by EcoRII restriction endonuclease. Biochemistry. 2005;44:1054–1066. doi: 10.1021/bi048130y. [DOI] [PubMed] [Google Scholar]
- 28.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 29.Maltseva DV, Baykov AA, Jeltsch A, Gromova ES. Impact of 7,8-dihydro-8-oxoguanine on methylation of the CpG site by Dnmt3a. Biochemistry. 2009;48:1361–1368. doi: 10.1021/bi801947f. [DOI] [PubMed] [Google Scholar]
- 30.Brennan CA, Van Cleve MD, Gumport RI. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986;261:7279–7286. [PubMed] [Google Scholar]
- 31.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 32.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon M, Webb EC, Thorne CJR, Tipton KF. Enzymes. 3rd ed. Longman Group Ltd.; London: 1979. pp. 140–148. [Google Scholar]
- 34.Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008;36:6656–6663. doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibirum and Steady-State Enzyme Systems. John Wiley & Sons; New York: 1993. [Google Scholar]
- 36.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Portland Press; London: 1995. [Google Scholar]
- 37.Lin IG, Han L, Taghva A, O'Brien LE, Hsieh CL. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol Cell Biol. 2002;22:704–723. doi: 10.1128/MCB.22.3.704-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 39.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (−)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti-[BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 41.Purdy MM, Holz-Schietinger C, Reich NO. Identification of a second DNA binding site in human DNA methyltransferase 3A by substrate inhibition and domain deletion. Arch Biochem Biophys. 2010;498:13–22. doi: 10.1016/j.abb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]