Abstract
The proliferation in genetic association studies, and the recurring failure of initially promising findings to robustly replicate, demonstrates the need for stringent standards to ensure the identification of credible associations. The Human Genome Epidemiology Network has recently published intermin guideliness on evidential criteria for genetic association studies. These are reviewed, and their value and importance discussed, as well as the impact these guidelines will have on the conduct of genetic association studies.
Keywords: Genetic association, replication, HuGENet, Meta-Analysis
Growth in genetic association studies
There has been astonishing growth in the use of molecular genetic technologies to investigate potential associations between genetic variants and a range of complex disease and trait phenotypes. All too often, unfortunately, initially promising findings have proved unreliable, and have subsequently failed to replicate. This has led to suggestions that the search for common genetic variants that are meaningfully associated with complex phenol-types is futile, and efforts should be redirected towards novel approaches [1]. Certainly, the pattern of initial excitement followed by subsequent disappointment, which appears to be endemic to the field, irrespective of the specific gene or phenotype, has led to both academic and public disillusionment. Much of this is due to grandiose claims following an initial report, over-optimistic interpretation of ambiguous results, and a loose definition of replication [2], often exacer-bated by small sample size, correspondingly low statistical power, and potential hetero-geneity between studies. It is therefore critical that stringent standards are adopted to ensure that credible associations are identified, and (perhaps more importantly) excessive claims are avoided.
Assessing strength of evidence
One method that has become popular for assessing the strength of evidence of genetic associations is meta-analysis [3]. This has been in part due to the proliferation of a large number of relatively small studies performed by disparate research groups, and the increasing requirement for replication in independent data sets. It is a potentially powerful tool for assessing effects of candidate genes on complex phenotypes and may provide evidence for previously unexpected diversity, for example by revealing heterogeneity in studies of apparently similar populations [4, 5]. However, the results are only as good as the data that go into the analysis in the first place, and the availability of and reporting of these data may constrain the extent to which a meta-analysis may be informative, or even be performed in the first place. Nevertheless, there has been considerable growth in the use of meta-analysis in genetic association studies, with methods having also been developed to apply these techniques to linkage and genome-wide association data [6]. As it has become increasingly apparent that individual genetic effects are likely to be very small, there has also been growing consensus that the vast majority of individual studies simply lack sufficient statistical power to detect these effects. While a single laboratory may not be able to obtain the requisite numbers, the combined world literature may.
Despite this growth in interest, and the poten-tial benefits offered through its application, meta-analysis is not a panacea for the problems of genetic association studies – differences in analytical methods, study selec-tion and other aspects of individual meta-analyses may lead to differing conclusions for the same gene-disease association [7]. This may be particularly dangerous, given the authoritative status that meta-analyses typically enjoy. The Human Genome Epidemiology Network (HuGENet) has developed “HuGE reviews” (typically systematic reviews, but including meta-analysis where possible) as an online resource containing the cumulative and changing information on epidemiologic as-pects of human gene-phenotype associations [8]. This has led to the development of guide-lines for the conduct of systematic reviews and meta-analyses of genetic association studies [9]. An extension of this initiative has been the establishment of a HuGENet Working Group on the Assessment of Cumulative Evidence, which sought to establish criteria for assessing the strength and credibility of evidence for putative gene-phenotype associations [10].
Criteria for strength of evidence
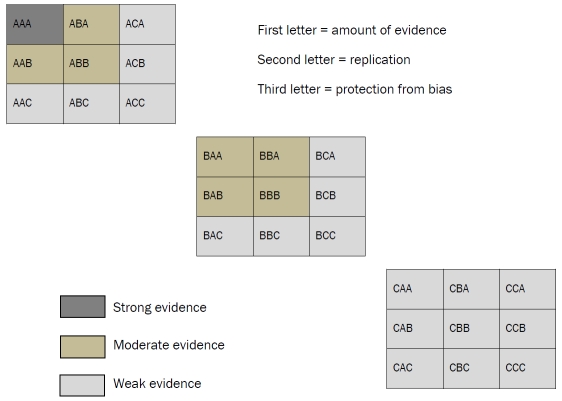
What criteria should be considered? The HuGENet Working Group proposes three: amount of evidence, replication, and protect-tion from bias, where each includes three levels of confidence (roughly, weak, moderate and strong). There are therefore 27 possible combinations across these three criteria, which are then simplified further into three levels of evidence (again, weak, moderate and strong). Only one cell in the resulting 3 × 3 × 3 matrix constitutes strong evidence (i.e., strong confidence for each of the three criteria), while seven constitute moderate evidence (mixed strong and moderate confidence, with no weak confidence, for each of the three criteria). The remaining cells (i.e., weak confidence in any of the three criteria) represent weak evidence.
The most important consideration is strong statistical evidence, amounting to a p-value of 10−7 or less to avoid an excessive false positive rate. Given this requirement for statistical stringency, and the likely small effect of individual genes, very large samples will be required. Extensive replication is also necessary, including at least one well-conducted meta-analysis with minimal heterogeneity (i.e., inconsistency) across studies. Finally, bias (for example due to potential confounding in individual studies, or selective reporting of individual studies in meta-analyses) should be minimal.
An appropriate, well-conducted meta-analysis is therefore regarded as critical for establishing strong evidence for a genetic association. In this context, “well-conducted” essentially means comprehensive and free from bias. Recently, major online initiatives have been developed to achieve this for two widely-studied phenotypes: Alzheimer disease (AlzGene: http://www.alzgene.org) and schizophrenia (SZGene: http://www.szgene.org). These constitute regularly updated online databases of all published genetic association studies for these phenotypes. For all polymorphisms having genotype data available in at least four independent case-control samples, random-effects meta-analyses using allelic contrasts are available [11]. A particular strength of these initiatives is the comprehensive and standardized nature of the meta-analysis – all associations are analyzed in the same way, using all available data, and between-study heterogeneity is assessed. AlzGene has now incorporated the interim guidelines of the HuGENet Working Group in their assessment of which candidate genes show the most promising evidence for association.
Optimism for the future
Unfortunately, initiatives like AlzGene and SZGene require considerable resources to establish and maintain, and there is an endless array of phenotypes for which such databases would be valuable. The situation will improve, but slowly and with uneven coverage. This means that individual meta-analyses will continue to be an important source of evidence in genetic epidemiology. Given their central place in determining the credibility of evidence for association, these should follow HuGENet guidelines for the conduct of systematic reviews and meta-analyses, and ideally include an appraisal of the strength of evidence for the genetic association tested in their conclusions, based on the HuGENet criteria. Of course, as well as establishing that a genetic association appears to be robust, it will be necessary to evaluate whether this association is likely to have any clinical relevance. Many effects may be so small that the practical implications are negligible, but this assessment should only be made after the evidence for an association becomes convincing. The interim guidelines of the HuGENet Working Group are to be welcomed as a first step towards establishing the standards for such efforts.
Figure 1.
HuGENet Evidential Criteria for Genetic Association Studies. The HuGENet Working Group [10] proposes three criteria to be used in the assessment of the credibility of putative genetic associations: amount of evidence, degree of replication, and protection from bias, with three levels of confidence for each. This matrix is used to gauge the overall level of evidence for a particular genetic association. At least one well-conducted meta-analysis is necessary before the evidence for a particular association can be regarded as strong.
References
- 1.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 2.Munafo MR, Flint J. Replication and heterogeneity in gene × environment interaction studies. Int J Neuropsychopharmacol. 2009;12:727–729. doi: 10.1017/S1461145709000479. [DOI] [PubMed] [Google Scholar]
- 3.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Ioannidis JP, Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003;361:567–571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- 5.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 6.Trikalinos TA, Salanti G, Zintzaras E, Ioannidis JP. Meta-analysis methods. Adv Genet. 2008;60:311–334. doi: 10.1016/S0065-2660(07)00413-0. [DOI] [PubMed] [Google Scholar]
- 7.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 8.Khoury MJ, Dorman JS. The Human Genome Epidemiology Network. Am J Epidemiol. 1998;148:1–3. doi: 10.1093/aje/148.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28. doi: 10.1371/journal.pmed.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, Balding DJ, Chokkalingam A, Dolan SM, Flanders WD, Higgins JP, McCarthy MI, McDermott DH, Page GP, Rebbeck TR, Seminara D, Khoury MJ. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 11.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of gen etic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]