Abstract
Interferon beta (IFNβ) production is an inaugural event in the innate immune response to viral infections, with relatively small fold changes in IFNβ expression resulting in the activation of important antiviral signaling cascades. In our rapid SIV/macaque model of HIV encephalitis, virus enters the CNS within four days of infection, accompanied by a marked IFNβ response that wanes as SIV replication is controlled. The centrality of IFNβ to the innate antiviral response in the CNS combines with the potential inflammatory damage associated with long-term activation of this pathway to suggest that IFNβ may be subject to regulatory fine-tuning in addition to well-established transcriptional and message stability mechanisms of regulation. Here, we present for the first time evidence that microRNAs, including miR-26a, -34a, -145, and let-7b, may directly regulate IFNβ in human and macaque cells. In primary primate macrophages, the main cell type implicated in HIV and SIV infection in the CNS, specific miRNAs reduce, while miRNA inhibitors enhance, IFNβ protein production. The potential biological significance of this regulation is supported by evidence of an apparent negative feedback loop, with increased expression of three IFNβ-regulating miRNAs by primate macrophages exposed to recombinant IFNβ or stimulated to produce IFNβ. Thus, miRNAs may contribute significantly to the regulation of IFNβ in innate immune responses.
Introduction
Acute retroviral infection provokes rapid and striking innate immune responses in what has been termed a “cytokine storm” (1). These responses are crucial in determining the course of disease, as a delicate balance must be achieved between pro- and anti-inflammatory processes. The former must be sufficiently ferocious to dampen viral replication and impede further infection, while the latter are needed to prevent the tissue damage inherent in chronic activation of the immune system. In our rapid SIV/macaque model of HIV encephalitis, we have shown that both virus and the innate response are present in the central nervous system early in acute infection (2-4). In addition, HIV and SIV infection of macrophages induces IFNβ, which in turn evokes downstream antiviral responses (2-8).
The cytokine IFNβ, with both pro- and anti-inflammatory roles, is the main Type I interferon induced during the initial innate response to retroviral infection of the central nervous system (9). IFNβ mRNA and protein levels rise two- and four-fold, respectively, in response to viral replication during the acute phase of infection, followed by a decline during the asymptomatic phase (2, 4). However, these relatively small fold-changes produce multiplicative effects on downstream effectors such as the antiviral protein MxA. IFNβ is also crucial for induction of anti-bacterial defenses (10, 11).
The pivotal nature of IFNβ and the magnified effects of its differential regulation suggest that intricate modulatory mechanisms have evolved to regulate its production. Indeed, over several decades, studies have elucidated numerous transcriptional and post-transcriptional strategies for IFNβ regulation (12). Maniatis and colleagues showed that an adenylate-uridylate-rich element (ARE)3 in the 3′UTR of the IFNβ mRNA was partly responsible for message degradation (13). The IFNβ ARE may be the binding site for destabilizing ARE binding proteins (AUBPs) such as tristetraprolin (14). Interestingly, the early work on these cis-acting elements in the 3′UTR also presaged an additional regulatory possibility: translational modulation by microRNAs (miRNAs)3.
miRNAs are small RNA regulatory molecules, on average 22 nucleotides in length, that, when integrated into protein complexes known as RNA-induced silencing complexes (RISCs), bind to partially complementary sequences in the 3′ untranslated regions of target mRNAs and thereby contribute to gene regulation by inhibiting translation and destabilizing transcripts (15, 16). The human genome may encode over one thousand potentially functional miRNAs (17), and each mature miRNA may have the ability to regulate the expression of many genes. An estimated one-half of all protein-coding transcripts are thought to be subject to miRNA regulation (15). Among several features predicting miRNA regulation of a given 3′UTR is the presence of an AU-rich sequence (18) such as that found in the transcripts of IFNβ and many other cytokines (19). AU-rich elements are relatively unstructured and may thereby enhance miRNA target site accessibility for miRNA-containing ribonucleoproteins. Recent work has identified several potential cytokine miRNA targets (19), with experimental confirmation reported for IL-10 (20) and the p35 subunit of IL-12 (21). Indirect effects of miRNAs on cytokines have also been reported (22, 23). IFNβ is intimately involved in miRNA regulation, modulating the expression of numerous miRNAs (24) as well as the miRNA processing enzyme Dicer (25). To date, no observations of direct effects of miRNAs on IFNβ have been published, although indirect effects of miR-146a on the Type I Interferons have been reported (26, 27).
The importance of IFNβ regulation in the innate immune response to HIV and SIV and the presence of an extensive adenylate-uridylate-rich region in the IFNβ 3′UTR prompted us to investigate the role of miRNA in direct regulation of IFNβ. We used miRNA target prediction algorithms to identify several miRNAs with potential recognition sites in the IFNβ 3′UTR, and we screened for miRNAs expressed and differentially regulated in primary macrophages following exposure to IFNβ. Of these miRNAs, four silenced through the 3′UTR in a reporter assay and affected secretion of IFNβ protein by primary macrophages. Treatment of primary macrophages with IFNβ upregulates the expression of miR-26a, -34a, and let-7b, suggesting a negative feedback loop for the regulation of IFNβ protein. Treatment with polyinosinic:polycytidylic acid (poly I:C), a dsRNA stimulator of innate immune responses including Type I interferons, similarly resulted in miRNA upregulation, and in a time frame consistent with IFNβ-mediated regulation. These findings may have significant implications for the fine-tuning of the innate immune response during retroviral infection and, potentially, for the therapeutic modulation of innate immune responses.
Materials and Methods
MicroRNA target predictions and in silico genomics
Sequences of known and predicted mature human and macaque miRNAs were obtained from the microRNA Registry at miRBase, http://www.mirbase.org/ (28-30). IFNβ sequences were from the NCBI Nucleotide database, http://www.ncbi.nlm.nih.gov/. The prediction algorithms miRanda (31), RNAhybrid (32), microInspector (33), and PITA (34) were used to search for miRNA target sites in the macaque and human IFNβ 3′UTRs. Where applicable, miRNA-target seed sequence matches were allowed to include G:U wobble and up to one mismatch. Human and macaque IFN-beta proteins were found to be 95% identical and 97% similar by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi for NP_001129267.1 and NP_002167.1). miRNA primary transcript sequences were obtained from the UCSC genome browser (http://genome.ucsc.edu/) (35). The presence of CpG islands and transcription factor binding sites was assessed with the UCSC browser tracks “CpG Islands” (36) and “HMR Conserved Transcription Factor Binding Sites” by Weirauch and Raney.
Cells and Reagents
Monocyte-derived macrophages were isolated from pigtailed macaques (Macaca nemestrina) and from human subjects as previously described (7). The Johns Hopkins Institutional Review Board reviewed and approved all human and animal studies, and all samples were obtained in accordance with IRB protocols. Human cells were used in experiments involving ELISA quantitation of IFNβ protein, as the ELISA reagents available are relatively insensitive to macaque IFNβ (in our hands). HEK-293T cells were acquired from ATCC. miRNA mimics and antagonists were purchased from Ambion or Dharmacon. Negative control RNA was the miRIDIAN microRNA Mimic Negative Control #1 from Dharmacon (Sequence: UCACAACCUCCUAGAAAGAGUAGA), screened against the human and macaque genomes by BLAT (37). Oligonucleotides for cloning were obtained from IDT (see Supplemental information for sequences). Recombinant IFNβ (PBL) and polyinosinic:polycytidylic acid (poly I:C, Amersham), were used at 100 U/ml and 50 ug/ml, respectively.
RNA isolation
Total RNA was isolated and purified by Trizol method (Invitrogen) or miRvana kit (Ambion) according to the manufacturer's protocol. Concentration and purity were measured using a NanoDrop spectrophotometer (ND-1000 V3.5.2 software) and denaturing RNA gels, which were imaged by an Eagle Eye detection system (Stratagene).
Vectors
The 3′UTR of the pigtailed macaque IFNβ mRNA was generated by PCR and inserted between the XhoI and NotI restriction sites downstream of the Renilla luciferase gene of the dual luciferase vector psiCHECK-2 (Promega). Sense oligonucleotides containing the predicted wild type and mutated microRNA recognition elements of the IFNβ 3′UTR (for miRNAs let-7b, miR-26a, miR-34a, and miR-145) were annealed with corresponding antisense oligos and inserted between the XhoI and KpnI restriction sites downstream of the in pEGFP-C1 (Clontech). Oligo sequences are provided in Supplemental information.
Luciferase assays
HEK-293T cells were co-transfected with psiCHECK-2 (with or without the IFNβ 3′UTR) and miRNA mimics using Lipofectamine 2000 (Invitrogen). Cell lysates were prepared 24 hours post-transfection and luciferase levels were measured using an Ascent Fluoroskan fluorometer and the Dual Luciferase Reporter Assay System (Promega).
Fluorescence assay
HEK-293T cells were co-transfected with specific pEGFP-C1 constructs and a transfection control, pdsRed-N1 (Clontech). Fluorescence expression was measured 24 hours later with a Typhoon scanner (Amersham-GE Healthcare) and analyzed by ImageQuant software (Amersham). For each transfection, green fluorescence intensities were normalized by red fluorescence.
IFNβ ELISA
Human macrophages were transfected with miRNA mimics or antagonists (Dharmacon, Qiagen, or Ambion) using HiPerFect (Qiagen). After six hours, cells were washed with PBS and refed with media containing 50 μg/ml polyinosinic:polycytidylic acid (poly I:C3, Amersham). Culture supernatants were collected after 24 hours, and IFNβ levels were measured by ELISA (FujiRebio - Invitrogen) according to the manufacturer's protocol but with an overnight primary incubation at 4°C to increase sensitivity. ELISA results were obtained using a Microplate Reader, Model 680 (Bio-Rad).
miRNA and IFNβ mRNA qRT-PCR
Macaque macrophages were treated with 100 U/ml of IFNβ (PBL), and cells were harvested at 2, 8, and 24 hours post-treatment for RNA isolation. Levels of mature miRNAs were measured by individual qRT-PCR assays (Applied Biosystems) per manufacturer's protocol, using 10 ng of total template RNA. Quantitative RT-PCR for IFNβ mRNA was performed as described previously (4).
Statistical analysis
Analysis was performed using Microsoft Office 2004 Excel, including the Data Analysis add-on, and Prism (GraphPad). For Student's t-test, two-tailed tests were performed and equal variance was not assumed. Confidence intervals were generated using Data Analysis (Excel).
Results
Predicted microRNA recognition elements in the IFNβ 3′UTR
To assess the potential for miRNA regulation of IFNβ, we sought miRNAs that could interact with miRNA recognition elements (MREs)3 in the IFNβ 3′UTR. We used multiple prediction algorithms, including miRanda (31), RNAhybrid (32), and microInspector (33), to search for target sites in the macaque and human IFNβ 3′UTRs. We also queried PITA (34), which incorporates 3′UTR folding parameters into its predictions. The different target prediction algorithms place varying emphases on seed sequence complementarity, heteroduplex free energy of binding, location and size of internal loops and bulges, and accessibility of the target site (as predicted by RNA folding).
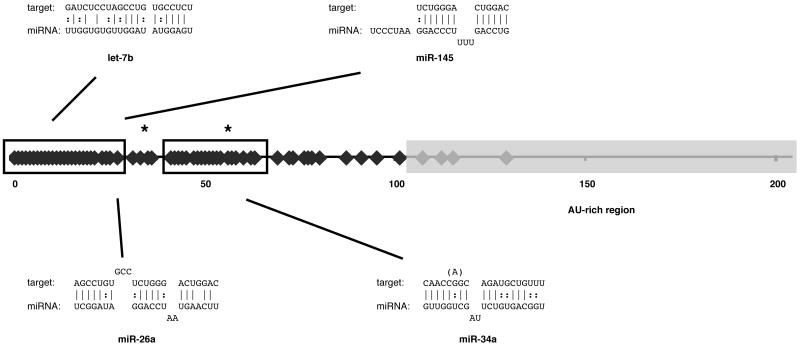
Over 200 total miRNA-mRNA target duplexes were predicted by these algorithms. Most target sites were concentrated in two clusters in the 5′ half of the IFNβ 3′UTR (Figure 1, boxes). To pare this list of predictions to an experimentally tractable size, we next considered only those predictions shared by three prediction programs, and we further reduced the number of candidates by selecting those likely to be expressed in macrophages, a cell type of central importance to the innate immune response, HIV/SIV CNS disease, and IFNβ production (38). The likelihood of expression in macrophages was assessed according to reported miRNA expression in CD14+ cells (39) and our own preliminary results from miRNA microarrays with both human and macaque macrophages (K. W. Witwer, unpublished). miRs -26a, -34a, -145, -181a, -198, and let-7b were selected for initial study.
FIGURE 1. Predicted miRNA recognition elements in the IFNβ 3′UTR.
miRNA prediction algorithms were used to evaluate the likelihood of miRNA binding in the IFNβ 3′UTR. Over 200 hits (3′ ends represented as red diamonds along the ∼200 nt UTR) were filtered by algorithm overlap and expression analysis, with six miRNAs (including the four forming the duplexes shown, which were later validated) selected for further analysis. The 5′ half of the 3′UTR contains most of the predicted miRNA recognition elements (MREs), including those for the four selected miRNAs, which are further concentrated in two areas as indicated by black-outlined boxes. The 3′ half of the UTR is AU-rich (shaded region) and contains few predicted target sites. MREs for miRs -145, -26a, and -let-7b overlap in the first box, while the predicted -34a MRE starts at nt 50 of the 3′UTR. From human to macaque, only two nucleotide changes are found in the target-rich region (asterisks). Neither affects the seed binding regions though to be needed for miRNA interaction. Duplexes are shown with the target sequence above the miRNA sequence. G:U wobble (‘:’) and Watson-Crick pairing (‘|’) are indicated. A macaque to human transition in the -34a binding site (parentheses) makes the human pairing marginally more favorable.
As is the case for most of the other predicted miRNAs, the MREs for macrophage-expressed miRNAs later shown to target the UTR (see below) are located in the target-rich 5′ half of the IFNβ 3′UTR (Figure 1). Importantly, only two nucleotide differences distinguish the human and macaque 3′UTRs in this region (positions indicated by asterisks in Figure 1). Neither of these nucleotides is predicted to bind the 5′ seed (6-8 nucleotides) of the targeting miRNA, although a G-A (macaque-human) difference in the miR-34a binding site changes a “wobble” G-U base pairing outside the seed region to a Watson-Crick interaction, potentially lending a marginally more favorable binding energy to the human miR-34a:target pair. This near-identity of the targeted sequences in human and macaque, combined with the high level of miRNA conservation in primates, suggests that observations made in one species translate well to the other.
miRNA mimics silence reporters containing IFNβ 3′UTR sequences
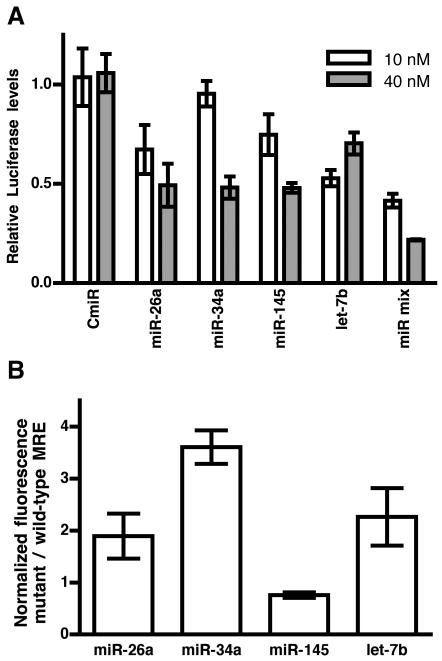
To examine the functional effects of these miRNAs, we used reporter assays. The macaque IFNβ 3′UTR sequence was cloned 3′ to a luciferase reporter gene in a dual luciferase expression vector and transfected into HEK-293T cells. Four of the candidate miRNAs (miRs -26a, -34a, -145, and let-7b), when added exogenously as miRNA mimics, reduced the expression of luciferase in HEK-293T cells (Figure 2a) compared with no-miRNA controls and a control C. elegans miRNA with nucleotide composition similar to that of the mimics of interest. At 10nM transfected mimic, let-7b and an equimolar mixture of the four miRNAs significantly reduced luciferase expression (p<0.001). At 40nM, all four miRNAs reduced expression significantly at p=0.01 or lower. miR-34a and the equimolar mixture achieved significant dose-dependent reduction in luciferase (p<0.01 and p<0.05, respectively). The apparent lack of dose dependence for other miRNAs may indicate that the lower concentration was sufficient to saturate native miRNAs and achieve maximal effect. The equimolar mixture (10nM=2.5nm each, 40nM=10nM each) effected a greater control than any single mimic, suggesting a cooperative effect of multiple microRNA recognition element occupancy, as has been reported previously (40). Predicted binding sites for let-7b, miR-26a, and miR-145 overlap in a region near the 5′ end of the UTR, while the miR-34a MRE is located in a separate region, just 5′ to the AU-rich region.
FIGURE 2. miRNAs directly target sequences in the IFNβ 3′UTR.
miRNA mimics reduce luciferase expression from an IFNβ 3′UTR-containing dual firefly/Renilla luciferase reporter vector (A). Normalized luciferase expression is reduced in HEK-293T cells transfected with specific miRNA mimics and equimolar mixtures of the four indicated miRNAs, but not in cells transfected with a control miRNA (CmiR). Each condition is presented relative to a no-miRNA control, set equal to one. Error bars are SEM from three independent experiments. Individual predicted IFNβ miRNA recognition elements cloned downstream of GFP suppressed GFP expression in HEK-293T cells, but three of four mutated MREs relieved this suppression (B). Fluorescence intensity is displayed as the ratio of mutant to wild-type MRE, normalized to red fluorescence (from pdsRed transfection control). Error bars represent SEM.
To confirm the specific interaction of native miRNAs with the predicted MREs, we designed defective MREs with mutations in three (miR-145) or four (-26a, -34a, let-7b) nucleotides in the 5′ seed-binding region of the MRE (see Supplemental Table I for sequences). Mutations were screened in silico (RNAHybrid) in the context of the full 3′UTR to avoid inadvertent introduction of a novel consensus sequence for another miRNA. Wild-type or seed-mutated MREs corresponding to the four miRNAs were then inserted downstream of a fluorescent reporter. These constructs and normalization controls were transfected into HEK-293T cells. MRE-containing constructs were silenced more efficiently than constructs with seed- mutated MREs (Figure 2b) for all but the miR-145 MRE constructs. This effect was significant (p<0.05) for the miR-34a MRE. The lower level of significance for the -26a and let-7b MREs and the opposite result for the miR-145 constructs suggest several possibilities: the mutations we introduced did not fully abrogate binding of the targeting miRNAs; additional native miRNAs may bind to the unmutated regions of the MREs; and/or the introduced mutations may have created seed binding regions for additional miRNAs that were not predicted by our screening methods. In addition, comparing the results from Figure 1A and B, additional sites for one or more of the four predicted miRNAs present in the full-length UTR, may not be represented in the MREs we examined in these experiments.
miRNA mimics reduce, while miRNA antagonists increase, stimulated secretion of IFNβ
The potential effect of the four identified miRNAs on IFNβ protein production was assessed in primary human macrophages. Human macrophages were chosen for these experiments for several reasons. As described above, the near-identity of known, expressed macaque and human miRNAs and their respective IFNβ 3′UTRs suggest that these miRNAs play the same regulatory roles in humans and macaques. Additionally, no IFNβ ELISAs we tested could reliably detect macaque IFNβ protein in our hands. For human protein, the ELISA we used was sensitive from about 1 through several hundred IU/ml with a slightly modified protocol (see Materials and Methods), and the human IFNβ response to poly I:C quickly reached the upper end of this range (Figure 3). Although macaque macrophages exhibit a robust IFNβ response as measured by qRT-PCR and downstream products of the IFNβ signaling pathway (7), only low levels of protein (in the 1-10 IU/ml range) were detected by ELISA in our experiments. Because the amino acid sequences of the respective proteins are highly similar, we hypothesize that species-specific glycosylation differences are responsible for the observed lack of sensitivity to macaque IFNβ protein. Indeed, changes in two potential N-glycosylation sites and one O-linked site appear to be among the macaque-human amino acid differences (glycosylation predictions made by NetNGlyc, http://www.cbs.dtu.dk/services/NetNGlyc/ and GPP, http://comp.chem.nottingham.ac.uk/glyco/, data not shown) (41).
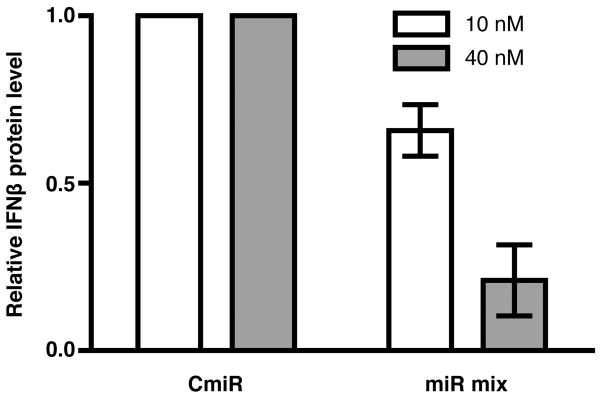
FIGURE 3. miRNA mimics inhibit IFNβ protein secretion by primary macrophages.
A 10 or 40 nM equimolar mixture of four miRNAs (miRs -26a, -34a, -145, and let-7b) or control miRNA was transfected into primary human macrophages, which were then treated with 50 ug/ml poly I:C to stimulate IFNβ production. 24 hours post-treatment, supernatants were collected from no-miRNA controls as well as miRNA- (miR mix) and control miRNA-transfected (CmiR) macrophages; IFNβ levels were measured by ELISA. Levels from control miRNA-treated samples are depicted normalized to poly I:C-treated, no-miRNA controls. Error bars indicate standard deviation.
Macrophages were treated or not with two concentrations of an equimolar mixture of miRs -26a, -34a, -145, and let-7b. The levels of transfected miRNAs were confirmed by qRT-PCR comparison of each miRNA in pre- and post-transfection macrophages (data not shown). Poly I:C RNA (50 ug/ml) was added to the media to stimulate production and secretion of IFNβ protein, and protein levels were measured by ELISA (Figure 3). At 24 hours post stimulation, the level of IFNβ protein secreted by macrophages treated with 10 or 40 nM miRNA mimics (normalized to no-miRNA, poly I:C-treated control) was reduced by approximately 35% and 80%, respectively, compared with stimulated controls treated with a control miRNA.
miRs -26a (42, 43), -34a (44-46), -145 (47), and let-7 family members (48, 49) have reported effects on cell death processes in cancer, and promotion of apoptosis by transfected cells is one a explanation for lower IFNβ production by miRNA-transfected cells. Although no excess cell death was observed in the macrophages transfected with miRNA mimics, the short time course of these experiments might have limited detection. Accordingly, we transfected macrophages with miRNA mimics and control RNA (CmiR) and measured cell death at three and 10 days post-transfection by trypan blue exclusion. No differences in live cell counts were observed between untreated cells, transfection reagent-treated, CmiR-, and miRNA-transfected cells (Supplemental Figure 1). Thus, it is unlikely that the apparent miRNA-mediated downregulation of IFNβ protein is due to promotion of apoptosis by the transfected miRNAs. We also conclude that the pro-apoptotic effects observed in the cancer literature may not triggered by these same miRNAs in the regulatory environment of healthy primary cells.
Beyond cell death, exogenous miRNA mimics could have other unintended indirect or off-target consequences (50), including saturation of the miRNA processing machinery, stimulation of intracellular signaling pathways, and miRNA-mediated up- or down-regulation of transcripts whose products could affect IFNβ production. Accordingly, we sought to inhibit native levels of the four identified miRNAs using miRNA antagonist oligos, chemically modified to enhance stability and reduce off-target effects. These antagomiRs bind to their cognate miRNAs and prevent association with target mRNAs.
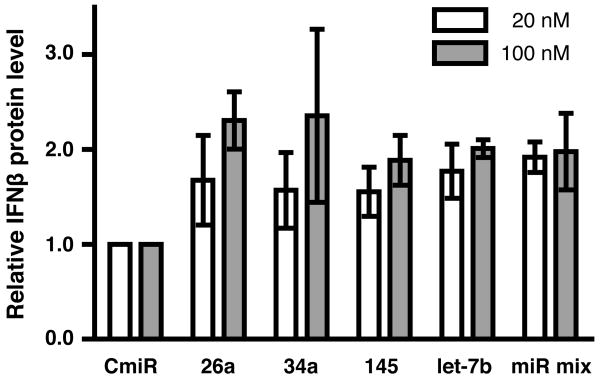
The transfected antagomiRs, both singly and in equimolar mixture, increased the amount of IFNβ secreted by primary macrophages exposed to poly I:C stimulation (Figure 4). At 20 nM added antagonist, the effects of miR-34a and let-7b\\ antagonists were significant, as was an equimolar mixture of the four miRNAs (all, p<0.05). Effects of miR-26a and miR-145 approached significance (p<0.07). At 100 nM, miR-26a, miR-34a, and let-7b produced significant effects (p<0.01), while miR-145 antagonist approached significance (p<0.06). We observed an apparent lack of dose-dependence with the equimolar mixture of antagonist, suggesting that a saturation of some native cognate miRNA may occur at the lower concentration.
FIGURE 4. miRNA antagonists relieve native miRNA inhibition of IFNβ secretion.
Primary human macrophages were transfected with specific or control antagomiRs, chemically modifed to enhance stability and to hinder recognition by intracellular RNA sensors and subsequent activation of the interferon pathway. Macrophages were treated with poly I:C (50 ug/ml). After 24 hours, supernatants were collected and secreted IFNβ protein levels were measured by ELISA. Results from four independent experiments with macrophages from three donors are shown relative to IFNβ levels of poly I:C treated, no-miRNA controls; error bars indicate standard deviation.
The antagonist-mediated increased levels of IFNβ protein do not appear to result from increased levels of IFNβ mRNA. Using quantitative real-time RT-PCR and a delta-delta ct normalization method as described previously (4), we compared IFNβ mRNA levels from primary macrophages treated or not with miRNA antagonists and poly I:C. No significant differences were found between transcript levels in antagonist-treated and -untreated macrophages (R2=0.0035; Supplemental Figure 2).
A negative feedback mechanism: IFNβ stimulates expression of modulating miRNAs
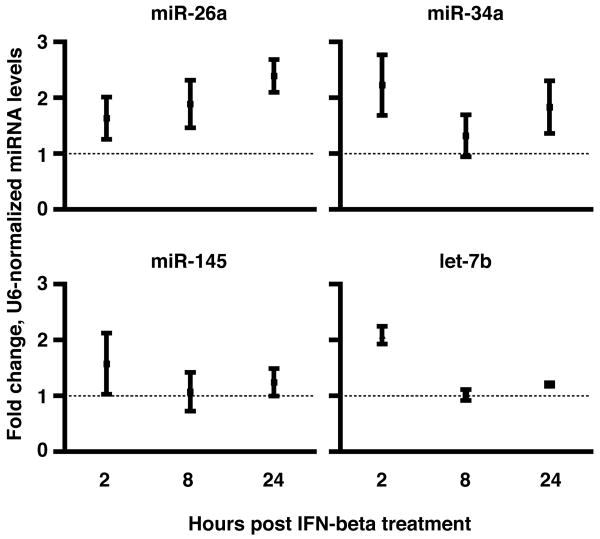
We next assessed the potential biological significance of these results by profiling miRNA expression in primary macaque macrophages treated or not with recombinant IFNβ. Preliminary results from miRNA microarrays had indicated upregulation of three of the four IFNβ-targeting miRNAs upon stimulation with recombinant IFNβ (data not shown). We used a quantitative and mature miRNA-specific method, stem-loop qRT-PCR (51), to measure the levels of miRNAs in primary macaque macrophages treated or not with recombinant IFNβ. Three of the four miRNAs were upregulated in response to IFNβ, and the results suggest the possibility of distinct regulation patterns for different miRNAs in response to IFNβ (Figure 5). miR-26a was upregulated by 2 hr post-IFN treatment, and levels continued to increase through 8 and 24 hr. In contrast, miR-34a increased by 2 hr, was lower at 8 hr, and increased again by 24 hr post-treatment. let-7b, initially increased dramatically, but decreased to background levels at subsequent time points. Consistent modulation was not observed with miR-145.
FIGURE 5. IFNβ treatment of primary macrophages modulates three of four IFNβ-regulating miRNAs.
Primary macrophages from two pigtail macaque donors were treated with 100U/ml recombinant IFNβ, with RNA collected at 2, 8, and 24 hr post-treatment. miRNA levels for miRNAs -26a, -145, -34a, and let-7b were measured on all samples in triplicate by stem-loop qRT-PCR, including no reverse transcriptase and no template controls. The results were analyzed by ΔΔCt, with normalization to U6 snRNA levels and untreated controls. Error bars are standard deviation.
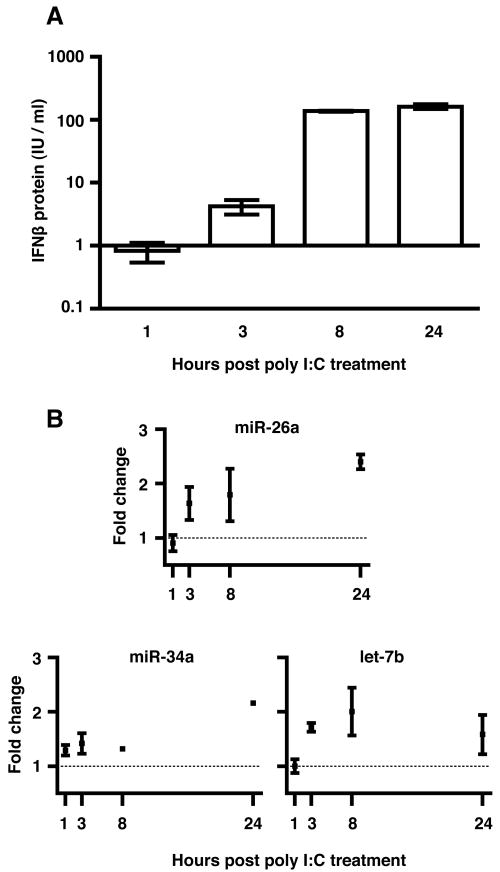
To address whether native IFNβ protein, produced in response to a stimulus such as exogenous dsRNA, could reproduce the effects seen with recombinant protein, we treated primary human macrophages with poly I:C and sampled culture supernatants and total cellular RNA at one, three, eight, and 24 hours post-treatment. ELISA for secreted IFNβ (Figure 6A) revealed that at one hour, protein levels were at or below the limit of detection and did not differ from those of untreated cells. IFNβ was detectable by three hours; a large increase was observed by eight hours. IFNβ remained elevated through 24 hours post-treatment.
FIGURE 6. poly I:C treatment of macrophages induces IFNβ and miRNA production consistent with IFNβ-mediated miR upregulation.
Primary human macrophages were treated with 50 ug/ml poly I:C. IFNβ response (A) was detectable by ELISA by three hours and increased through 24 hours post-treatment. Limit of detection (about 1 IU/ml) is shown as a line. Results are from two donors, measured in duplicate. miRs -26a, -34a, and let-7b were quantitated by stem-loop qRT-PCR (B). Results are fold change of treated over untreated macrophages at each time point, normalized to U6 snRNA. Error bars indicate standard deviation.
Modulation of the three miRNAs that responded to recombinant IFNβ was measured by stem-loop qRT-PCR (Figure 6B). Increased miR abundance in poly I:C-treated cells, compared with untreated cells, occurred only at or after the first detection of IFNβ protein, providing further evidence that IFNβ mediates the abundance of IFNβ-targeting miRNAs by a negative feedback mechanism. miR-26a follows a steady increase like that elicited by recombinant IFNβ. Similarly, let-7b increases initially, then declines. For both -26a and let-7b, early upregulation in the presence of initially low IFNβ protein levels suggests exquisite sensitivity. The 34a response, in contrast, does not appear until much later, when IFNβ protein is present at over 100 IU/ml.
Discussion
Our results suggest that miRNAs -26a, -34a, -145, and let-7b may modulate expression of IFNβ, thereby influencing innate immunity from the earliest responses to viral infection. For -26a, -34a, and let-7b, this modulation may be exerted directly through miRNA recognition elements in the IFNβ 3′UTR. Although we used macaque IFNβ sequences to evaluate direct interactions, our results appear to apply as well to human IFNβ, as supported by ELISA experiments with human macrophages. Based upon target predictions and the sequences of human and macaque IFNβ, we observe that although the human and macaque 3′UTRs have only 94% sequence identity, all but two of the nucleotide differences are found in the relatively MRE-devoid AU-rich 3′ half of the UTR. Also, miRNA binding is thought to be determined largely by perfect or near-perfect target complementarity to a “seed” region of six to eight nucleotides at the 5′ end of the miRNA, and neither of the two macaque-human nucleotide differences in the 5′ half of the IFNβ 3′UTR is found in a relevant predicted seed-binding sequence. The human and macaque miRNA sequences we have examined are identical. Many miRNAs and IFNβ are conserved in vertebrates, implying that the identified miRNA-IFNβ interactions may not be restricted to primates.
The potential biological significance of miRNA regulation of IFNβ in primates receives in vivo support from our SIV/macaque model of HIV encephalitis, as three of the four putative IFNβ-modulating miRNAs are upregulated in the CNS at 42 days post SIV infection (K. W. Witwer, unpublished observations) and may contribute to maintaining the low levels of IFNβ measured at this time point (4). Moreover, biological significance is also supported by the apparent presence of a negative feedback loop. Three of the identified IFNβ-modulating miRNAs are upregulated in response to both recombinant IFNβ and poly I:C-stimulated production of native IFNβ in primary macrophages, suggesting that a biological feedback mechanism may govern the interaction of miRNAs and IFNβ: IFNβ triggers production of miRNAs capable of binding the IFNβ transcript and interfering with protein production.
To confirm that IFNβ itself is necessary for upregulation of miRs -26a, -34a, and let-7b, we performed several experiments with neutralizing antibody to IFNβ. Curiously, the neutralizing antibody appeared to potentiate, not abrogate, the IFNβ response (data not shown), a result consistent with a recent report in this journal (52) wherein IFN neutralizing antibodies are shown to elicit or potentiate Type I IFN responses in endothelial cells and PBMCs. The reported effects are dependent on IFN binding and the presence of the Fc portion of the antibody. In light of these results, future experiments using Fab neutralizing antibody fragments may further clarify the role of IFNβ in miRNA regulation.
Since both miR-26a and let-7b are expressed at relatively and constitutively high levels in many cell types, these miRNAs may exert a constant inhibitory pressure on IFNβ levels even in the absence of miRNA upregulation (53).
Of note, the two IFNβ-regulating miRNAs, -26a and -34a, that are increased from 2 through 24 hours post IFNβ treatment of primary macrophages, are both implicated in cancer and have been studied in the clinic or as potential treatment targets. miR-26a has recently been shown to affect cell cycle progression and used as a therapy for liver cancer in an animal model (42). It may be modulated diurnally (54), and among its targets are PTEN and the Ezh2 histone methyltransferase (55-58). miR-34a has been characterized as a p53-regulated miRNA involved in cell cycle progression and apoptosis (59) and as part of a positive feedback loop involving p53 and SIRT1, a direct target of miR-34a (60). Like miR-26a, -34a is implicated in cancers, and miR-34a expression has been studied in a clinical trial of chronic lymphocytic leukemia (61). Our results indicate an additional mode of action for these important miRNAs.
The mechanisms governing transcriptional and post-transcriptional regulation of IFNβ-regulating miRNAs demand experimental study, but we note here that the genomic context of the primary transcripts for miR-26a, miR-34a, and let-7b suggests several regulatory strategies. The start sites and promoters for these transcripts are found in or near CpG islands (35, 36), implying epigenetic control. Numerous predicted and several experimentally validated transcription factor binding sites are also present and could contribute to regulation.
Of particular interest are two transcription factors, p53 and Stat3. The connection between IFNβ and p53 signaling has recently been characterized (62), and the transcripts for both miR-34a (44) and let-7b (35) contain p53 binding sites. It is thus possible that p53 mediates the IFNβ-stimulated upregulation of these two miRNAs.
We further propose that rapid downregulation of let-7b following a brief post-IFNβ surge, may be effected by Stat3, a transcription factor involved in interferon signaling. Stat3 has been reported as an activator for miR-21 expression (63), but a single transcription factor can have opposite effects in different settings (64). Stat3 has been implicated as a negative regulator of IFN-mediated antiviral responses (65) and, reminiscent of its reported role in downregulating E-Cadherin expression (66), Stat3 may be responsible for repressing let-7b following initial IFNβ-mediated transcriptional upregulation. Interestingly, the predicted Stat3 binding site is actually within the sequence encoding the mature let-7b.
Regulation of innate immune responses, and particularly of IFNβ, has been demonstrated to occur at multiple levels, however, miRNAs have not previously been implicated directly in this regulation. We demonstrate for the first time that secretion of IFNβ from primary macrophages is reduced by miRNAs that recognize sequences in the 3′UTR of the mRNA.
The regulation of IFNβ by miRNAs as demonstrated in these studies is consistent with our broad hypothesis that a cytokine pivotal in downstream signaling for innate immunity requires regulation at every level of expression. IFNβ is among the first genes induced in response to, e.g., retroviral infection, and it then regulates expression of signaling pathways, transcriptional regulation and the induction of antiviral cascades. Two to four fold changes in the level of IFNβ mRNA and protein initiates signaling cascades that ultimately magnify the IFNβ effect thousands to ten thousands fold. Because these downstream changes include potent inflammatory cytokines, the cell has evolved highly sensitive controls, both positive and negative (67), to keep them in careful balance, and previously unknown mechanisms continue to be discovered (68). miRNA regulation of the IFNβ protein is now added to transcriptional control, message stability, and protein stability as an effector of IFNβ regulation.
We note that the miRNAs reported here are unlikely to be the only miRNA species to affect the IFNβ 3′UTR, for at least two reasons. First, despite considerable advances and much thoughtful work in the development of prediction algorithms, no single method or combination of methods is yet completely reliable. Second, we observe that most studies in the current literature make the same assumption that guided our selection of candidate IFNβ regulatory miRNAs: namely, that regulation of a message by a small RNA presupposes (or is suggested by) regulation of the miRNA itself. This is probably an oversimplification. Modulation of gene expression by miRNA is unlikely to stand alone, but rather to be part of a constellation of regulatory mechanisms. A change in any one of the parts of this system could place a greater burden on another arm of the system without corresponding up- or downregulation of the respective components. Since miRNA-mediated regulation without differential expression of the regulating miRNAs is passed over by candidate screening such as in our methods, it is possible that additional small RNA regulators of IFNβ await discovery.
With an increasing number of innate immune system components showing evidence of miRNA targeting, the demonstrated potential of miRNA-based therapeutics (42, 69) provides a promising new possibility for modulation of the body's first line of defense against viral infections.
Supplementary Material
Acknowledgments
We thank: Dr. Joshua T. Mendell for invaluable advice and comments; Brandon T. Bullock for expert technical assistance; and all members of the Molecular and Comparative Pathobiology Retrovirus Laboratory for helpful discussions.
Footnotes
Disclosures: The authors have no financial conflict of interest.
This work was supported by National Institutes of Health Grant MH70306 (to J.E.C.).
Abbreviations used in this paper: ARE (adenylate-uridylate-rich element), miRNA, (microRNA), MRE, (microRNA recognition element), and poly I:C, (polyinosinic:polycytidylic acid)
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, Self SG, Borrow P. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber SA, Herbst DS, Bullock BT, Gama L, Clements JE. Innate immune responses and control of acute simian immunodeficiency virus replication in the central nervous system. J Neurovirol. 2004;10 1:15–20. doi: 10.1080/753312747. [DOI] [PubMed] [Google Scholar]
- 3.Barber SA, Gama L, Li M, Voelker T, Anderson JE, Zink MC, Tarwater PM, Carruth LM, Clements JE. Longitudinal analysis of simian immunodeficiency virus (SIV) replication in the lungs: compartmentalized regulation of SIV. J Infect Dis. 2006;194:931–938. doi: 10.1086/507429. [DOI] [PubMed] [Google Scholar]
- 4.Witwer KW, Gama L, Li M, Bartizal C, Queen SE, Varrone J, Brice A, Graham DR, Mankowski JL, Zink MC, Clements JE. Coordinated Regulation of SIV Replication and Immune Responses in the CNS. PLoS ONE. 2009 doi: 10.1371/journal.pone.0008129. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honda Y, Rogers L, Nakata K, Zhao BY, Pine R, Nakai Y, Kurosu K, Rom WN, Weiden M. Type I interferon induces inhibitory 16-kD CCAAT/enhancer binding protein (C/EBP)beta, repressing the HIV-1 long terminal repeat in macrophages: pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J Exp Med. 1998;188:1255–1265. doi: 10.1084/jem.188.7.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woelk CH, Ottones F, Plotkin CR, Du P, Royer CD, Rought SE, Lozach J, Sasik R, Kornbluth RS, Richman DD, Corbeil J. Interferon gene expression following HIV type 1 infection of monocyte-derived macrophages. AIDS Res Hum Retroviruses. 2004;20:1210–1222. doi: 10.1089/aid.2004.20.1210. [DOI] [PubMed] [Google Scholar]
- 7.Dudaronek JM, Barber SA, Clements JE. CUGBP1 is required for IFNbeta-mediated induction of dominant-negative CEBPbeta and suppression of SIV replication in macrophages. J Immunol. 2007;179:7262–7269. doi: 10.4049/jimmunol.179.11.7262. [DOI] [PubMed] [Google Scholar]
- 8.Henderson AJ, Calame KL. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4(+) T cells. Proc Natl Acad Sci U S A. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin DE. Immune responses to RNA-virus infections of the CNS. Nat Rev Immunol. 2003;3:493–502. doi: 10.1038/nri1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi VD, Kalvakolanu DV, Chen W, Zhang L, Kang TJ, Thomas KE, Vogel SN, Cross AS. A role for Stat1 in the regulation of lipopolysaccharide-induced interleukin-1beta expression. J Interferon Cytokine Res. 2006;26:739–747. doi: 10.1089/jir.2006.26.739. [DOI] [PubMed] [Google Scholar]
- 11.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 12.Vilcek J. Interferon research BC (before cloning) Curr Top Microbiol Immunol. 2007;316:9–22. doi: 10.1007/978-3-540-71329-6_2. [DOI] [PubMed] [Google Scholar]
- 13.Whittemore LA, Maniatis T. Postinduction repression of the beta-interferon gene is mediated through two positive regulatory domains. Proc Natl Acad Sci U S A. 1990;87:7799–7803. doi: 10.1073/pnas.87.20.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paste M, Huez G, Kruys V. Deadenylation of interferon-beta mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur J Biochem. 2003;270:1590–1597. doi: 10.1046/j.1432-1033.2003.03530.x. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 17.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 18.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alsaleh G, Suffert G, Semaan N, Juncker T, Frenzel L, Gottenberg JE, Sibilia J, Pfeffer S, Wachsmann D. Bruton's tyrosine kinase is involved in miR-346-related regulation of IL-18 release by lipopolysaccharide-activated rheumatoid fibroblast-like synoviocytes. J Immunol. 2009;182:5088–5097. doi: 10.4049/jimmunol.0801613. [DOI] [PubMed] [Google Scholar]
- 23.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 27.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 35.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 37.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckner CM, Luers AJ, Calderon TM, Eugenin EA, Berman JW. Neuroimmunity and the blood-brain barrier: molecular regulation of leukocyte transmigration and viral entry into the nervous system with a focus on neuroAIDS. J Neuroimmune Pharmacol. 2006;1:160–181. doi: 10.1007/s11481-006-9017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamby SE, Hirst JD. Prediction of glycosylation sites using random forests. BMC Bioinformatics. 2008;9:500. doi: 10.1186/1471-2105-9-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Moller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 44.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 47.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, Thomas-Tikhonenko A, Mendell JT. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, Davuluri R, Croce CM, Mills G, Negrini M, Calin GA. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moll HP, Freudenthaler H, Zommer A, Buchberger E, Brostjan C. Neutralizing type I IFN antibodies trigger an IFN-like response in endothelial cells. J Immunol. 2008;180:5250–5256. doi: 10.4049/jimmunol.180.8.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seitz H. Redefining MicroRNA Targets. Curr Biol. 2009 doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 54.Shi L, Ko ML, Ko GY. Rhythmic expression of MicroRNA-26a (mir-26a) regulates the L-type voltage-gated calcium channel {alpha}1C subunit (VGCC{alpha}1C) in chicken cone photoreceptors. J Biol Chem. 2009 doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 58.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 59.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 60.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A, Sarno A, Groner S, Mertens D, Busch R, Hallek M, Dohner H, Stilgenbauer S. Detailed analysis of p53 pathway defects in fludarabine-refractory CLL: dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009 doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 62.Zhang F, Sriram S. Identification and characterization of the interferon-beta-mediated p53 signal pathway in human peripheral blood mononuclear cells. Immunology. 2009;128:e905–918. doi: 10.1111/j.1365-2567.2009.03104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 64.Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- 65.Wang WB, Lee CK. STAT3 is a negative regulator of type I IFN-induced antiviral responses. Cytokine. 2008;43:266–267. [Google Scholar]
- 66.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savarin C, Bergmann CC. Neuroimmunology of central nervous system viral infections: the cells, molecules and mechanisms involved. Curr Opin Pharmacol. 2008;8:472–479. doi: 10.1016/j.coph.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green NM, Laws A, Kiefer K, Busconi L, Kim YM, Brinkmann MM, Trail EH, Yasuda K, Christensen SR, Shlomchik MJ, Vogel S, Connor JH, Ploegh H, Eilat D, Rifkin IR, van Seventer JM, Marshak-Rothstein A. Murine B cell response to TLR7 ligands depends on an IFN-beta feedback loop. J Immunol. 2009;183:1569–1576. doi: 10.4049/jimmunol.0803899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.