Abstract
Bile acids have long been implicated in the etiology of colorectal carcinogenesis by their genotoxicity as well as cytotoxicity. Cholesterol 7-alfa-hydroxylase (CYP7A1) is the rate-limiting enzyme that converts cholesterol into cholesterol 7-alfa-hydroxycholesterol in the first step of the classical pathway of bile acid synthesis. Recently, an association between a polymorphism (−204A>C, rs3808607) in CYP7A1 and proximal colon cancer/adenoma has been reported, which was not observed with distal colon or rectal cancer/adenoma. In this case-control study, we examined the association between haplotypes of CYP7A1 and proximal or distal colon/rectal cancer risk in a Japanese population. Subjects were 96 cases of proximal colon cancer, 357 of distal colon/rectal cancer and 961 age- and sex-matched non-cancer controls at Aichi Cancer Center. We examined five loci, including rs3808607, and evaluated the impact of haplotype on risk. In locus-specific analyses, we saw no association with rs3808607 for any site. Haplotype analyses revealed that the TAAGG haplotype was positively associated with proximal colon cancer [confounder-adjusted odds ratio: 1.72 (95% confidence interval: 1.10-2.71), p=0.018] but not with distal colon and rectal cancer combined. This association was consistently observed in analyses stratified by potential confounders. Our results indicate that CYP7A1 plays a role in the carcinogenesis of colorectal cancer specifically in the proximal colon. Confirmation of this association in other epidemiologic studies and biological evaluation of the TAAGG haplotype are warranted.
Keywords: CYP7A1, polymorphisms, proximal colon cancer, Japanese
Introduction
Colorectal cancer (CRC) remains a major cancer worldwide [1], accounting for 9.4% of all cancers diagnosed in 2007 and 7.9% of cancer deaths. Although numerous epidemiological and biological studies have revealed risk/protective factors for CRC, present knowledge is still insufficient to allow the disease to be overcome, and the struggle to elucidate mechanisms is ongoing.
Bile acids have long been implicated in the etiology of CRC [2]. The primary bile acids are the major products of cholesterol metabolism in the liver, and play an important role in the digestion and absorption of lipids in the small intestine. More than 95% of primary bile acids passing through the ileum are reabsorbed and return to the liver through the portal vein. Bile acids not absorbed in the ileum are further metabolized in the large bowel by anaerobic bacterial flora and finally converted to seconddary bile acids. The acidity of bile acids is sufficiently strong to implicate them in colorectal carcinogenicity. Although results consistent with this have also been shown in animal experiments [3, 4], epidemiologic evidence remains controversial.
Cholesterol 7-alfa-hydroxylase (CYP7A1) is the rate-limiting enzyme that converts cholesterol into cholesterol 7-alfa-hydroxycholesterol in the first step of the classical bile acid synthesis pathway [5]. Animal experimental studies suggest that CYP7A1 enzymes play significant roles in controlling both the choles-terol and bile acid pool [6, 7]. In humans, a polymorphism in the promoter region of CYP7A1 (−278A >C, also denoted as −204A>C or −203A>C, which is anti-sense strand of rs3808607: T->G) was suggested to have sufficient functionality to change plasma concentrations of total or low density lipo-protein cholesterol [8], albeit that subsequent results were controversial [9-11]. Recent epidemiologic studies examining the association between CYP7A1 promoter region polymer-phism and the risk of colorectal cancer [12] or colorectal adenoma [13] have shown a significant association with lesions in the proximal colon but not in the distal colon or rectum, suggesting that the potential involvement of CYP7A1 variation in colorectal carcinogenesis acts via a mechanism involving bile acid production.
Given functional controversies about CYP7A1 promoter polymorphism in human populations [8-11] and the heterogeneity of haplotype structure across ethnicities [14], a more comprehensive investigation of CYP7A1 polymorphisms and colorectal cancer risk is required. Here, we conducted a case-control study to investigate the impact of CYP7A1 haplotype on CRC risk, with special reference to the proximal colon.
Subjects and methods
Subjects
Cases were 453 patients who were histologycally diagnosed with CRC, consisting of 96 with proximal colon cancer (International Classification of Diseases version 10 (ICD-10), C18.0 – C184), 123 with distal colon cancer and 234 with rectal cancer (ICD-10, C18.5 – C18.7, C19.9 and C20.9) between January 2001 and November 2005 at Aichi Cancer Center Hospital (ACCH) and who had no prior history of cancer. Controls were 961 frequency-matched first-visit outpatients at ACCH during the same period that was confirmed to have no cancer or prior history of neoplasia. The cases and controls were selected from the database of the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC). The framework of HERPACC has been described elsewhere [15, 16]. Briefly, all outpatients aged 20–79 years were asked at first visit to fill out a questionnaire regarding their lifestyle and provide 7 ml of blood. Case/control status was determined by data linkage between hospital-based cancer registration and HERPACC database. Controls were defined as those who enrolled HERPACC and were not confirmed to have cancer within one year from the first visit. Cases were defined as incident cases that had been diagnosed as colorectal cancer within one year from the first visit. A trained interviewer checked the completion of each questionnaire. Approxi-mately 95% of eligible subjects completed the questionnaire and 55% provided blood samples. Some 30% of first-visit outpatients were diagnosed at ACCH as having cancer. Under the assumption that the non-cancer population within HERPACC will visit ACCH if they develop cancer in the future, we defined non-cancer first-visit outpatients as those from among whom such cases may arise. Our previous study confirmed that the lifestyle patterns of first-visit outpatients matched the profile of a group randomly selected from the general population of Nagoya City, conferring external validity on the study [17]. Written informed consent was obtained from all subjects and the study was approved by the ethics committee of ACC.
Determination of CYP7A1 loci genotype
The loci examined in this study were rsll786580, rs3747809, rs2162459, rs3824260 and rs3808607. Rs3808607 (anti-sense strand of −204A>C) was selected because it is reported to be functional in the promoter region [8] and was examined in previous studies [12, 13]. The other four loci were selected as tagSNPs based on the HapMap database with the setting MAF>0.05 and pairwise R2>0.8.
DNA of each subject was extracted from the buffy coat fraction with a Blood Mini Kit (Qiagen K.K., Tokyo, Japan) and assessed using the polymerase chain reaction (PCR) TaqMan method [18] with the 7500 Fast Realtime PCR system (Applied Biosystems, Foster City, CA, USA). rs3808607 was assessed by PCR-RFLP as previously described [13]. The quality of genotyping was assessed by duplicate analysis of 5% of samples, which showed an agreement rate of 100%.
Exposure data
Cumulative smoking dose was evaluated as pack-years, the product of the number of packs consumed per day and years of smoking. Smoking habit was classified into the three categories of never, pack-years <20 (low-moderate) and ≥20 pack years (heavy). Consumption of types of alcoholic beverages (Japanese sake, beer, shochu, whiskey and wine) per occasion was determined with reference to the average number of drinks per day, which was then converted into a Japanese sake (rice wine) equivalent (one unit sake=23g ethanol) [19]. Daily ethanol consumption was estimated as the product of the frequency of alcohol beverage consump-tion and average ethanol consumption per occasion, and drinking habit was classified into the four categories of non-drinker, low (<5g/day), moderate (<23g/day) and heavy (≥23g/day). Consumption of folate was deter-mined using a semi-quantitative food frequ-ency questionnaire (SQFFQ) as described in detail elsewhere [20]. Briefly, the SQFFQ consisted of 47 single food items with the eight frequency categories of never or seldom, 1-3 times/month, 1-2 times/week, 3-4 times/week, 5-6 times/week, once/day, twice/day, and 3+ times/day. Average daily intake of nutrients was estimated by multiplying the food intake (in grams) or serving size by the nutrient content per 100 grams of food as listed in the Standard Tables of Food Composition in Japan, 5th edition. Consumption of supplemental folate was not considered in total consumption because the questionnaire for multi-vitamins was not quantitative. Energy-adjusted intake of nutrients was calculated by the residual method [21]. The SQFFQ was validated by reference to a 3-day weighted dietary record as a standard, which showed the reproducibility and validity to be acceptable [22, 23]. The de-attenuated correlation coefficients for energy-adjusted intakes of folate were 0.36 in men and 0.38 in women. Body mass index (BMI) was calculated as the self-reported weight in kilograms divided by the square of the self-reported height in meters. A family history of CRC in a first-degree relative was based on self-reporting, as described elsewhere [24]. The questionnaire also covered the regularity of physical exercise: subjects were asked to report the frequency and intensity of recreational exercise, with average daily exercise hours in any intensity calculated and categorized into the three levels of none, and <0.5 and ≥0.5 hours/day.
Statistical analysis
Odds ratios (ORs) and 95% confidence intervals (CIs) for assessment of the impact of each CYP7A1 locus, included in the model as an ordinal score (1 to 3), were calculated using muInvariable polytomous logistic regression models (log-additive model). Dominant or recessive models were also examined. Potential confounders included in the model included age, sex, regular exercise (none, <0.5 hour/day, and ≥0.5 hour/day as indicator variables), folate consumption by tertile (T1-3 as indicator variables), BMI (<22.5, <25< <27.5 and ≥27.5 kg/m2, as indicator variables), total energy consumption, smoking status (never, former, current moderate, and heavy as indicator variables), drinking habit (non, low, moderate, and heavy as indicator variables), and family history of colorectal cancer (yes or no). The impact of CYP7A1 haplotype was also examined by the haplologit procedure [25] in STATA (Stata Corp., College Station, TX) by applying modified retrospective semi-parametric profile-likelihood methods [26, 27]. Potential confounders considered in the haplologit procedure were the same as those in the analyses for individual loci. Stratified polytomous logistic regression analyses by potential confounders were conducted to explore possible effect modification, as well as to confirm the consistency of the association between a risk haplotype and risk.
To assess possible discrepancies between expected and observed haplotypes, accordance with the Hardy-Weinberg equilibrium (HWE) was checked for controls with the χ2 test. Statistical analyses were performed using STATA version 10, with P-values <0.05 considered statistically significant.
Results
Table 1 shows baseline characteristics of the 96 proximal colon cancer case subjects, 357 distal CRC case subjects, and 961 control subjects. Proximal CRC cases were signifycantly older than distal CRC cases and controls. Heavy smoking was more prevalent in the distal CRC cases, while drinking showed no statistically significant difference. BMI, folate consumption and recreational exercise were not significantly different across subjects.
Table 1.
Characteristics of cases and controls
| Variable | Proximal Colon | Distal Colon + Rectum | Controls | p-value | |||
|---|---|---|---|---|---|---|---|
| Total | 96 | 357 | 961 | ||||
| Sex | 0.83 | ||||||
| Male | 57 | 59.0% | 224 | 63.0% | 599 | 62.0% | |
| Female | 39 | 41.0% | 133 | 37.0% | 362 | 38.0% | |
| Age(years) | 0.013 | ||||||
| <40 | 1 | 1.0% | 19 | 5.0% | 39 | 4.0% | |
| 40-49 | 9 | 9.0% | 41 | 11.0% | 105 | 11.0% | |
| 50-59 | 23 | 24.0% | 136 | 38.0% | 327 | 34.0% | |
| 60-69 | 38 | 40.0% | 112 | 31.0% | 353 | 37.0% | |
| 70- | 25 | 26.0% | 49 | 14.0% | 137 | 14.0% | |
| Mean age (SD) | 62.6 (9.0) | 58.4 (10.5) | 59.0 (9.9) | ||||
| Smoking | 0.025 | ||||||
| Non or low (<5 pack-years) | 53 | 55.0% | 149 | 42.0% | 493 | 51.0% | |
| Moderate (<20 pack-years) | 13 | 14.0% | 42 | 12.0% | 116 | 12.0% | |
| Heavy (≥20 pack-years) | 30 | 31.0% | 162 | 45.0% | 344 | 36.0% | |
| Unknown | 0 | 0.0% | 4 | 1.0% | 8 | 1.0% | |
| Drinking | 0.452 | ||||||
| Non | 41 | 43.0% | 136 | 38.0% | 383 | 40.0% | |
| Low (<5g ethanol/day) | 17 | 18.0% | 44 | 12.0% | 125 | 13.0% | |
| Moderate (<23g ethanol/day) | 18 | 19.0% | 64 | 18.0% | 195 | 20.0% | |
| High (≥23g ethanol/day) | 20 | 21.0% | 107 | 30.0% | 243 | 25.0% | |
| Unknown | 0 | 0.0% | 6 | 2.0% | 15 | 2.0% | |
| Daily folate consumption | 0.143 | ||||||
| T1 (- 270.2 μg/day) | 24 | 25.0% | 141 | 39.0% | 315 | 33.0% | |
| T2 (- 352.3 μg/day) | 33 | 34.0% | 105 | 29.0% | 315 | 33.0% | |
| T3 (352.4- μg/day) | 37 | 39.0% | 107 | 30.0% | 314 | 33.0% | |
| Unknown | 2 | 2.0% | 4 | 1.0% | 17 | 2.0% | |
| Body-Mass Index (BMI) kg/m2 | 0.961 | ||||||
| <22.5 | 42 | 44.0% | 150 | 42.0% | 397 | 41.0% | |
| < 25 | 29 | 30.0% | 120 | 34.0% | 313 | 33.0% | |
| < 27.5 | 17 | 18.0% | 52 | 15.0% | 162 | 17.0% | |
| ≥ 27.5 | 7 | 7.0% | 33 | 9.0% | 79 | 8.0% | |
| Unknown | 1 | 1.0% | 2 | 1.0% | 10 | 1.0% | |
| Family history of colorectal cancer in the first degree relatives | 0.026 | ||||||
| No | 88 | 92.0% | 340 | 95.0% | 931 | 97.0% | |
| Yes | 8 | 8.0% | 17 | 5.0% | 30 | 3.0% | |
| Average recreational exercise | 0.149 | ||||||
| None | 29 | 30.0% | 151 | 42.0% | 349 | 36.0% | |
| < 0.5 hour/day | 42 | 44.0% | 138 | 39.0% | 397 | 41.0% | |
| 0.5- hour/day | 25 | 26.0% | 68 | 19.0% | 215 | 22.0% | |
Family history in a first-degree relative was significantly more common in cases than controls.
Genotype distributions and minor allele frequencies for CYP7A1 loci are shown in Table 2. Among controls, all CYP7A1 loci were accordant with the HWE. The minor allele frequency for each locus were closely similar with those in the HapMap JPT data [28]. Regarding linkage disequilibrium (LD) across the five loci, pairwise D′ showed complete LD, while R2 indicated moderate LD between rs2162459 and rs3824260 (Figure 1).
Table 2.
Genotypes distribution of CYP7A1 polymorphisms among subjects
| Genotype distribution | Minor allele freuqencies | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Proximal colon | Distal colon + Rectum | HapMap-JPT | Controls | Proximal colon | Distal colon + Rectum | HWE p-value in CNT | |||
| Location in choromosome # | Description | |||||||||
| rs11786580 | 59568494 | Intron 4, tag SNP | ||||||||
| CC/CT/TT | 797/160/4 | 71/22/3 | 291/64/2 | 0.091 | 0.087 | 0.146 | 0.095 | 0.18 | ||
| rs3747809 | 59569538 | Intron 4, tag SNP | ||||||||
| AA/AG/GG | 855/100/6 | 88/8/0 | 323/31/3 | 0.058 | 0.058 | 0.042 | 0.052 | 0.11 | ||
| rs2162459 | 59573596 | Intron 1, tagSNP | ||||||||
| GG/GA/AA | 321/465/175 | 34/38/24 | 119/181/57 | 0.375 | 0.424 | 0.448 | 0.413 | 0.77 | ||
| rs3808607 (−204A>C) | 59575415 | 5′ near gene | ||||||||
| GG/GT/TT*2 | 246/492/223 | 27/41/28 | 94/182/81 | 0.430 | 0.488 | 0.505 | 0.482 | 0.45 | ||
| rs3824260 | 59575744 | 5′ near gene, tagSNP | ||||||||
| AA/AG/GG | 245/493/223 | 27/41/28 | 94/181/82 | 0.433 | 0.489 | 0.495 | 0.517 | 0.41 | ||
*1 NA indicates not available.; *2 Genotypes are CC/CA/AA based on −204A>C.
Figure 1.
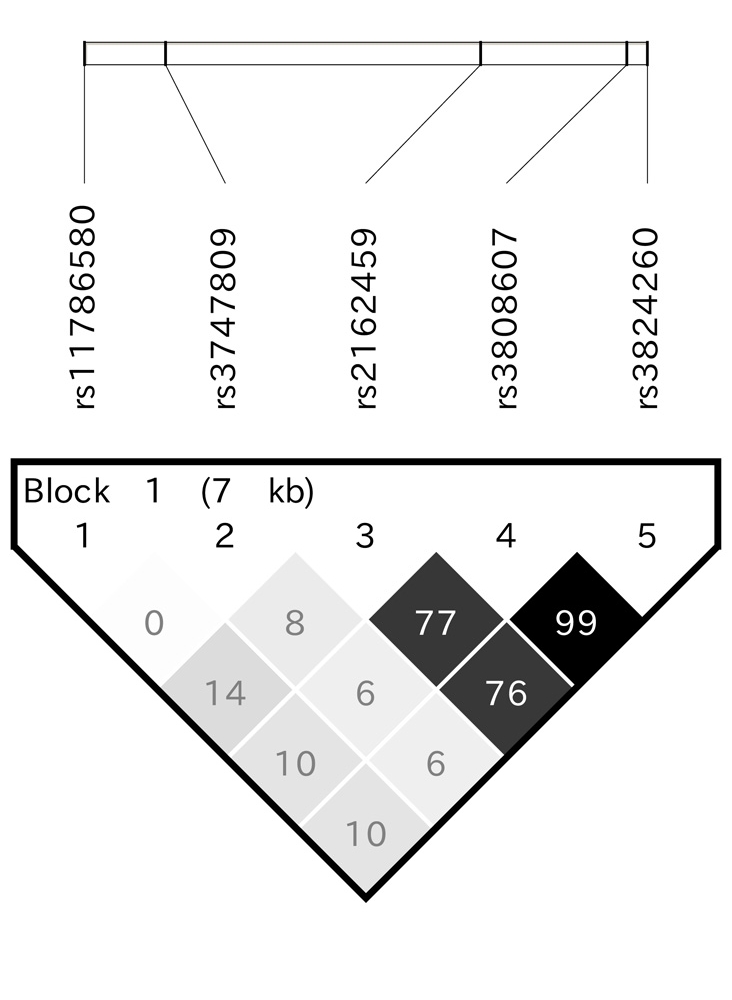
Linkage disequilibrium within five CYP7A1 gene polymorphisms. Increasing shading indicates a high degree of correlation. Numbers in the panel indicate pair-wise D′ values.
Table 3 shows adjusted ORs for each locus in log-additive, dominant and recessive models. rs11786580 showed a statistically significant positive association with proximal colon cancers. OR in the log-additive model was 1.72 (95%CI: 1.10-2.71) for T-allele. This positive association was remarkably increased in the recessive model. In contrast, this locus showed no association with distal colon and rectal cancer combined, or in separate analyses of distal colon cancer and rectal cancer (data not shown). Other loci, including A-204C, showed no association with either site. Table 4 shows haplotype frequencies constructed from CYP7A1 loci examined in each group and their ORs and 95%Cls. We identified five haplotypes in one LD block, and the number of haplotypes and their frequencies in controls were consistent with those previously reported [14]. Similar to the loci-based analyses, a haplotype TAAGG which could be tagged by rsll786580 (JPT-B1H4 haplotype) showed a significantly increased risk for proximal colon cancer only, and not for distal colon or rectal cancer. Table 5 shows stratified analyses by potential confounders. Associations were consistently observed for proximal colon cancer but not for distal colon or rectal cancer. For proximal colon cancer, several stratification factors, namely recreational exercise, BMI, folate consumption, and alcohol drinking, showed a difference in point estimates of ORs, suggesting possible effect modification, but the statistical evaluation of interactions was limited by the small number of subjects, particularly with proximal colon cancer.
Table 3.
Genotypes distribution of CYP7A1 polymorphisms and their odds ratios for colorectal cancer risk
| Proximal colon | Distal colon + Rectum | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log additive model | Dominant model | Recessive model | Log additive model | Dominant model | Recessive model | |||||||||||||
| OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | |
| rs11786580 | 1.72 | 1.10-2.71 | 0.018 | 1.62 | 0.99-2.66 | 0.056 | 6.88 | 1.39-34.0 | 0.018 | 1.11 | 0.82-1.51 | 0.49 | 1.12 | 0.81-1.55 | 0.485 | 1.15 | 0.20-6.49 | 0.874 |
| rs3747809 | 0.71 | 0.34-1.46 | 0.345 | 0.72 | 0.34-1.54 | 0.402 | NE*2 | - | - | 0.92 | 0.63-1.33 | 0.65 | 0.88 | 0.58-1.33 | 0.544 | 1.32 | 0.32-5.44 | 0.696 |
| rs2162459 | 0.82 | 0.61-1.10 | 0.188 | 0.56 | 0.35-0.89 | 0.015 | 1.02 | 0.66-1.59 | 0.926 | 1.03 | 0.86-1.22 | 0.755 | 1.13 | 0.81-1.56 | 0.476 | 0.98 | 0.76-1.28 | 0.908 |
| rs3808607(−204A>C) | 0.98 | 0.73-1.32 | 0.888 | 0.75 | 0.48-1.17 | 0.204 | 1.30 | 0.81-2.07 | 0.276 | 0.96 | 0.81-1.15 | 0.67 | 0.96 | 0.73-1.27 | 0.773 | 0.94 | 0.70-1.26 | 0.679 |
| rs3824260 | 0.98 | 0.73-1.31 | 0.874 | 0.74 | 0.47-1.16 | 0.193 | 1.30 | 0.81-2.08 | 0.274 | 0.97 | 0.81-1.15 | 0.705 | 0.96 | 0.72-1.26 | 0.744 | 0.96 | 0.71-1.28 | 0.769 |
Adjusted for age as continuous variable, drinking (non, low, moderate, heavy, and unknown), smoking (non, moderate, heavy, unknwon), BMI (<22.5, <25, <27.5, 27.5-, unknown), folate in tertile (T1, T2, T3, and unknown), total energy intake, family history of colorectal cancer, average recreational exercise (none, <0.5 hour/day, 0.5-hour/day) in polytomous logistic regression models.
NE indicates not estimated.
Table 4.
Genotypes distribution of CYP7A1 polymorphisms and their odds ratios for coiorectai cancer risk
| Haplotype frequencies | Proximal colon | Distal colon + Rectum | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Log additive model | Dominant mode | Recessive model | Log additive model | Dominant model | Recessive model | ||||||||||||||||
| Proximal | Distal | Controls | ||||||||||||||||||||
| Haplotype | Annotation of haplotype*2 | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | OR*1 | 95% CI | p-value | |||
| CAGTA | JPT-B1H1 | 0.494792 | 0.516807 | 0.511446 | 1.00 | Reference | 1.00 | Reference | 1.00 | Reference | 1.00 | Reference | 1.00 | Reference | 1.00 | Reference | ||||||
| CAAGG | JPT-B1H2 | 0.260417 | 0.267507 | 0.278356 | 0.97 | 0.68-1.38 | 0.85 | 0.88 | 0.57-1.36 | 0.559 | 1.11 | 0.53-2.34 | 0.774 | 1.00 | 0.73-1.37 | 0.993 | 1.23 | 0.83-1.82 | 0.293 | 0.40 | 0.15-1.09 | 0.074 |
| TAAGG | JPT-B1H4 | 0.145833 | 0.095238 | 0.087409 | 1.72 | 1.10-2.71 | 0.018 | 1.64 | 1.00-2.71 | 0.052 | 3.88 | 1.19-12.7 | 0.025 | 0.98 | 0.60-1.60 | 0.942 | 1.09 | 0.65-1.81 | 0.75 | NE*3 | - | - |
| CAGGG | JPT-B1H3 | 0.057292 | 0.068627 | 0.063996 | 0.92 | 0.48-1.77 | 0.813 | 0.92 | 0.47-1.81 | 0.819 | NE*3 | - | - | 0.89 | 0.50-1.60 | 0.707 | 0.89 | 0.48-1.64 | 0.701 | 2.04 | 0.28-15.0 | 0.485 |
| CGAGG | JPT-B1H5 | 0.041667 | 0.05042 | 0.058273 | 0.74 | 0.35-1.56 | 0.426 | 0.73 | 0.34-1.56 | 0.413 | NE*3 | - | - | 0.84 | 0.45-1.57 | 0.588 | 0.73 | 0.37-1.46 | 0.375 | 5.17 | 1.22-21.8 | 0.025 |
Adjusted for age as continuous variable, drinking (non, low, moderate, heavy, and unknown), smoking (non, moderate, heavy, unknwon), BMI (<22.5, <25, <27.5, 27.5-, unknown), folate in tertile (Tl, T2, T3, and unknown), total energy intake, family history of colorectal cancer, average recreational exercise (none, <0.5 hour/day, 0.5-hour/day) in the models.
Annotations of haplotype were based upon Reference 16 (Nakamoto S et al.)
NE indicates not estimated.
Table 5.
Impact of the TAAGG haplotype stratified by potential confounders
| Controls | Proximal colon | Distal colon + Rectum | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diplotypes | Log additive model | Log additive model | |||||||||||||
| Non-TAAGG*1 | Heterozygous | Homozygous | Non-TAAGG*1 | Heterozygous | Homozygous | OR*2 | 95% CI | p-value | Non-TAAGG*1 | Heterozygous | Homozygous | OR*2 | 95% CI | p-value | |
| Sex | |||||||||||||||
| Male | 497 | 99 | 3 | 41 | 14 | 2 | 1.90 | 1.07-3.36 | 0.028 | 186 | 37 | 1 | 0.98 | 0.66-1.47 | 0.932 |
| Female | 300 | 61 | 1 | 30 | 8 | 1 | 1.63 | 0.73-3.66 | 0.233 | 105 | 27 | 1 | 1.23 | 0.75-2.03 | 0.410 |
| Age groups | |||||||||||||||
| <60 | 387 | 81 | 3 | 22 | 10 | 1 | 2.41 | 1.17-4.96 | 0.017 | 162 | 32 | 2 | 0.99 | 0.65-1.50 | 0.956 |
| ≥60 | 410 | 79 | 1 | 49 | 12 | 2 | 1.68 | 0.90-3.11 | 0.102 | 129 | 32 | 0 | 1.19 | 0.76-1.88 | 0.448 |
| Recreational exercise | |||||||||||||||
| No | 292 | 55 | 2 | 22 | 5 | 2 | 2.39 | 1.04-5.51 | 0.041 | 124 | 26 | 1 | 1.12 | 0.69-1.80 | 0.656 |
| Yes | 505 | 105 | 2 | 49 | 17 | 1 | 1.76 | 1.00-3.09 | 0.048 | 167 | 38 | 1 | 1.13 | 0.76-1.70 | 0.541 |
| Body mass index*3 | |||||||||||||||
| <25 kg/m2 | 586 | 121 | 3 | 53 | 16 | 2 | 1.67 | 0.97-2.87 | 0.065 | 222 | 46 | 2 | 1.03 | 0.72-1.47 | 0.863 |
| ≥ 25 kg/m2 | 202 | 38 | 1 | 17 | 6 | 1 | 2.20 | 0.87-5.55 | 0.095 | 68 | 17 | 0 | 1.30 | 0.69-2.45 | 0.418 |
| Folate consumption*4 | |||||||||||||||
| Tertile 1 | 261 | 54 | 0 | 18 | 3 | 3 | 2.46 | 1.07-5.64 | 0.034 | 119 | 21 | 1 | 0.93 | 0.55-1.58 | 0.797 |
| Tertile 2/3 | 527 | 99 | 3 | 51 | 19 | 0 | 1.72 | 0.97-3.03 | 0.062 | 169 | 42 | 1 | 1.30 | 0.88-1.92 | 0.188 |
| Drinking*5 | |||||||||||||||
| < 23g/day | 581 | 119 | 3 | 57 | 18 | 1 | 1.49 | 0.87-2.54 | 0.144 | 201 | 41 | 2 | 1.10 | 0.76-1.61 | 0.603 |
| ≥ 23g/day | 202 | 40 | 1 | 14 | 4 | 2 | 3.02 | 1.22-7.51 | 0.017 | 88 | 19 | 0 | 0.99 | 0.55-1.79 | 0.983 |
| Smoking*6 | |||||||||||||||
| Pack-year < 20 | 505 | 101 | 3 | 48 | 17 | 1 | 1.72 | 0.98-3.03 | 0.059 | 150 | 39 | 2 | 1.32 | 0.89-1.96 | 0.167 |
| Pack-year≥ 20 | 285 | 58 | 1 | 23 | 5 | 2 | 1.85 | 0.84-4.06 | 0.126 | 137 | 25 | 0 | 0.86 | 0.52-1.43 | 0.565 |
| Family history of colorectal cancer in first degree relatives | |||||||||||||||
| No | 778 | 150 | 3 | 65 | 20 | 3 | 1.84 | 1.15-2.94 | 0.010 | 277 | 61 | 2 | 1.19 | 087-1.62 | 0.289 |
| Yes | 19 | 10 | 1 | 6 | 2 | 0 | NE*7 | - | - | 14 | 3 | 0 | NE*7 | - | - |
Non-TAAGG indicates diplotypes not including the TAAGG haplotypes
Adjusted for age as continuous variable, drinking (non, low, moderate, heavy, and unknown), smoking (non, moderate, heavy, unknwon), BMI (<22.5, <25, <27.5, 27.5-, unknown), folate in tertile (T1, T2, T3, and unknown), total energy intake, family history of colorectal cancer, average recreational exercise (none, <0.5 hour/day, 0.5-hour/day) in polytomous logistic regression models. A stratification factor was excluded from the models
Ten controls, one proximal colon cancer case and two distal colorectal/rectal cancer cases were excluded from analysis because of lack of body mass index information
Seventeen controls, two proximal colon cancer cases, and four distal colorectal/rectal cancer cases were excluded from analysis because of lack of folate information.'
Fifteen controls and six distal colorectal/rectal cancer cases were excluded from analysis because of lack of drinking information
Eight controls and four distal colorectal/rectal cancer cases were excluded from analysis because of lack of smoking information.
NE indicates not evaluable due to lack of cases.
Discussion
In this study, we found that the CYP7A1 TAAGG haplotype tagged by rs11786580 was associated with a significantly increased risk of proximal colon cancer but not distal colon or rectal cancer in a Japanese population. This finding indicates that bile acid metabolism as defined by genetic variation in CYP7A1 may be involved in carcinogenesis in the proximal colon.
To date, only two studies have examined the association between polymorphisms in a promoter region, namely A-204C (rs3808607), and colorectal neoplasms, cancer and adenoma in proximal colon, both of which were by the same research group [12, 13]. Here, we did not observe a significant association between this locus and proximal colon cancer risk, although the log additive OR was below unity. Instead, we found a significant association with the TAAGG haplotypes tagged by rs 11786580. Although it is difficult to conclude which locus in CYP7A1 is responsible, frequencies of the risk haplotypes or genotype may provide a clue. In our study, frequency of the TAAGG haplotype (JPT-B1H4) was 0.087, versus 0.242 for the corresponding haplotypes (CEU-B1H3 and CEU-B1H4) reported in Caucasians [14]. In contrast, the allele frequency of rs3808607 (−204A>C) in Japanese was 0.482 versus 0.609 in Caucasians [14]. Supposing that the difference in risk haplotype/allele frequency corresponds to the difference in incidence, our results appear a reasonable explanation for the difference in incidence of CRC between Japanese and Caucasians [1]. Alternatively, considering the controversy over the functional relevance of the rs3808607 (−204A>C) polymorphism [9-11], either finding may be due to chance. In any case, further studies in various populations with markers better covering CYP7A1 are warranted, coupled with functional evaluation.
We exploratorily observed possible effect modification for several factors in the association between TAAGG haplotype and proximal colon cancer risk. We observed higher ORs for those expected to be at higher risk, namely those who do not exercise regularly, have a BMI of 25 or more, consume less folate, and drink more alcohol (23 or more g/day), although it is not clear how those factors might interact with the CYP7A1 TAAGG haplotype. Moreover, the limited number of subjects hampered evaluation of effect modification. Replication of these findings in other studies is therefore essential.
The major strength of this study is its limited information bias in terms of CYP7A1 polymorphism. Subjects and researchers were not aware of genotype before enrollment. Given the similarity in minor allele frequency between our controls and that in the HapMap database for Japanese, it is reasonable to assume the external validity of our study results to the general population. With regard to limitations, the use of non-cancer patients at our hospital as controls may be a limitation, given the likelihood that our cases arose within this population base. Second, as with other case-control studies, this study may have suffered from information bias, albeit that the questionnaires were completed before diagnosis in our hospital. Third, limited number of cases for subsite was another limitation. Post-hoc power was 0.55, indicating needs for replication in larger studies. Caution is therefore needed in interpreting the lack of interaction between CYP7A1 haplotype and questionnaire-based factors.
In conclusion, our present investigation showed that CYP7A1 was an independent risk factor for proximal colon cancer, but not distal colon or rectal cancer, in a Japanese population. The locus showing statistical significance was not the A-204C polymorphism, but rather rs11786580 located in intron 4, indicating a potential functional region around this locus. Further studies to clarify the biological mechanisms of this association are warranted.
Acknowledgments
The authors are grateful to the many doctors, nurses, and technical and administration staff of Aichi Cancer Center Hospital for the daily administration of the HERPACC study.
This study was supported by a Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan, for Cancer Research from the Ministry of Health, Labour and Welfare of Japan, and for the Third Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare of Japan.
The authors declare that they have nothing to disclose regarding financial issues as well as conflict of interest.
References
- 1.Parkin DM, Whelan SL, Ferlay J, Teppo L, Thomas DB. Cancer Incidence in Five Continents. Lyon: 2003. [Google Scholar]
- 2.Nagengast FM, Grubben MJ, van Munster IP. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31A:1067–1070. doi: 10.1016/0959-8049(95)00216-6. [DOI] [PubMed] [Google Scholar]
- 3.Narisawa T, Magadia NE, Weisburger JH, Wynder EL. Promoting effect of bile acids on colon carcinogenesis after intrarectal instillation of N-methyl-N′-nitro-N-nitrosoguanidine in rats. J Natl Cancer Inst. 1974;53:1093–1097. doi: 10.1093/jnci/53.4.1093. [DOI] [PubMed] [Google Scholar]
- 4.Reddy BS, Narasawa T, Weisburger JH, Wynder EL. Promoting effect of sodium deoxycholate on colon adenocarcinomas in germfree rats. J Natl Cancer Inst. 1976;56:441–442. doi: 10.1093/jnci/56.2.441. [DOI] [PubMed] [Google Scholar]
- 5.Schwarz M, Lund EG, Russell DW. Two 7 alpha-hydroxylase enzymes in bile acid biosynthesis. Curr Opin Lipidol. 1998;9:113–118. doi: 10.1097/00041433-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Spady DK, Cuthbert JA, Willard MN, Meidell RS. Adenovirus-mediated transfer of a gene encoding cholesterol 7 alpha-hydroxylase into hamsters increases hepatic enzyme activity and reduces plasma total and low density lipoprotein cholesterol. J Clin Invest. 1995;96:700–709. doi: 10.1172/JCI118113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7alpha-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- 8.Wang J, Freeman DJ, Grundy SM, Levine DM, Guerra R, Cohen JC. Linkage between cholesterol 7alpha-hydroxylase and high plasma low-density lipoprotein cholesterol concentrations. J Clin Invest. 1998;101:1283–1291. doi: 10.1172/JCI1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couture P, Otvos JD, Cupples LA, Wilson PW, Schaefer EJ, Ordovas JM. Association of the A-204C polymorphism in the cholesterol 7alpha-hydroxylase gene with variations in plasma low density lipoprotein cholesterol levels in the Framingham Offspring Study. J Lipid Res. 1999;40:1883–1889. [PubMed] [Google Scholar]
- 10.Han Z, Heath SC, Shmulewitz D, Li W, Auerbach SB, Blundell ML, Lehner T, Ott J, Stoffel M, Friedman JM, Breslow JL. Candidate genes involved in cardiovascular risk factors by a family-based association study on the island of Kosrae, Federated States of Micronesia. Am J Med Genet. 2002;110:234–242. doi: 10.1002/ajmg.10445. [DOI] [PubMed] [Google Scholar]
- 11.Abrahamsson A, Krapivner S, Gustafsson U, Muhrbeck O, Eggertsen G, Johansson I, Persson I, Angelin B, Ingelman-Sundberg M, Bjorkhem I, Einarsson C, van't Hooft FM. Common polymorphisms in the CYP7A1 gene do not contribute to variation in rates of bile acid synthesis and plasma LDL cholesterol concentration. Atherosclerosis. 2005;182:37–45. doi: 10.1016/j.atherosclerosis.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara T, Kono S, Yin G, Toyomura K, Nagano J, Mizoue T, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Yasunami Y, Maekawa T, Takenaka K, Ichimiya H, Imaizumi N. Genetic polymorphism in cytochrome P450 7A1 and risk of colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Res. 2005;65:2979–2982. doi: 10.1158/0008-5472.CAN-04-3872. [DOI] [PubMed] [Google Scholar]
- 13.Tabata S, Yin G, Ogawa S, Yamaguchi K, Mineshita M, Kono S. Genetic polymorphism of cholesterol 7alpha-hydroxylase (CYP7A1) and colorectal adenomas: Self Defense Forces Health Study. Cancer Sci. 2006;97:406–410. doi: 10.1111/j.1349-7006.2006.00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamoto K, Wang S, Jenison RD, Guo GL, Klaassen CD, Wan YJ, Zhong XB. Linkage disequilibrium blocks, haplotype structure, and htSNPs of human CYP7A1 gene. BMC Genet. 2006;7:29. doi: 10.1186/1471-2156-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tajima K, Hirose K, Inoue M, Takezaki T, Hamajima N, Kuroishi T. A Model of Practical Cancer Prevention for Out-patients Visiting a Hospital: the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC) Asian Pac J Cancer Prev. 2000;1:35–47. [PubMed] [Google Scholar]
- 16.Hamajima N, Matsuo K, Saito T, Hirose K, Inoue M, Takezaki T, Kuroishi T, Tajima K. Gene-environment Interactions and Polymorphism Studies of Cancer Risk in the Hospital-based Epidemiologic Research Program at Aichi Cancer Center II (HERPACC-II) Asian Pac J Cancer Prev. 2001;2:99–107. [PubMed] [Google Scholar]
- 17.Inoue M, Tajima K, Hirose K, Hamajima N, Takezaki T, Kuroishi T, Tominaga S. Epidemiological features of first-visit outpatients in Japan: comparison with general population and variation by sex, age, and season. J Clin Epidemiol. 1997;50:69–77. doi: 10.1016/s0895-4356(96)00297-1. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo K, Wakai K, Hirose K, Ito H, Saito T, Tajima K. Alcohol dehydrogenase 2 His47Arg polymorphism influences drinking habit independently of aldehyde dehydrogenase 2 Glu487Lys polymorphism: analysis of 2,299 Japanese subjects. Cancer Epidemiol Biomarkers Prev. 2006;15:1009–1013. doi: 10.1158/1055-9965.EPI-05-0911. [DOI] [PubMed] [Google Scholar]
- 20.Matsuo K, Ito H, Wakai K, Hirose K, Saito T, Suzuki T, Kato T, Hirai T, Kanemitsu Y, Hamajima H, Tajima K. One-carbon metabolism related gene polymorphisms interact with alcohol drinking to influence the risk of colorectal cancer in Japan. Carcinogenesis. 2005;26:2164–2171. doi: 10.1093/carcin/bgi196. [DOI] [PubMed] [Google Scholar]
- 21.Willett W, Stampfer MJ. Total energy intake: implications for epidemiologic analyses. Am J Epidemiol. 1986;124:17–27. doi: 10.1093/oxfordjournals.aje.a114366. [DOI] [PubMed] [Google Scholar]
- 22.Imaeda N, Goto C, Tokudome Y, Hirose K, Tajima K, Tokudome S. Reproducibility of a short food frequency questionnaire for Japanese general population. J Epidemiol. 2007;17:100–107. doi: 10.2188/jea.17.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokudome Y, Goto C, Imaeda N, Hasegawa T, Kato R, Hirose K, Tajima K, Tokudome S. Relative validity of a short food frequency questionnaire for assessing nutrient intake versus three-day weighed diet records in middle-aged Japanese. J Epidemiol. 2005;15:135–145. doi: 10.2188/jea.15.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Matsuo K, Wakai K, Hiraki A, Hirose K, Sato S, Ueda R, Tajima K. Effect of familial history and smoking on common cancer risks in Japan. Cancer. 2007;109:2116–2123. doi: 10.1002/cncr.22685. [DOI] [PubMed] [Google Scholar]
- 25.Marcheko YV CR, Lin DY, Amos Cl, Guierrez RG. Semiparametric analysis of case-control genetic data in the presence of enrionmental factors. The Stata Journal. 2008;8:305–333. [Google Scholar]
- 26.in DY, Zeng D. Likelihood-based inference on haplotype effects in genetic association studies. J Am Stat Assoc. 2006;101:89–118. [Google Scholar]
- 27.Spinka C, Carroll RJ, Chatterjee N. Analysis of case-control studies of genetic and environmental factors with missing genetic information and haplotype-phase ambiguity. Genet Epidemiol. 2005;29:108–127. doi: 10.1002/gepi.20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]


