Abstract
The production of lytic enzymes in Trichoderma is considered determinant in its parasitic response against fungal species. A mitogen-activated protein kinase encoding gene, tvk1, from Trichoderma virens was cloned, and its role during the mycoparasitism, conidiation, and biocontrol was examined in tvk1 null mutants. These mutants showed a clear increase in the level of the expression of mycoparasitism-related genes under simulated mycoparasitism and during direct confrontation with the plant pathogen Rhizoctonia solani. The null mutants displayed an increased protein secretion phenotype as measured by the production of lytic enzymes in culture supernatant compared to the wild type. Consistently, biocontrol assays demonstrated that the null mutants were considerably more effective in disease control than the wild-type strain or a chemical fungicide. In addition, tvk1 gene disruptant strains sporulated abundantly in submerged cultures, a condition that is not conducive to sporulation in the wild type. These data suggest that Tvk1 acts as a negative modulator during host sensing and sporulation in T. virens.
Many species of Trichoderma have been used as potent biocontrol agents for a variety of soil-borne phytopathogenic fungi (1). The response of Trichoderma to the presence of a potential host includes production of antibiotic compounds, formation of specialized structures, and degradation of the host's cell wall followed by the assimilation of its cellular content, namely mycoparasitism (2). This last phenomenon has been proposed as the central mechanism accounting for the antagonistic activity of Trichoderma species. Hydrolytic enzymes produced by Trichoderma, such as chitinases, β-1-3 glucanases, β-1-6 glucanases, and proteases (2), facilitate penetration of the host. Strains of Trichoderma transformed to overexpress hydrolytic enzymes have been shown to be better biocontrol agents than their corresponding parental strains (2). Consequently, most of these hydrolytic enzyme-encoding genes have been designated as mycoparasitism-related genes (MRGs). The ability of Trichoderma to sense and respond to different environmental conditions, including the presence of a potential host, is essential for successful colonization of soil, organic material, and developing plant roots. Sensing of such environmental conditions may occur through a variety of transduction pathways, which determine the adequate cellular response. Mitogen-activated protein kinase (MAPK) pathways transduce a large variety of signals, including those associated with pathogenesis. Parasitism by Trichoderma resembles in many aspects the interaction of phytopathogenic fungi with their host. In this sense, MAPKs have been directly implicated in pathogenicity in Magnaporthe grisea (Pmk1, Pms1), Botrytis cinerea (Bmp1), Fusarium oxysporum (Fmk1), Cochliobolus heterostrophus (Cmk1), and Ustilago maydis (Ubc3/Kpp2) (3). In several fungal systems, including phytopathogenic fungi, homologues to the MAPK Kss1 from Saccharomyces cerevisiae have been implicated in the expression of cell-degrading enzymes. In F. oxysporum, the induction of the pectate lyase-encoding gene (pl1) was abolished in the fmk1 null mutant (4). Similarly, Bmp1 from B. cinerea positively controls the expression of those enzymes involved in plant penetration (5). In Trichoderma atroviride, a species closely related to Trichoderma virens, transcription of a proteinase-encoding gene (prb1) in response to nitrogen limitation was blocked by the addition of a specific inhibitor of MAP kinase kinases (6).
To test the hypothesis that MAPKs are directly involved in the establishment of the parasitic relationship between Trichoderma and its hosts, we have cloned a MAPK from T. virens that is similar to Pmk1. Here we report the role of this gene in several aspects of the life cycle of T. virens, including growth, conidiation, expression of MRGs, secretion of cell wall-degrading enzymes, and biocontrol activity.
Materials and Methods
Fungal Strains. T. virens Gv29-8 (wild type) and arginine auxotrophic Tv10.4 strains were used in this study (7). Rhizoctonia solani AG-4 and Pythium ultimum were used as hosts. These plant pathogens were isolated from the roots of dying cotton seedlings. The fungal strains were maintained on Potato Dextrose Agar (Difco), unless otherwise indicated.
Bacterial Strains and Plasmids. Escherichia coli strains DH5α (Bethesda Research Laboratories) and JM103 (Invitrogen) were used for all DNA manipulations. The plasmids used were pBluescript (Stratagene) and pCB1004 (Fungal Genetics Stock Center). All PCR products were cloned in pCR2.1 (Invitrogen). Probes used for Northern blot analysis were obtained as follows. A 1.3-kb HindIII/BamHI fragment of Tv-prb1 cDNA was cut from pPOE. A 1.4-kb HindIII/XbaI fragment corresponding to Tv-cht1 was removed from pCOE plasmid (8). From pSZD2, a 0.52-kb PstI/XhoI fragment corresponding to Tv-bgn2 was excised. Two fragments corresponding to Tv-cht2 (from position 988-1417) and Tv-nag1 (from position 513-1608) were obtained from T. virens genomic DNA by PCR by using oligonucleotides designed based on the sequences reported by Kim et al. (9).
DNA and RNA Manipulations. Plasmid DNA was isolated by using a commercial kit (Qiagen). DNA from Trichoderma was obtained as described (10). Total RNA was isolated by using phenol:chloroform extraction according to the protocol of Jones et al. (11). Southern and Northern blots were performed by using Hybond-N+ membranes (Amersham Biosciences) according to the manufacturer's recommendations.
Western Blot Analysis. Proteins extracts were prepared as described (6), and protein concentration was determined by using the Bradford assay (Bio-Rad) with BSA as a standard. Equivalent amounts of protein (25 μg) from each sample were resuspended in Schägger 2× buffer (12) and boiled after adding 2-mercaptoethanol (5%). Proteins were fractionated by SDS/PAGE on 10% gels according to Schägger and Von Jagow (12). Gels were transferred to Hybond-C extra membranes (Amersham Biosciences) and probed according to the instructions of the Phospho Plus p42/p44 MAP Kinase (Thr202/Tyr204) Antibody kit (Cell Signaling Technology, Beverly, MA).
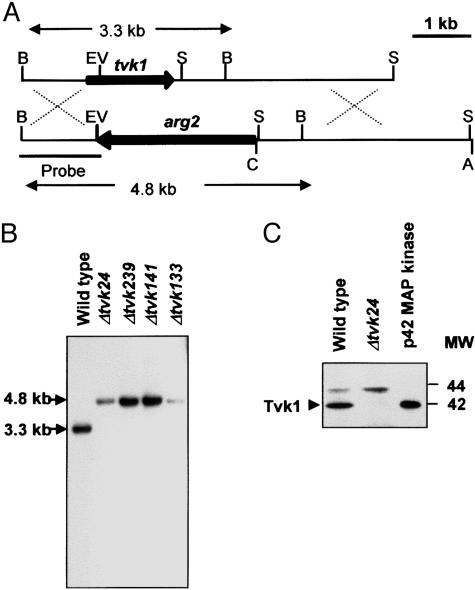
Cloning and Sequencing of tvk1. Genomic DNA of T. virens Gv29-8 was used as a template for PCR reactions with the primers described in ref. 13. The PCR product was cloned and sequenced by the dideoxynucleotide chain termination method of Sanger (14) with a Sequenase kit (Version 2.0, United States Biochemical). A fragment that had high similarity with the pmk1 gene from M. grisea was selected and used as a probe to screen a cosmid DNA library of T. virens Gv28-9. Three clones were identified and one was selected for sequencing. Southern analysis allowed the identification of a 3.3-kb BamHI fragment containing the tvk1 gene, which was subcloned into the BamHI site of plasmid pCB1004 (pDXG35). This fragment was sequenced entirely, followed by a BLAST DNA–protein sequence database search.
Construction of the Gene Replacement Vector pTVK1::arg2. To construct the tvk1 gene replacement vector, a 1.48-kb EcoRV/SalI fragment containing part of the tvk1 coding region (from amino acid 88 until the end of the protein) was replaced by a 3.2-kb SmaI/EcoRV fragment of the T. virens arg2 gene (7). Briefly, a BamHI/EcoRV fragment of pDGX35 was subcloned into BamHI/EcoRV sites of pBluescript SK(-) to generate plasmid pAM1. The pAM1 plasmid was digested with EcoRV and ligated to an EcoRV/SmaI fragment of the arg2 gene (pAM2). To obtain the C-terminal region of Tvk1, we used a 0.4-kb SalI/BamHI fragment from pDGX35 to probe cosmid DNA digested with several enzymes. A 3.2-kb SalI fragment was cloned in pBluescript KS(+) and then subcloned as a ClaI/ApaI fragment into pAM2. The resulting vector (pTVK1::arg2) was linearized and used for transformation of the T. virens arginine auxotrophic strain Tv10.4 (Fig. 1A).
Fig. 1.
Strategy for tvk1 gene disruption and analysis of transformants. (A) Schematic representation of Tvk1 replacement. Thick arrows represent the tvk1 and arg2 coding regions. Lines represent the 5′ and 3′ regions of the tvk1 gene. Crossover events are indicated by dotted lines. EV, EcoRV; B, BamHI; S, SalI; C, ClaI; A, ApaI. (B) Southern analysis of transformants. Genomic DNA was digested with BamHI. Lanes 2–5 represent four independent transformants; lane 1 is the wild type. The blot was hybridized with the probe indicated in A. (C) Immunoblot analysis of crude extracts from T. virens (wild-type and Δtvk24 strains) with an ERK1/ERK2-specific polyclonal antibody. The arrow indicates the signal corresponding to Tvk1. p42 MAPK is a recombinant MAPK protein used as positive control.
Fungal Transformation. Preparation and transformation of T. virens protoplast were performed according to the method described in ref. 7. Prototrophic transformants were selected on Vogel's minimal medium containing sucrose as a sole carbon source (VMS). Disruption of the tvk1 gene in selected transformants was confirmed by Southern and Western blot analysis.
Submerged Culture Analysis. Spores (1 × 106 spores per ml) of the wild-type and Δtvk1 mutants (Δtvk24 and Δtvk133) were inoculated into Potato Dextrose Broth (Difco), Vogel's medium (VMS), or minimal medium (MM) (15) and incubated for 72 h at 28°C. Then samples were analyzed by using a light microscope (BX60, Olympus, Melville, NY). Images were captured and modified by using the programs IMAGE-PRO PLUS 4.0 (Media Cybernetics, Silver Spring, MD) and PHOTOSHOP (Adobe Systems, Mountain View, CA), respectively.
Simulated Mycoparasitism Assay. Trichoderma spores (1 × 106 spores per ml) were germinated and grown for 48 h in VMS. Mycelia were then harvested and transferred to fresh media. Vogel's minimal medium without carbon or nitrogen source (VM or VM-N, respectively) was used to evaluate the effect of nutrient limitation; VMS plus 0.5% R. solani cell walls (VMSR) and Vogel's medium without nitrogen or carbon source plus 0.5% R. solani cell walls (VM-NR or VMR, respectively) were used to simulate a mycoparasitic condition. VMS was used as control. Samples were collected after 3, 6, and 24 h of incubation, frozen in liquid nitrogen, and stored at -70°C until used. For analysis of enzymatic activities, the culture filtrate was recovered and frozen at -20°C until used.
Confrontation Assays. Trichoderma strains were subjected to confrontation assays without contact by using R. solani as a host, as described by Cortés et al. (15). Confrontation was carried out on modified VMS agar (mVMS) containing 0.75 g/liter sucrose and 0.45 g/liter NH4NO3. Trichoderma mycelia were collected from the zone of interaction between the fungi.
Biocontrol Assays. Assays were conducted as described (8). Briefly, cotton seeds (cultivar 112, Stoneville, Memphis, TN) were coated with Trichoderma strains and planted into a non-sterile soilless medium (Metro Mix, 366, Scotts, Marysville, OH) infested with R. solani or P. ultimum. Seeds planted into non-infested medium and seeds treated with the commercial fungicide Apron XL LS (Syngenta, Guelph, ON, Canada; active against P. ultimum) were used as positive controls. Healthy, surviving seedlings were counted after a 10-day incubation at 25°C in a growth chamber. Additionally, the extension of the disease symptoms in the root system was evaluated in R. solani-infected plants by using an arbitrary scale of 0 (no symptoms) to 5 (entire root system discolored and decayed) with a maximum of 6 for nongerminated/dead seeds. Each treatment was replicated six times, with 10 seeds each, and the entire experiment was repeated twice.
Results
Isolation of tvk1, a Gene Encoding a MAPK from T. virens. Southern blot analysis indicated that tvk1 is present as a single copy in T. virens. The cloned tvk1 gene contains four exons interrupted by three introns as reported for other MAPK encoding genes in fungi (GenBank accession no. AY162318). The deduced protein sequence of the gene has 360 amino acids with an estimated molecular mass of 41.6 kDa and a pI of 6.44. Alignment of the sequence by using the MEGALIGN-CLUSTAL program (DNAS-TAR, Madison, WI) indicated that Tvk1 corresponds to the recently reported TmkA protein from T. virens IMI306092 (16), sharing 98% identity with Tmk1 from T. atroviride, 95% identity to Cmk1, Pmk1, and Fmk1 from F. oxysporum, M. grisea, and Colletotrichum lagenarium, respectively (4, 13, 17), and 54% identity with Kss1 from S. cerevisiae. The region comprised between residues 58 and 160 contains the typical sequence observed in several members of the MAPK family: F-X (10)-R-E-X (72–82)-R-D-X-K-X (9)-C (18). Tvk1 also contains the residues T (184)-E-Y (186) required for its phosphorylation and activation by a MAP kinase kinase homologue.
Generation of tvk1 Null Mutants. Disruption of tvk1 in 20 stable transformants was evaluated by Southern blotting with a 1.4-kb BamHI/EcoRV fragment from plasmid pDGX35 as a probe. A 3.3-kb hybridizing fragment was expected in the wild type, a 4.8-kb fragment was expected in null mutants, and both bands were expected in case of ectopic integration events (Fig. 1B). Seven transformants with the expected hybridization pattern were identified and further analyzed by Southern blot to verify that no additional integration events had occurred. None of the selected transformants showed additional bands other than that corresponding to the replacement event. Transformants designated Δtvk24 and Δtvk133 were arbitrarily chosen for further phenotypic and physiological studies.
To verify that Tvk1 was not produced in the gene disruptants, total protein extracts from T. virens wild type and Δtvk24 strains were probed with a polyclonal antibody that recognizes the p42/p44 MAPKs from mammalian cells (Fig. 1C). As expected, both 42- and 44-kDa proteins were detected in the wild-type strain (Fig. 1C, lane 1). In contrast, only the signal corresponding to the 44-kDa MAPK was found in Δtvk24 (Fig. 1C, lane 2). These results demonstrated that the Δtvk24 strain did not produce a p42 MAPK homologue. Similar results were obtained after the analysis of other gene disruptants.
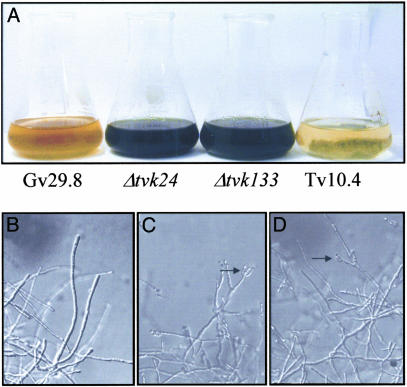
Tvk1 Mutants Show Altered Vegetative Growth and Conidiation. Colonies of Δtvk1 mutants showed a reduction in the rate of colony growth and development of aerial hyphae on solid media. Conidial suspensions of wild type showed an intense green color compared with the pale green color of suspensions of the mutant conidia. In addition, null mutants produced two times less conidia than the wild type when grown on Potato Dextrose Agar with no significant changes in conidiophore morphology. When growth was analyzed in liquid media, all Δtvk1 mutants formed mycelial pellets smaller than those generated by the wild-type strain. Surprisingly, Δtvk1 mutants conidiated massively in late submerged cultures (72 h), whereas no conidia appeared in cultures of the parental strain even after 7 days of culture. Fig. 2A shows liquid cultures of T. virens wild type (Gv29-8) (leftmost flask), Δtvk1 mutants (Δtvk24 and Δtvk133) (two center flasks), and the parental strain (Tv10.4) (rightmost flask) after 72 h of incubation in VMS. The ability of Δtvk1 mutants to conidiate in liquid culture seems to be independent of the media used, because conidiation was evident either in VMS or Potato Dextrose Broth. Microscopic observations of samples from Δtvk24 and Δtvk133 sporulating liquid cultures showed normal conidiophore development, resembling those produced in aerial hyphae (Fig. 2 C and D). In contrast, the WT strain did not produce conidia in the same media (Fig. 2B).
Fig. 2.
Conidiation in submerged culture. (A) Overview of T. virens parental and mutant strains liquid cultures after 72 h in Potato Dextrose Broth. (B–D) Microscopic analysis of T. virens wild type (Gv29.8) (B), Δtvk24 (C), and Δtvk133 (D). Conidiophores are indicated by arrows.
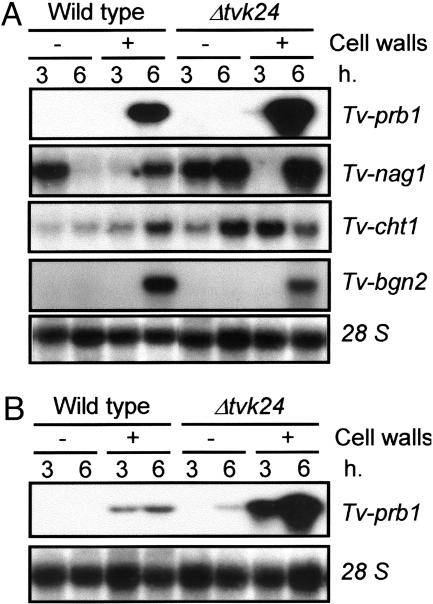
Increased Expression of MRGs in Δtvk24 Under Simulated Mycoparasitism. Northern blot analysis showed that in the wild type the N-acetylglucosaminidase-encoding gene Tv-nag1 was expressed in medium with no carbon source 3 h after transfer but no message was detected after 6 h. Whereas in the mutant the expression was detected after 3 h and increased by 6 h. In medium containing R. solani cell walls the gene was expressed only after 6 h for both the wild-type and the mutant strains but reached much higher levels in the mutant (Fig. 3A, Tv-nag1). The second chitinase-encoding gene analyzed (Tv-cht1) was expressed in both strains in the absence of carbon source but reached higher levels in Δtvk24 by 6 h. Tv-cht1 was clearly induced by cell walls in the wild-type strain with maximum expression by 6 h. In contrast, no obvious induction could be observed in the mutant strain, which reached the same level observed under carbon limitation conditions, except that this occurred earlier (Fig. 3A, Tv-cht1). The endoglucanase-encoding gene Tv-bgn2 was induced by cell walls by 6 h, but no difference in expression was observed for the wild type and the mutant, and the gene was not expressed in medium with no carbon source (Fig. 3A, Tv-bgn2). The pattern of expression of a third chitinase gene (Tv-cht2) was similar to that of Tv-cht1, except that maximum expression was reached after 24 h under simulated mycoparasitism. Consistent with the previous expression pattern of other MRGs analyzed, Δtvk24 showed a much more pronounced induction of Tv-cht2 than the wild-type strain. No expression of these genes was detected in minimal media supplemented with primary nitrogen and carbon sources (data not shown).
Fig. 3.
Expression of MRGs in simulated mycoparasitism. (A) Northern analysis of the genes Tv-prb1, Tv-nag1, Tv-cht1, and Tv-bgn2 under carbon limitation with and without R. solani cell walls in the wild type and the Δtvk24 mutant. Numbers indicate time of induction. (B) Northern analysis of Tv-prb1 under nitrogen limitation and simulated mycoparasitism. + and -, presence and absence of R. solani cell walls, respectively. Fifteen micrograms of total RNA was loaded in each lane. 28S ribosomal RNA was used as load control.
Expression of the protease-encoding gene, Tv-prb1, was determined under simulated mycoparasitism both in the absence of an alternative carbon source (Fig. 3A, Tv-prb1) and in VM-NR that contains no ammonia (Fig. 3B, Tv-prb1). Induction by cell walls was observed for both strains, although higher transcript levels were detected under carbon starvation than under nitrogen limitation conditions. In both cases, induction was much higher for Δtvk24 than for the wild-type strain. Δtvk24 showed the greatest level of expression after 6 h, 7- to 10-fold higher than the expression in wild type. The mutant showed detectable levels of Tv-prb1 expression after 6 h when grown in VM-N (Fig. 3B, Tv-prb1). Accordingly, in-gel protease, endochitinase, N-acetylglucosaminidase, and glucanase activity analyses of the culture filtrates showed up to 10-fold higher activity levels in the mutants than in the wild type. Interestingly, the presence of an additional endochitinase activity band in the null mutants under carbon source limitation was observed (Fig. 6, which is published as supporting information on the PNAS web site).
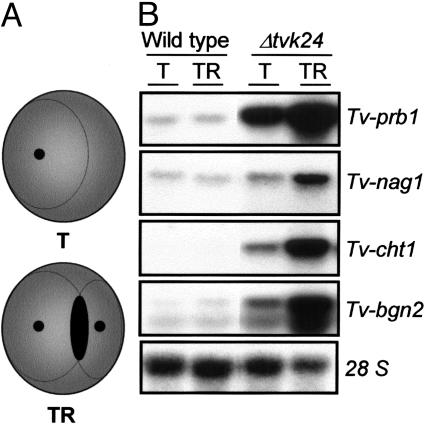
High Levels of MRGs Are Detected in Δtvk24 During Confrontation with R. solani. Expression analysis of the same set of genes was carried out in Δtvk24 and wild-type strains during confrontation assays with mVMS plates (Fig. 4A). When Δtvk24 overgrew the colony of R. solani (TR), a clear induction of Tv-prb1, Tv-nag1, Tv-cht1, and Tv-bgn2 was detected as compared with the control condition where the mutant strain was growing alone (T) (Fig. 4B). Interestingly, there was a detectable level of expression of all genes in the control (T). Transcript accumulation of all MRGs in Δtvk24 was not only increased in the presence of the host but also reached much higher levels than for the wild-type strain. Unexpectedly, at the time point sampled, no difference in the expression of MRGs was detected when the wild-type strain was growing alone (T) or in the presence of R. solani (TR) (Fig. 4B).
Fig. 4.
Expression of MRGs in direct confrontation with R. solani. (A) Schematic representation of the interaction between T. virens strains and R. solani. T, T. virens strain growing alone in VMS plates; TR, T. virens strain growing in presence of R. solani. The interaction zone is colored black. (B) Expression of MRGs in T. virens strains. Samples were collected when T. virens strains grew 1.0 cm over the colony of R. solani. The probes used for the Northern blot analysis were the same as those indicated in Fig. 3. Fifteen micrograms total RNA were loaded in each lane.
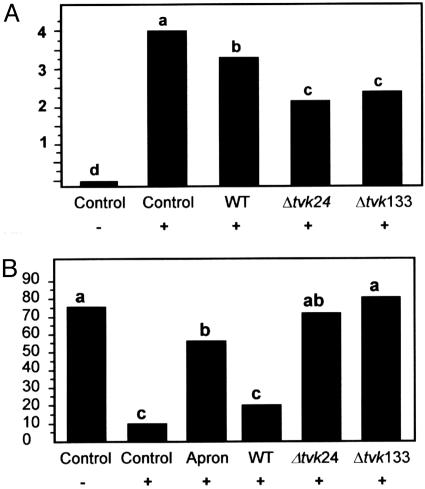
Biocontrol Activity. The biocontrol ability of Δtvk24 and Δtvk133 was tested in vivo against two different root pathogens, R. solani and P. ultimum. Plant mortality was very low in treatments with R. solani (10%). Although protection by Δtvk24 and Δtvk133 was higher (mortality 0% and 5%, respectively) than with wild type (mortality 8%), the differences were not statistically significant. However, when the root systems of these plants were evaluated for disease symptoms, significant differences were detected (Fig. 5A). The disease index was significantly less for the treatments with the Δtvk1 mutant strains than for the wild type. When cotton plants were infected with Pythium, only 10% of the nontreated seeds survived. However, survival increased to 70% and 80% in treatment with Δtvk24 and Δtvk133, respectively, compared to only 20% in wild-type strain. This level of protection afforded by the two mutant strains was comparable and even significantly higher than the protection obtained by treatment of the seeds with the commercial fungicide Apron (Fig. 5B).
Fig. 5.
Biocontrol activity of wild type, Δtvk24, and Δtvk133. (A) Disease index of the root systems from cotton plants infected with R. solani. (B) Percentage of healthy plants after P. ultimum treatment. Columns with a letter in common did not differ significantly according to Fisher's protected least significant difference test at a significance level of 5%. + and -, presence and absence of the pathogen, respectively.
Discussion
The morphological and molecular responses observed in Trichoderma when confronted by a host depend on the capacity of Trichoderma to sense and respond to external stimuli and to adjust its intracellular activities accordingly. In this study, we isolated and characterized a MAPK-encoding gene from T. virens, tvk1. The deduced protein sequence of tvk1 showed high similarity with other MAPKs reported for phytopathogenic fungi, especially Pmk1 from M. grisea (13). The tvk1 gene contains three intron sequences also reported for other MAPK genes from fungi. Even though tmkA and tvk1 encode the same protein in different strains of T. virens, tmkA appears to have lost a conserved intron found in diverse genes belonging to Pmk1 homologues in filamentous fungi, including tvk1 (16), suggesting that tmkA from T. virens IMI306092 has suffered recent evolutionary changes. Tvk1 belongs to a family of kinases regulated by external signals (extracellular-regulated kinases, or ERKs), which constitute part of the MAPK superfamily. The signature sequence present in the Tvk1 protein indicates that it is related to the YERK1 family (yeast and fungal ERK1) (19).
MAPK pathways have been implicated in controlling cellular growth in a variety of eukaryotic organisms. Tvk1 null mutants showed a reduction in growth rate as reported for tmkA mutant strains (16). In addition, Δtvk1 mutants produced less conidia than the wild-type strain on solid media (data not shown). Similar morphological alterations have been reported for mutants in homologues of tvk1 in other fungi. The analysis of MAPK null mutants from C. lagenarium, U. maydis, and C. heterostrophus has established that these tvk1 homologues are required for sporulation (17, 20, 21). In contrast, mutants in the corresponding gene in M. grisea, F. oxysporum, and B. cinerea showed no alterations in spore production (4, 5, 13). All Δtvk1 strains analyzed showed reduced conidial pigmentation with loss of the characteristic dark green color observed in the wild type. The pigment associated with the spore surface of T. viride, a closely related species to T. virens, has been reported to be a nonindolic melanin-like polyphenol (22). In C. lagenarium, the expression of genes involved in melanin synthesis is regulated by the Cmk1-MAPK. Mutations in cmk1 result in the production of albino spores. In T. virens, Δtvk1 strains produced spores with decreased pigmentation on solid media, without becoming albinos. In contrast, all Δtvk1 strains produced abundant conidia that developed the characteristic dark green color of the wild type in submerged cultures independently of the media used. These results suggest that Tvk1 may differentially regulate melanin biosynthesis as reported for C. lagenarium (17).
In general, filamentous fungi grow by hyphal elongation in submerged cultures, but nutrient depletion may induce conidiation, as in Neurospora crassa (23). Growth of Δtvk1 strains under nutrient limitation led to the early production of spores (24 h) when compared with the 72 h required to detect conidia in complete medium. However, no change in vegetative hyphal growth pattern was observed in the wild-type strain when grown under starvation conditions. Additionally, sporulation in Δtvk1 strains is completely dependent on the availability of nutrients, because continuous replacement of fresh medium to the culture blocked conidial formation.
As a mycoparasite, Trichoderma depends on the production of hydrolytic enzymes for the colonization of host fungi. Recently, Xu (3) suggested that Pmk1 and its homologues might be involved in the positive regulation of the expression of cell wall-degrading enzymes. In support of this hypothesis, a Δkss1 mutant of S. cerevisiae and a Δbmp1 mutant of B. cinerea were unable to trigger the expression of the endopolygalacturonases PGU1 and BCPG1, respectively (5, 24), and Δfmk1 mutants of F. oxysporum showed considerably reduced transcript levels of pl1, a gene encoding pectate lyase (4). To test this hypothesis for a mycoparasite, we analyzed the expression of lytic enzymes associated with the parasitic process in the wild type and the Δtvk1 mutants of T. virens. Unexpectedly, Δtvk24 highly expressed most of the lytic enzyme-encoding genes selected as compared with the wild-type strain. These results are in clear contrast to the hypothesis suggested by Xu (3). However, mpk4 null mutants from Arabidopsis thaliana showed constitutive overexpression of several chitinase and glucanase genes, suggesting a negative regulation role for this MAPK in the expression of the genes (25). In contrast, Δtvk1 mutants still require nutrient limitation combined with the presence of cell walls to express the hydrolytic enzymes. Among the MRGs analyzed in the Δtvk24 strain, the expression pattern of the Tv-prb1 gene illustrated several interesting features. In T. atroviride prb1 is subjected to nitrogen catabolic repression (NCR) as well as being induced by the presence of R. solani cell walls, but this response is completely dependent on the absence of a primary nitrogen source (6). Interestingly, the expression of Tv-prb1 seems to be equally dependent on NCR in both wild-type and Δtvk24 strains, because no transcript was detected when the strains were grown in VMSR. Tv-prb1 expression associated with the sole lack of a nitrogen source was detected for the Δtvk24 mutant, whereas for the wild-type strain no signal of the corresponding mRNA was observed (Fig. 4B). Thus, Δtvk1 mutants seem to be more sensitive to nitrogen limitation than the wild type in terms of Tv-prb1 expression. These data suggest that Tvk1 may be involved in nitrogen repression of Tv-prb1. Recently, we (6) proposed a positive role for a phosphorylated MAPK similar to either Kss1 or Slt2 on the expression of prb1 under similar growth conditions in T. atroviride. In contrast, in this study we showed that Tvk1 acts as a negative element in the expression of Tv-prb1, the T. virens homologue of prb1, under nitrogen limitation or simulated mycoparasitism. This apparent contradiction may be explained by a differential regulation depending on the state of phosphorylation of the kinase, as in the case of Kss1 in S. cerevisiae, which has inhibiting and activating functions depending on the phosphorylation status of the protein (26). On the other hand, expression of the chitinases Tv-nag1 and Tv-cht1 driven by the absence of carbon source seems to be more pronounced in Δtvk24. Overall, regulation of MRGs in T. virens appears to be complex, but it seems clear that they share common elements including Tvk1.
In contrast with the recently suggested decrease in the production of chitinase and cellulase activity in ΔtmkA mutants (16), chitinase and protease in-gel activities were higher in Δtvk24 than in the wild type, correlating with the elevated transcript levels. Furthermore, even though expression of Tv-bgn2 was not increased, total β-1,3-glucanase activity did notably increase in Δtvk1 mutants as compared with the wild type. This suggests that additional glucanase genes could be under negative modulation by Tvk1. The observed increase in the production of enzymes correlated with an increase in secretion of proteins in liquid media (seven times more in Δtvk1 mutant strains in VM-NR and four times more in VMR).
In direct confrontation assays of Trichoderma with a host, expression of Tv-prb1, Tv-nag1, Tv-gln2, and Tv-cht1 strongly increased compared with Trichoderma growing alone in the Δtvk1 mutants (Fig. 4). Surprisingly, in the wild-type strain no induction of MRGs was detected in the presence of R. solani. Host-mediated induction in the wild type may be delayed compared to that observed for null mutants. The detectable level of transcription of the genes when Δtvk24 grew alone may be indicative of derepression, driven by carbon and/or nitrogen limitation. The facts that in Δtvk1 mutants all MRGs followed the same pattern of expression in the direct confrontation assays and that this expression reached very high levels suggest that Tvk1 plays a major role in the coordinated regulation of MRGs during the actual Trichoderma–host interaction.
Considering the increased production of lytic enzymes observed in the T. virens Δtvk1 strains and the relevance of these enzymes in the biocontrol activity of Trichoderma, we examined the potential use of the mutants generated as biocontrol agents. We hypothesized that the robust induction of lytic enzymes in the Δtvk1 strains might lead to a more effective biocontrol. The null mutants showed a greater capacity to control and reduce damage by R. solani and P. ultimum when compared to the wild-type strain. T. virens has been successfully used in combination with several fungicides including metalaxyl (Apron FL) (27). Surprisingly, Δtvk1 strains were more effective than metalaxyl against P. ultimum (Fig. 5B). This enhanced biocontrol ability appears to be associated with the high level of expression of lytic enzymes observed in direct confrontation assays because both strains formed hooks and coils around R. solani and produced antibiotics to the same extent. These results point in the opposite direction of those recently reported by Mukherjee and coworkers (16), who suggested that deletion of tmkA reduces the biocontrol efficacy of T. virens, based on direct confrontations assays. This apparent contradiction may be explained by the fact that their experiments were carried out on Potato Dextrose Agar medium, where expression of most MRGs is repressed because of the high levels of glucose present in that medium (9). In fact, similar experiments carried out by our group with several phytopathogenic fungi, including Sclerotium rolfsii and R. solani, indicated varying behavior of the mutant as compared to the wild type, going from slightly decreased overgrowth capacity to increased growth inhibition, depending on the host (Fig. 7, which is published as supporting information on the PNAS web site). These data suggest that Trichoderma uses different mechanisms to control different hosts. An additional consideration that must be made is that some Trichoderma species are capable of inducing defense responses in plants, as shown for T. virens (28). To the best of our knowledge, this is the first report indicating that the deletion of a MAPK gene generates a more aggressive parasite and, consequently, a better biocontrol agent.
Supplementary Material
Acknowledgments
We thank Dr. Charles Howell for assistance with the analysis of antibiotics from fungal strains, Dr. Raúl Rodriguez for suggestions in biocontrol assays, and Dr. John Délano for critical reading of the manuscript. A.M.-M. is indebted to Consejo Nacional de Ciencia y Technología (Mexico) for a doctoral fellowship. This work was supported by U.S. Department of Agriculture–National Research Initiative Competitive Grants Program Grant 99-35316-7940 (to C.M.K.) from the Texas Cotton BioTechnology Initiative and grants from the International Center for Genetic Engineering and Biotechnology (Italy) (CRP/MEX99-02 to A.H.-E.) and the Texas A&M University Collaborative Research Grant Program (to A.H.-E. and C.M.K.).
Abbreviations: ERK, extracellular-regulated kinase; MAPK, mitogen-activated protein kinase; MRG, mycoparasitism-related gene.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY162318).
References
- 1.Papavizas, G. C. (1985) Annu. Rev. Phytopathol. 23, 23-54. [Google Scholar]
- 2.Herrera-Estrella, A. & Chet, I. (2003) in Handbook of Fungal Biotechnology, ed. Arora, D. (Dekker, New York), in press.
- 3.Xu, J.-R. (2000) Fungal Genet. Biol. 31, 137-152. [DOI] [PubMed] [Google Scholar]
- 4.Di Pietro, A., García-Maceira, F. I., Méglecz, E. & Roncero, I. G. (2001) Mol. Microbiol. 39, 1140-1152. [PubMed] [Google Scholar]
- 5.Zheng, L., Campbell, M., Murphy, J., Lam, S. & Xu, J.-R. (2000) Mol. Plant–Microbe Interact. 13, 724-732. [DOI] [PubMed] [Google Scholar]
- 6.Olmedo-Monfil, V., Mendoza-Mendoza, A., Gómez, I., Cortés, C. & Herrera-Estrella, A. (2002) Mol. Gen. Genomics 267, 703-712. [DOI] [PubMed] [Google Scholar]
- 7.Baek, J.-M. & Kenerley, C. M. (1998) Fungal Genet. Biol. 23, 34-44. [DOI] [PubMed] [Google Scholar]
- 8.Baek, J.-M., Howell, C. R. & Kenerley, C. M. (1999) Curr. Genet. 35, 41-50. [DOI] [PubMed] [Google Scholar]
- 9.Kim, D.-J., Baek, J.-M., Uribe, P., Kenerley, C. & Cook, D. R. (2002) Curr. Genet. 40, 374-384. [DOI] [PubMed] [Google Scholar]
- 10.Reader, U. & Broda, P. (1985) Lett. Appl. Microbiol. 1, 17-20. [Google Scholar]
- 11.Jones, J., Dunsmuir, P. & Bedbrook, J. (1985) EMBO J. 4, 2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schägger, H. & Von Jagow, G. (1987) Anal. Biochem. 166, 368-379. [DOI] [PubMed] [Google Scholar]
- 13.Xu, J.-R. & Hamer, J. E. (1996) Genes Dev. 10, 2696-2706. [DOI] [PubMed] [Google Scholar]
- 14.Sanger, F., Nicklen, S. & Coulson, A. R. (1977) Proc. Natl. Acad. Sci. USA 74, 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortés, C., Gutierrez, A., Olmedo, V., Chet, I. & Herrera-Estrella, A. (1998) Mol. Gen. Genet. 260, 218-225. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee, P. K., Latha, J., Hadar, R. & Horwitz, B. A. (2003) Eukaryotic Cell 2, 446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takano, Y., Kikuchi, T., Kubo, Y., Hamer, J. E., Mise, K. & Furusawa, I. (2000) Mol. Plant–Microbe Interact. 13, 374-383. [DOI] [PubMed] [Google Scholar]
- 18.Dorin, D., Alano, P., Boccaccio, I., Ciceron, L., Doerig, C., Sulpice, R., Parzy, D. & Doerig, D. (1999) J. Biol. Chem. 274, 29912-29920. [DOI] [PubMed] [Google Scholar]
- 19.Kultz, D. (1998) J. Mol. Evol. 46, 571-588. [DOI] [PubMed] [Google Scholar]
- 20.Muller, P., Alchinger, C., Feldbrugge, M. & Kahmann, R. (1999) Mol. Microbiol. 34, 1007-1017. [DOI] [PubMed] [Google Scholar]
- 21.Lev, S., Sharon, A., Hadar, R., Ma, H. & Horwitz, B. A. (1999) Proc. Natl. Acad. Sci. USA 96, 13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benitez, T., Villa, T. G. & García-Acha, I. (1976) Can. J. Microbiol. 22, 318-321. [DOI] [PubMed] [Google Scholar]
- 23.Springer, M. L. & Yanofsky, L. (1992) Genes Dev. 6, 1052-1057. [DOI] [PubMed] [Google Scholar]
- 24.Madhani, H. D., Galitski, T., Lander, E. S. & Fink, G. R. (1999) Proc. Natl. Acad. Sci. USA 96, 12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., Nielsen, H. B., Lacy, M., Austin, M. J., Parker, J. E., et al. (2000) Cell 103, 1111-1120. [DOI] [PubMed] [Google Scholar]
- 26.Cook, J. K., Bardwell, L. & Thorner, J. (1997) Nature 390, 85-88. [DOI] [PubMed] [Google Scholar]
- 27.Howell, C. R., Stipanovic, R. D. & Lumsden, R. D. (1993) Biocontrol Sci. Tech. 3, 435-441. [Google Scholar]
- 28.Howell, C. R., Hanson, L. E., Stipanovic, R. D. & Puckhaber, L. S. (2000) Phytopathology 90, 248-252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.