Abstract
Previously, we reported a strong association of the high activity SULT1A1*1 allele and overall survival of patients receiving tamoxifen therapy, indicating that sulfation of 4-hydroxytamoxifen (4-OHT) via SULT1A1 may contribute to the therapeutic efficacy of tamoxifen treatment. In most, but not all cases, sulfation is considered to be an elimination pathway; therefore we sought to define the biological mechanism by which increased sulfation of tamoxifen could provide a therapeutic benefit. We compared the antiproliferative and apoptotic responses between MCF7-SULT1A1 expressing cells and control MCF7 pcDNA3 cells when treated with 4-OHT. We observed a greater than 30% decrease in cell proliferation in MCF7-SULT1A1 expressing cells at physiological concentrations of 4-OHT, and significant cell death in SULT1A1-expressing cells treated with 2µM 4-OHT for 48 hours compared to control cells (p<0.05). Within 24 hours of drug treatment, an 80% increase in apoptosis in SULT1A1-expressing cells was apparent when compared to similarly treated cells that did not express SULT1A1. We also observed an increase in endonuclease G, the primary endonuclease expressed in ER-dependent breast cancer cells, which participates in caspaseindependent apoptosis. These data confirm that SULT1A1-mediated biotransformation of 4-OHT is important in the efficacy of 4-OHT cytotoxicity in breast tumors, and reveals a potential role for sulfated metabolites in the efficacy of tamoxifen therapy.
Keywords: SULT1A1, tamoxifen, apoptosis, genotype
Introduction
Tamoxifen has been the principal agent used in endocrine therapy for post-menopausal women diagnosed with estrogen receptor (ER) positive breast cancer for more than 30 years. Its use has contributed significantly to the decline in breast cancer mortality [1], as well as reducing the incidence of ER positive breast cancer by 48% when used as a chemopreventive agent in high risk women [2]. In a recent report published by the Early Breast Cancer Trialist’s Collaborative Group (EBCTCG), it was noted that adjuvant tamoxifen therapy for 5 years reduced the annual death rate in women diagnosed with ER positive breast cancer by 31% regardless of age [3]. However, 30% of women treated with tamoxifen experience relapse within the first 5 years of therapy. Clearly, not all women diagnosed with ER positive breast cancer benefit from tamoxifen, and additional prognostic markers aside from ER, and progesterone status are needed [4].
The pharmacology and metabolism of tamoxifen is complex and it is postulated that differences in patient outcomes could be the result of individual genetic variation in tamoxifen’s extensive metabolic pathways. For example, cytochrome P450 2D6 (CYP2D6) plays a critical role in the production of tamoxifen’s active metabolites, endoxifen and 4-OHT. Several studies have demonstrated that CYP2D6 genotypes have a significant impact on efficacy of tamoxifen therapy, as well as on drug interactions with therapeutic agents that are also substrates of CYP2D6 [5-7]. While P450-mediated biotransformation is critical to tamoxifen’s efficacy, tamoxifen metabolites also undergo secondary conjugation reactions that can also be expected to play a role in the pharmacogenetics of tamoxifen therapy.
Sulfotransferase isoform 1A1 (SULT1A1) is a phase II enzyme that is the primary sulfotransferase isoform responsible for the sulfation of 4OHT in breast tumors [8, 9]. A genetic polymorphism in exon 7 of the SULT1A1 gene results in the substitution of a histidine for an arginine at position 213 of the translated protein, which in turn produces an enzyme with much lower catalytic activity and decreased thermal stability [10, 11]. Previously, we conducted a retrospective study examining the relationship between SULT1A1 genotype and overall survival of a cohort of women diagnosed with breast cancer and treated with tamoxifen [12]. We hypothesized that the impaired enzymatic activity and decreased thermal stability conferred by the SULT1A1*2 allele would result in decreased clearance of 4-OHT via sulfation and therefore increase the efficacy of tamoxifen treatment. Contrary to our original hypothesis, we found a strong association between survival and the high activity SULT1A1*1 allele [12]. This association was not observed among women who did not receive adjuvant tamoxifen therapy, indicating that variability in prognosis is dependent on SULT1A1 expression and the subsequent sulfation of 4-OHT. Other studies have reported the same trend of overall survival associated with tamoxifen treatment and the SULT1A1*1 gene [13]. Although these findings suggest that the sulfated metabolite, 4-sulfoxy-tamoxifen (SO4-TAM) has some therapeutic value, the biological basis of this association is not clear. The current study demonstrates that expression of SULT1A1 in breast cancer cells significantly increases cytotoxicity and apoptotic response when these cells are exposed to 4-OHT. Apoptosis was associated with increased expression of cell death endonuclease G (EndoG), which has recently been associated with cell injury in breast, head and neck, and prostate cancer cells [14-16]. These data demonstrate that SULT1A1-mediated sulfation of 4-OHT in breast tumors is not a process of elimination, but of activation, that enhances the well-characterized cytotoxic effects of 4-OHT in tamoxifen therapy.
Materials and methods
Chemicals
(z)-4-OHT was purchased from Sigma-Aldrich. (e) -SO4-TAM was prepared as previously described [17] by reacting (z)-4-OHT with SO3.Py, and precipitated as a salt by the exchange of the pyridinium ion for potassium. Working stocks were made in dimethylsulfoxide (DMSO). Chemicals used in HPLC analysis were purchased from Fisher Scientific (Waltham, MA). The [2, 43H] E2 (26.5 Ci/mmol) used in the ER competi-tive-binding assays was purchased from Sigma-Aldrich (St. Louis, MO) and the [methyl-3H] thymidine (6.7 Ci/mmol) used in the cell proliferation assays was purchased from Perkin Elmer (Waltham, MA).
Estrogen receptor competitive binding assay
The specificity of SO4-TAM for either recombinant ER α or β (PanVera, Madison, WI) was determined by a method described previously [18]. Each competitor was assayed in three independent experiments, all within a standard error of 10%. Data for each competitor and the E2 standard curve were plotted as a percent 3[H] E2 bound versus molar concentrations and the IC50 for each competitor determined. The relative binding affinity for each competitor was determined by dividing the IC50 of E2 by the IC50 of the competitor and expressed as a percent (E2 =100%).
Steroid sulfatase assay
Inhibition of estrone sulfatase by SO4-TAM was analyzed by incubation of estrone sulfate (200 µM), SO4-TAM (100 µM) with human liver microsomes containing sulfotransferases (2.5 mg of protein) in a Tris-HCl buffer [50 mM Tris-HCl, pH 7.5, 15 mM MgCl2], total volume 250µl, for 15 minutes at 37°C, shaking at 450 rpm in a thermomixer. The reaction was terminated with 250µl ice-cold acetonitrile and proteins were precipitated by centrifugation [22,000 × g, 5 minutes, 4°C]. The supernatant was filtered using an Ultrafree-MC LH PTFE 0.45µm spin column (Millipore, Billerica, MA) and then analyzed by HPLC using a YMC AM column (50 × 4 mm I.D., C-18 3 μm) as previously described [19].
Cell culture and generation of cell lines expressing SULT1A1
Cell lines were obtained from the American Type Culture Collection (Rockville, MD). MCF7 cells were maintained at 37°C under 95% humidity and 5% CO2; cells were cultured in EMEM medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and 0.01 mg/ml bovine insulin. MDA MB 231 cells were cultured in RPMI supplemented with 10% FBS and 1% glutamine, and maintained at 37°C and 5% CO2. The SULT1A1*1 cDNA, a kind gift from Dr. Shogo Ozawa (Iwate Medical University, Japan), was subcloned into the EcoRI/HindIII sites of the pcDNA3 (-) vector. Cells were transfected according to manufacturer’s instructions (Invitrogen, Carlsbad, CA) and expression of active SULT1A1 was assessed by Western blot and enzymatic activity assays. Stable clones were routinely cultured in the appropriate media with the addition of G418.
Western blotting
Cell pellets from MCF7 pcDNA3 and SULT1A1 cell lines were lysed [50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP40, 50 µg/ml phenylmethanesulfonyl fluoride] and proteins were separated by SDS-polyacrylamide gel electrophoresis. Blotted proteins were incubated with the polyclonal anti-SULT1A1 antibody (1:1000) overnight at 4°C, rinsed and incubated in rabbit anti-goat secondary (1:1000) for 1 hour before chemiluminescense detection using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL). For a loading control, actin was also probed using a monoclonal antibody (1:5000) (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoreactive bands were analyzed by an AlphaImager 8900 gel doc system (San Leandro, CA). A commercially available human liver cytosol pool was used as the positive control for SULT1A1 expression (Gentest, BD Biosciences, Woburn, MA). Matched pairs of infiltrating ductal carcinoma lysates and their corresponding normal tissue lysates were purchased from Protein Biotechnologies (San Diego, CA).
Sulfotransferase enzymatic assays
MCF7 pcDNA3 and SULT1A1 expressing cells were lysed in lysis buffer [50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP40, 50 µg/ml phenylmethanesulfonyl fluoride] for 30 minutes on ice. Supernatants were recovered after a 1 hour spin in an ultracentrifuge (100,000g, 4°C), removing intact cells and most subcellular membrane contaminants. Cytosols were assayed for SULT1A1 activity using a colorimetric assay as previously described [20]. Commercially available human liver cytosol pool was used as the positive control for SULT1A1 activity (Gentest, BD Biosciences, Woburn, MA). Results were reported as nmol/min/mg protein. Controls without 3’-phosphoadenosine-5’-phosphosulfate (PAPS), and protein were used in each assay set to assess time-dependent changes in optical density. Protein concentrations were measured by a microscale Bradford method using bovine serum albumin as the reference standard.
Production of SO4-TAM in human hepatocytes exposed to 4-OHT
Cryopreserved human hepatocytes were thawed according to manufacturer’s instructions (BD Biosciences, San Jose, CA) and incubated with increasing concentrations of 4-OHT for 1 to 2 hours after which the cells were harvested and lysed in buffer [50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% NP40, 50 µg/ml phenylmethanesulfonyl fluoride], and filtered using an Ultrafree-MC LH PTFE 0.45 µm spin column (Millipore, Billerica, MA). Sulfated-4-OHT metabolites were analyzed by Micro triple quadrupole mass spectrometer (Waters Associates, Manchester, UK) as previously described [21].
3[H]-Thymidine incorporation assays
MCF7 parent cells and MCF7 cells expressing pcDNA3 or SULT1A1 were deprived of estrogens for 48 hours in media containing charcoal-stripped FBS, after which cells were seeded at a density of 1-3 × 104 cells per well in a 96-well plate and treated with vehicle (<0.1% DMSO), or increasing concentrations of estradiol or 4-OHT for 24 hours. Cells were labeled using 3[H] thymidine, 0.25 µCi/ well, for 1-3 hours, then harvested and collected onto a glass fiber filter using a Microbeta FilterMate 96 harvester (Perkin Elmer, Waltham, MA) as per manufac-turer’s instructions. Proliferation was measured using a Microbeta Trilux 1450 LSC and luminescence counter (Perkin Elmer, Waltham, MA). Each experiment was performed in triplicate and expressed as a percentage of the vehicle control which was set at 100%.
Cell viability assays
In the MCF7 parent cells, viability was determined by crystal violet staining. In brief cells were deprived of estrogen for 48 hours in media containing charcoal-stripped fetal bovine serum, seeded at 10,000 cells/well in a 96-well plated and treated with vehicle (<0.1% DMSO) or increasing concentrations of 4-OHT (1-30 µM) for 24 hours after which the cells were washed with 1X PBS and stained with crystal violet [0.5% crystal violet, 30% ethanol, 3% formaldehyde], washed again with 1X PBS and lysed using a 1% SDS solution. Dye uptake into live cells was measured at 570 nm using a microplate reader. Additionally, cell viability was assessed by clonal dilution assays. Cells expressing pcDNA3 or SULT1A1 were deprived of estrogen for 48 hours, seeded at a density of 2 × 105 in a 24-well plate, and treated with vehicle (<0.1% DMSO), 2 µM tamoxifen or 2 µM 4-OHT for 48 hours, after which the cells were serially diluted in EMEM with 10% fetal bovine serum and incubated for 5 days. On day 5, colonies were fixed and stained using a crystal violet solution (0.5% crystal violet, 30% ethanol, 3% formaldehyde) for 10 minutes at room temperature. Excess stain was washed away with 1 × PBS, and colonies were counted using the Kodak ID image analysis software (Carestream Molecular Imaging, New Haven, CT). Colonies were counted at the 103 dilution, which averaged 31 colonies in vehicle treated cells. This experiment was performed in triplicate. The percent of colony formation was calculated by dividing the average of the treated cells by the vehicle which was set to 100%.
TUNEL assay and EndoG immunocytochemistry
Cells expressing SULT1A1 or pcDNA3 were seeded at a density of 3-4 × 104 cells in a 8-well glass chamber slide, and treated with vehicle, 10 µM 4-OHT, or SO4-TAM in phenol-red free medium supplemented with 5% charcoal stripped fetal bovine serum for 24 hours. Cells were fixed (4% paraformaldehyde, 0.012% saponin, 1× PBS), washed, and probed with either polyclonal anti-EndoG antibody (dilution 1:400) (Millipore, Bedford, MA) at 4°C overnight. Primary antibodies were detected with anti-rabbit IgG-AlexaFluor 594 (Invitrogen, Carlsbad, CA). Cells were then subjected to terminal deoxynucleotidyl transferase mediated dUTP nick end labeling (TUNEL) staining using the In Situ Cell Death Detection Kit from Roche Diagnostics (Indianapolis, IN). After washing and DNA counterstaining with 4,6-diamidino-2-phenylindole (DAPI) cells were mounted under coverslips using the Antifade kit (Invitrogen) and analyzed under an Olympus IX-81 microscope (Olympus, Center Valley, PA) with green, red or blue filter sets from Chroma Technology Corp. (Rockingham, VT). Images and acquisitions were done using digital camera HAMAMATSUORCA (Hamamatsu, Bridgewater, NJ) and software SlideBook 4.2 (Intelligent Imaging Innovations, Denver, CO). Images were developed, masked and analyzed using the SlideBook software. For evaluation of TUNEL all images were automatically masked by DAPI and TUNEL. The areas of the designated masks were measured and percentage of TUNEL-positive nuclei was calculated for each field of view. Expression of the EndoG was acquired by evaluation of the integral optical density (IOD) per either every available cell or TUNEL-positive cell in the given field of view. The experiment was repeated in 8 replicates. At least 10 different fields of view per replicate were analyzed.
Statistical analysis
Statistical analysis was performed using ANOVA and Student's t test. Results were expressed as mean ± SEM. P-values < 0.05 were considered to be significant.
Results
To define the mechanism of the association between the high activity SULT1A1*1 allele and overall survival in patients receiving tamoxifen, we investigated several hypotheses pertaining to putative biological activity of SO4-TAM, including affinity for the estrogen receptor, potential as a substrate or inhibitor for sulfatase, and capacity to induce apoptosis. Although it was expected that SO4-TAM, in common with sulfated estrogens, would possess no activity toward nuclear receptors, we tested this empirically. Recombinant ERα and ERβ were incubated with 3H-Estradiol and increasing concentrations of unlabeled estradiol, 4-OHT or SO4TAM as described in the Materials and Methods.
Results of these assays are summarized in Table 1. Under these experimental conditions, estradiol and 4-OHT bound the ERα receptor with equal affinities, consistent with previous reports [22], while 4-OHT binding affinity for ERβ was calculated as 40% that of estradiol. SO4TAM had very low or no affinity for either ERα or ERβ.
Table 1.
Interaction of tamoxifen metabolites with estrogen receptors and steroid sulfatase
| Estrogen Receptor Binding | Inhibition of Steroid Sulfatase Activity | ||||
|---|---|---|---|---|---|
| Ligand | * % Binding affinity for ERα (%, SD) | * % Binding affinity for ERβ (%, SD) | Incubation conditions | Estrone (pmol min-1 mg protein-1) | 4-OH-TAM (pmol min-1 mg protein-1) |
| Estradiol | 100 (ref) | 100 (ref) | Estrone-sulfate, no enzyme | nd | nd |
| 4-OH-TAM | 153.8 ± 12.3 | 59.2 ± 4.7 | Estrone-sulfate + HLM | 123.1 ± 7.7 | nd |
| SO4-TAM | 1.8 ± 0.4 | nd | Estrone-sulfate + HLM + 100 μM SO4-TAM | 118 ± 15.6 | nd |
percent compared to estradiol; SD from 3 independent measurements.
nd = not detectable
Sulfated compounds have been shown to inhibit the activity of sulfatase, an enzyme that contributes to the steroid pool in tumors by deconjugating circulating sulfated steroids. Reaction mixtures composed of hepatic microsomal fractions were incubated with estrone-sulfate with or without the addition of SO4-TAM and analyzed by HPLC. Incubations with estrone-sulfate resulted in substantial regeneration of estrone (Table 1) however the addition of SO4-TAM up to 100 µM did not inhibit sulfatase activity. Likewise, no regeneration of 4-OHT was detected in these reactions, suggesting that SO4-TAM is not a substrate nor inhibitor of sulfatase activity.
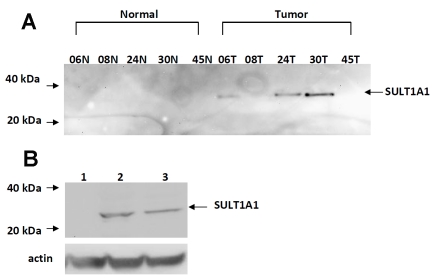
Since TAM-associated apoptosis has been reported in the literature, we also examined the capability of SO4-TAM to induce apoptosis in ER-dependent MCF7 cells. In the literature, it has been reported that SULT1A1 expression is low in normal breast epithelial compared to breast tumors [22]. Therefore, we screened for SULT1A1 expression in 5 matched specimens of infiltrating ductal carcinomas and their corresponding normal adjacent breast tissue by Western. As seen in Figure 1A, SULT1A1 expression was detected using a polyclonal antiSULT1A1 antibody in breast tumors (60%), but was not detected in the normal breast tissue. MCF7 cells, however, endogenously express very little SULT1A1 and so to test whether SO4TAM induces apoptosis, we stably transfected SULT1A1 into this cell line (Figure 1B).
Figure 1.
Characterization of SULT1A1 expression in breast cancer tumors and cell lines. (A) Western blot analysis of SULT1A1 expression in matched specimens of infiltrating ductal carcinoma lysates (100 μg) and their corresponding normal breast tissue controls. (B) stably-transfected ER-dependent MCF7 cells, Lane 1, MCF7 pcDNA3 (50 μg), Lane 2, MCF7 SULT1A1 (50 μg), Lane 3, human liver cytosol (50 μg). Lysates were separated by SDS-polyacrylamide gel electrophoresis, blotted, and probed with anti-SULT1A1 antibody (1:1000) developed by Open Biosystems (Huntsviiie, AL) as described in the Materials & Methods.
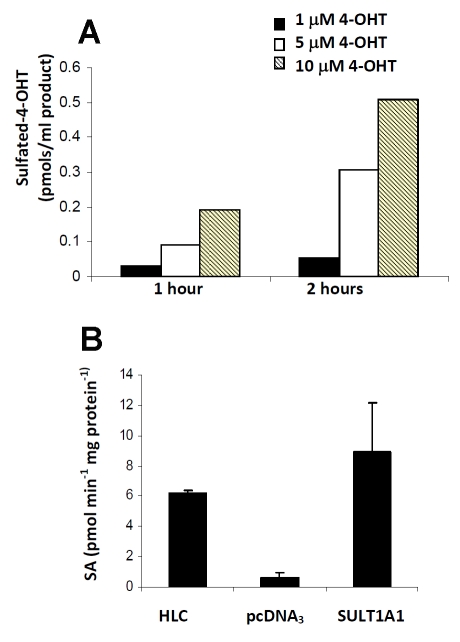
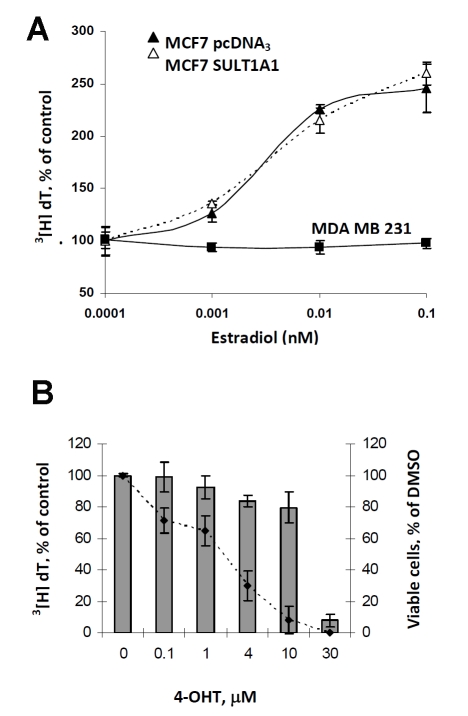
SULT1A1 is highly expressed in the liver [23], therefore, we sought to verify the capability of human hepatocytes to produce SO4-TAM on exposure to 4-OHT. In hepatocytes, we see an accumulation of SO4-TAM following short exposures to 4-OHTAM (Figure 2A). Cytosols prepared from hepatocytes are enzymatically active (6.23 nmol/mg/min) and activity in cytosols prepared from MCF7-SULT1A1 cells were comparable to liver cytosols (Figure 2B), and exhibited a 5-fold increase in activity compared to the MCF7 pcDNA3 cytosols. Thus, the level of SULT1A1 produced by MCF7-SULT1A1 was consistent with physiological levels found in human liver, strengthening the physiological relevance of our subsequent studies. SULT1A1 has little affinity for estradiol at physiological concentrations [8], so to further confirm that the expression of SULT1A1 was representative of physiological conditions, we examined whether SULT1A1 expression altered the estrogen-dependent cell proliferation of the MCF7 cells. As show in Figure 3A, increasing concentrations of estradiol (0-0.1 nM) added to estrogen-deprived MCF7 SULT1A1 cells stimulated cell growth at physiological concentrations of estradiol. At higher concentrations (µM), we observed inhibition of estrogen-dependent proliferation in the SULT1A1-expressing cells, consistent with in vitro experiments at non-physiological concentrations [24], therefore the following series of experiments were conducted in estrogen-free media.
Figure 2.
SULT1A1 is responsible for sulfation of 4-OHT. (A) Following short exposures to 4-OHT (1-10 µM), SO4-TAM accumulates in primary human liver hepatocytes as measured by mass spectrometry. (B) SULT1A1 activity is also monitored indirectly through the synthesis of 2-napthylsulfate and the PAPS regeneration system that generates p-nitrophenol which can be quantified colorimetrically at 405 nm [20]. SULT1A1 activity in MCF7 SULT1A1 cells correlates with protein expression as determined by Western blot.
Figure 3.
Expression of SULT1A1 does not alter estrogen-stimulated growth in ER positive cells. (A) Estrogen deprived MCF7 pcDNA3 (▴) and MCF7 SULT1A1 expressing cells (Δ) were treated with increasing concentrations of estradiol for 24 hours and the cells were labeled using 3[H]-thymidine. For control purposes, the ER-independent cell line MDA MB 231 (▪) was also treated with increasing concentrations of estradiol. (B) Estrogen-deprived MCF7 cells were treated with increasing concentrations of 4-OHT for 24 hours. Antiproliferation was measured by 3[H]-thymidine and plotted as a line-graph from the left y-axis. Cytotoxicity was also measured and plotted in a bar graph. In both experiments, samples were assayed in triplicate and expressed as a percentage of the vehicle control (DMSO) which was set at 100%.
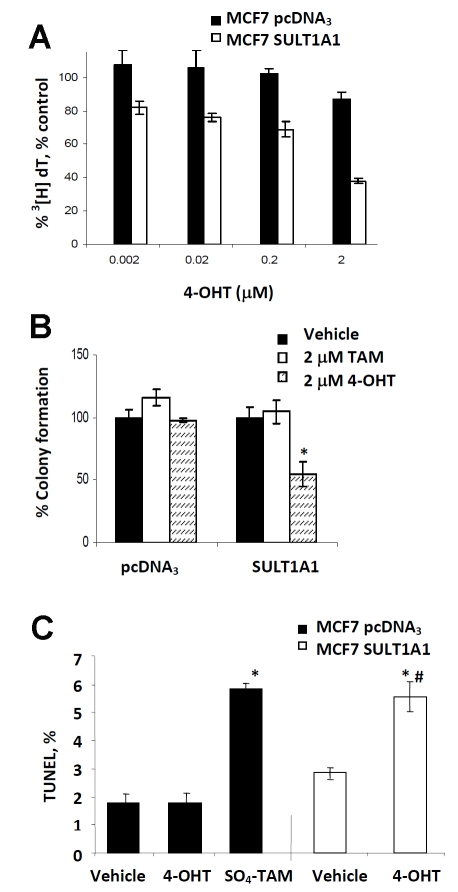
In ER-dependent breast cancer cell lines, 4-OHT treatment can result in an antiproliferative effect that is either cytostatic or cytotoxic; both are dependent on drug concentration and duration of treatment [25]. Therefore, we tested the effect of 4-OHT in our parent MCF7 cells. Estro-gen-deprived MCF7 cells were treated with increasing concentrations of 4-OHT for 24 hours at which time cell growth inhibition was measured by 3[H]-thymidine incorporation and cell viability measured by crystal violet. As seen in Figure 3B, we observed a steady decrease in proliferation with increasing concentrations of drug. At 10 µM, we observed ≥ 90% inhibition of cell growth, however, this antiproliferative effect did not correspond to cytotoxicity which was less than 20% at 10 µM of 4-OHT. Based on these observations, we treated estrogen deprived MCF7 pcDNA3 and SULT1A1 expressing cells with a low range of 4-OHT concentrations, which experimentally should have little effect on cell growth and measured growth inhibition by 3[H]-thymidine incorporation. As expected, in Figure 4A, we did not see substantial antiproliferative effect of 4-OHT treatment in the pcDNA3 cells at 24 hours, but there was a steady decrease in cell proliferation with increasing 4-OHT concentrations in the SULT1A1 expressing cells; this decreaseranged from 25-55% compared to the pcDNA3 expressing cells (p<0.05). In Figure 4B, at the highest concentration (2 µM), 4-OHT-mediated growth arrest leads to cell death as measured by colony formation. MCF7 pcDNA3 and SULT1A1 expressing cells were treated with 4-OHT for 48 hours, then serially diluted into drug-free complete media. Following 5 days of growth, we observed a significant decrease in the colonies formed in the 4-OHT-treated SULT1A1 cells compared to similarly treated pcDNA3 expressing cells (p<0.05). No observable difference was seen between pcDNA3 and SULT1A1 expressing cells following treatment with the parent compound, tamoxifen, which is not sulfated by SULT1A1, thereby supporting the observation that hydroxylation of the parent compound is necessary for SULT1A1 to exert an effect.
Figure 4.
SULT1A1 expression enhances 4-OHT anti-proliferative effects and cytotoxicity in estrogen-deprived MCF7 cells. (A) In the MCF7 SULT1A1 (□) expressing cells, 4-OHT mediated cell cycle arrest was greater for all concentrations (0-2 µM), *p < 0.05, at 24 hours, compared to the MCF7 pcDNA3 cells (▪). (B) The cytotoxic effect of 4-OHT treatment was measured in MCF7 pcDNA3 or SULT1A1 expressing cells by colony formation. Following 5 days of growth of 4-OHT treated SULT1A1 expressing cells was significantly hindered compared similarly treated pcDNA3 control cells, *p<0.05. (C) Apoptosis is also observed in MCF7 SULT1A1 expressing cells treated with 4-OHT (10 µM) for 24 hours compared to similarly treated pcDNA3 control cells, *p<0.05 compared to the appropriate vehicle; #p<0.05 compared to pcDNA3 expressing cells. TUNEL assay was performed as described in Materials and Methods
It has been established in the literature that micromolar concentrations of 4-OHT induce apoptosis in ER-dependent and ER-independent breast cancer cell lines [25, 26]. Based on the results in Figure 3B, pcDNA3 and SULT1A1 expressing cells were treated with 10 µM 4-OHT for 24 hours, a time point at which the ER-mediated 4-OHT apoptotic response would not factor into the apoptotic response associated with the accumulation of SO4-TAM in the SULT1A1 expressing cells, then measured apoptosis by TUNEL. At 24 hours, there was no observable increase in DNA fragmentation between the vehicle-treated and 4-OHT-treated MCF7 pcDNA3 cells (Figure 4C). However, in the MCF7 SULT1A1 expressing cells, we observed an 87% increase in apoptosis as measured by TUNEL following 4-OHT treatment compared to its vehicle control (p<0.05). MCF7 pcDNA3 cells treated with synthetic SO4TAM (10 µM) also induced apoptosis by DNA fragmentation (Figure 4C). Given the results in Figure 4A, it would be reasonable to hypothesize an additive or synergistic effect of both 4-OHT and SO4-TAM in the SULT1A1 expressing cells as measured by apoptosis (TUNEL), however, the MCF7 cells are caspase-3 deficient [27], resulting in a lower apoptotic response compared to cell lines with caspase-3 expression, which may effect our ability to observe an effect between the two metabolites. Although 4-OHT’s primary mechanism of action is ER-mediated, other ER-independent mechanisms have been reported via several different apoptotic pathways [22, 24]. At this time, it is not clear whether apoptosis observed for SO4-TAM- treated pcDNA3 cells and 4-OHT-treated MCF7-SULT1A1 cells share a common apoptotic pathway.
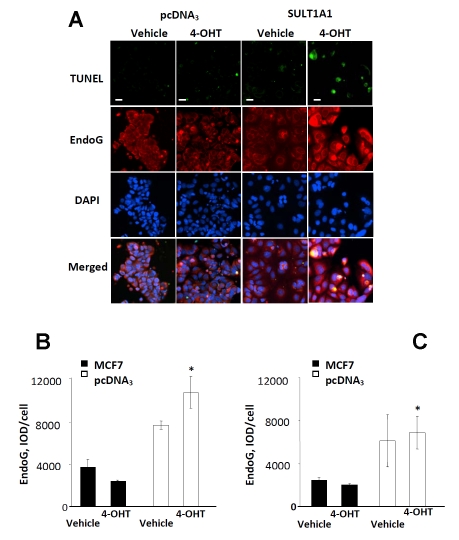
Finally, the effect of SULT1A1 expression on the expression of EndoG, the endonuclease that was previously shown to be important in breast cancer cell apoptosis [14] was examined (Figure 5A). Among TUNEL-positive SULT1A1 expressing cells, we observed a 4-fold increase in EndoG expression (p<0.05) compared to the TUNEL-positive pcDNA3 expressing cells treated with 4OHT (Figure 5B) and a trend for increased EndoG expression, which did not reach statistical significance in when compared to its vehicle control. In the total cell population, our data was similar to findings in TUNEL-positive cells, but had lower values and higher deviations due to the strong cellular diversity (Figure 5C).
Figure 5.
EndoG expression increases in 4-OHT treated SULT1A1 expressing cells, representative images (A). EndoG protein expression in apoptotic subpopulation (B) and entire population (C) of pcDNA3- and SULT1A1-expressing MCF7 cells treated with vehicle or 4-OHT for 24 hours. Immunocytochemistry and TUNEL assay were performed as described in Materials and Methods. n=8, #p<0.05 compared to pcDNA3 expressing cells. n=8, scale bar – 30 µm.
Discussion
The primary biological function of SULTs is to catalyze the transfer of the sulfonyl group from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) to the hydroxyl, sulfhydryl, amino or N-oxide groups of various substrates, thus increasing their solubility and facilitating their excretion. Of these, SULT1A1 is the most highly expressed hepatic sulfotransferase, and is widely distributed in other tissues, including breast tumor tissue [24]. Given that SULT1A1 is expressed in breast tumors (Figure 1A), but not in normal breast tissue and is the primary sulfotransferase responsible for sulfation of 4-OHT, it is plausible that changes in SULT1A1 activity may influence therapeutic outcome. In a retrospective study, we were the first to identify an association between the high activity allele of SULT1A1 and overall survival of breast cancer patients receiving tamoxifen [12]. This result was counter to our a priori hypothesis since the direction of the response favored patients who rapidly metabolized the active metabolite (4OHT) into what was believed to be an inactive sulfated form. Yet, subsequent studies have supported our initial observation [13]. Therefore, we investigated the possibility that SO4TAM is biologically active and contribute to the overall efficacy of tamoxifen therapy.
Sulfated steroids are generally considered to be receptor-inactive. Therefore, it was not surprising that SO4-TAM has little affinity for either ERα or ERβ (Table 1). We also concluded that SO4TAM is not a steroid sulfatase inhibitor and does not reduce the levels of synthesized estrogen in mammary tissues [28, 29]. SO4-TAM is not a substrate for sulfatase, which would provide a reservoir of circulating SO4-TAM for the regeneration of the active metabolite, 4-OHT in breast tissues, similar to the regeneration of dehydroepiandrosterone (DHEA) from DHEA-S [30]. However by introducing SULT1A1 into MCF7 cells, the antiproliferative and cytotoxic response associated with 4-OHT treatment in breast cancer cell lines at intra-tumor concentrations was significantly improved [25, 31-33]. These findings demonstrated that SO4-TAM is biologically active within the tumor cell, mediating an apoptotic response that is dependent on SULT1A1 expression, and is associated with increased expression of EndoG, which to our knowledge has not been reported previously in 4-OHT-mediated apoptosis. EndoG belongs to a family of cell death endonucleases normally described as downstream effectors of apoptotic cascades, though capable of acting alone following activation or overexpression in cell lines [34, 35]. In several cancers, EndoG has been identified as a key enzyme in caspaseindependent apoptosis following treatment with different chemotherapeutic drugs [14-16]. In breast epithelium and breast cancer cell lines, EndoG is the major endonuclease expressed, and endogenous expression is higher in well-differentiated ER-positive cells [14].
Although the number of TUNEL-positive SULT1A1 expressing cells treated with vehicle was lower than treated with 4-OHT, the EndoG expression in TUNEL-positive SULT1A1 expressing cells was insignificantly elevated as compared to pcDNA3 expressing cells and was not statistically different from EndoG expression in TUNEL-positive SULT1A1 expressing cells. This observation may be explained by the universal role of EndoG at sites of DNA degradation and cell injury that was shown in our other models [14] [36]. Perhaps, the ER-mediated 4-OHT anti-proliferative effects observed in the pcDNA3 expressing cells is also capable of inducing EndoG and therefore could be an interest for future studies.
In the liver, cytochrome P450 enzymes, CYP 3A4/3A5 and CYP2D6 are responsible for producing the active tamoxifen metabolites, 4-OHT and endoxifen, that bind to the ER, thereby inhibiting estradiol-driven cellular proliferation in tumors, which is considered the primary mechanism of action of tamoxifen therapy. Recent studies have identified several CYP2D6 genetic polymorphisms (CYP 2D6*3, *4, *5, *10), and CYP2D6 inhibitors routinely prescribed to tamoxifen patients to relieve depression and hot-flashes that significantly reduce endoxifen plasma concentrations, limiting the effectiveness of tamoxifen therapy, and increasing the risk of recurrence [5, 6, 37]. While CYP2D6 is critical in the hepatic production of endoxifen, our data suggests that SULT1A1 expression and activity may be equally important in eliciting an apoptotic response in breast tumor cells.
In conclusion, we found a mechanism of action that corroborates our report and other reports that high activity SULT1A1*1 allele is beneficial to patients receiving tamoxifen [12, 13]. Based on these data, it appears that SULT1A1 expression in breast tumor cells enhances the existing antiproliferative/apoptotic responses of ta-moxifen’s active metabolite, 4-OHT, and reveals a unique role for SULT1A1 in patient response and efficacy of tamoxifen therapy.
Acknowledgments
This work was supported in part by a grant from Susan G. Komen for the Cure BCTR0707584 and R01CA128897, and by a research grant from Fundação para a Ciência e a Technologia (FCT), Portugal and FEDER (PPCDT/QUI? 57110/2004). We thank Nathan Twaddle for his assistance in analyzing sulfated-4-hydroxytamoxifen by mass spectrometry.
References
- 1.Love RR, Mazess RB, Barden HS, Epstein S, Newcomb PA, Jordan VC, Carbone PP, De-Mets DL. Effects of tamoxifen on bone mineral density in postmenopausal women with breast cancer. N Engl J Med. 1992;326:852–856. doi: 10.1056/NEJM199203263261302. [DOI] [PubMed] [Google Scholar]
- 2.Cuzick J, Powles T, Veronesi U, Forbes J, Edwards R, Ashley S, Boyle P. Overview of the main outcomes in breast-cancer prevention trials. Lancet. 2003;361:296–300. doi: 10.1016/S0140-6736(03)12342-2. [DOI] [PubMed] [Google Scholar]
- 3.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM. Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol. 2003;21:1973–1979. doi: 10.1200/JCO.2003.09.099. [DOI] [PubMed] [Google Scholar]
- 5.Borges HL, Linden R. Gamma irradiation leads to two waves of apoptosis in distinct cell populations of the retina of newborn rats. J Cell Sci. 1999;112(Pt 23):4315–4324. doi: 10.1242/jcs.112.23.4315. [DOI] [PubMed] [Google Scholar]
- 6.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 7.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 8.Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 9.Adjei AA, Weinshilboum RM. Catecholestrogen sulfation: possible role in carcinogenesis. Biochem Biophys Res Commun. 2002;292:402–408. doi: 10.1006/bbrc.2002.6658. [DOI] [PubMed] [Google Scholar]
- 10.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 11.Ozawa S, Tang YM, Yamazoe Y, Kato R, Lang NP, Kadlubar FF. Genetic polymorphisms in human liver phenol sulfotransferases involved in the bioactivation of N-hydroxy derivatives of carcinogenic arylamines and heterocyclic amines. Chem Biol Interact. 1998;109:237–248. doi: 10.1016/s0009-2797(97)00135-x. [DOI] [PubMed] [Google Scholar]
- 12.Nowell S, Sweeney C, Winters M, Stone A, Lang NP, Hutchins LF, Kadlubar FF, Ambrosone CB. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J Natl Cancer Inst. 2002;94:1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 13.Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist LE, Wingren S. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basnakian AG, Apostolov EO, Yin X, Abiri SO, Stewart AG, Singh AB, Shah SV. Endonuclease G promotes cell death of non-invasive human breast cancer cells. Exp Cell Res. 2006;312:4139–4149. doi: 10.1016/j.yexcr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JS, Lee JH, Jeong WW, Choi DH, Cha HJ, Kim do H, Kwon JK, Park SE, Park JH, Cho HR, Lee SH, Park SK, Lee BJ, Min YJ, Park JW. Reactive oxygen species-dependent EndoG release mediates cisplatin-induced caspaseindependent apoptosis in human head and neck squamous carcinoma cells. Int J Cancer. 2008;122:672–680. doi: 10.1002/ijc.23158. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Tryndyak V, Apostolov EO, Yin X, Shah SV, Pogribny IP, Basnakian AG. Sensitivity of human prostate cancer cells to chemotherapeutic drugs depends on EndoG expression regulated by promoter methylation. Cancer Lett. 2008;270:132–143. doi: 10.1016/j.canlet.2008.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beland FA, Churchwell MI, Hewer A, Phillips DH, da Costa GG, Marques MM. Analysis of ta-moxifen-DNA adducts in endometrial explants by MS and 32P-postlabeling. Biochem Biophys Res Commun. 2004;320:297–302. doi: 10.1016/j.bbrc.2004.05.168. [DOI] [PubMed] [Google Scholar]
- 18.Blair RM, Fang H, Branham WS, Hass BS, Dial SL, Moland CL, Tong W, Shi L, Perkins R, Sheehan DM. The estrogen receptor relative binding affinities of 188 natural and xenochemicals: structural diversity of ligands. Toxicol Sci. 2000;54:138–153. doi: 10.1093/toxsci/54.1.138. [DOI] [PubMed] [Google Scholar]
- 19.Tolleson WH, Doerge DR, Churchwell MI, Marques MM, Roberts DW. Metabolism of biochanin A and formononetin by human liver microsomes in vitro. J Agric Food Chem. 2002;50:4783–4790. doi: 10.1021/jf025549r. [DOI] [PubMed] [Google Scholar]
- 20.Frame LT, Ozawa S, Nowell SA, Chou HC, DeLongchamp RR, Doerge DR, Lang NP, Kadlubar FF. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab Dispos. 2000;28:1063–1068. [PubMed] [Google Scholar]
- 21.Williams LD, Twaddle NC, Churchwell MI, Doerge DR. Quantification of tamoxifen and metabolites and soy isoflavones in human plasma using liquid chromatography with electrospray ionization tandem mass spectrometry. J AOAC Int. 2006;89:1168–1173. [PubMed] [Google Scholar]
- 22.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 23.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- 24.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56:1551–1555. [PubMed] [Google Scholar]
- 25.Mandlekar S, Kong AN. Mechanisms of tamoxifen-induced apoptosis. Apoptosis. 2001;6:469–477. doi: 10.1023/a:1012437607881. [DOI] [PubMed] [Google Scholar]
- 26.Obrero M, Yu DV, Shapiro DJ. Estrogen receptor-dependent and estrogen receptor-independent pathways for tamoxifen and 4-hydroxytamoxifen-induced programmed cell death. J Biol Chem. 2002;277:45695–45703. doi: 10.1074/jbc.M208092200. [DOI] [PubMed] [Google Scholar]
- 27.Janicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 28.Numazawa M, Tominaga T, Watari Y, Tada Y. Inhibition of estrone sulfatase by aromatase inhibitor-based estrogen 3-sulfamates. Steroids. 2006;71:371–379. doi: 10.1016/j.steroids.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Purohit A, Williams GJ, Howarth NM, Potter BV, Reed MJ. Inactivation of steroid sulfatase by an active site-directed inhibitor, estrone-3-O-sulfamate. Biochemistry. 1995;34:11508–11514. doi: 10.1021/bi00036a025. [DOI] [PubMed] [Google Scholar]
- 30.Longcope C. Metabolism of dehydroepiandrosterone. Ann N Y Acad Sci. 1995;774:143–148. doi: 10.1111/j.1749-6632.1995.tb17378.x. [DOI] [PubMed] [Google Scholar]
- 31.Daniel P, Gaskell SJ, Bishop H, Campbell C, Nicholson RI. Determination of tamoxifen and biologically active metabolites in human breast tumours and plasma. Eur J Cancer Clin Oncol. 1981;17:1183–1189. [PubMed] [Google Scholar]
- 32.Lien EA, Wester K, Lonning PE, Solheim E, Ueland PM. Distribution of tamoxifen and metabolites into brain tissue and brain metastases in breast cancer patients. Br J Cancer. 1991;63:641–645. doi: 10.1038/bjc.1991.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacCallum J, Cummings J, Dixon JM, Miller WR. Concentrations of tamoxifen and its major metabolites in hormone responsive and resistant breast tumours. Br J Cancer. 2000;82:1629–1635. doi: 10.1054/bjoc.2000.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieser RJ, Eastman A. The cloning and expression of human deoxyribonuclease II. A possible role in apoptosis. J Biol Chem. 1998;273:30909–30914. doi: 10.1074/jbc.273.47.30909. [DOI] [PubMed] [Google Scholar]
- 35.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 36.Yin X, Apostolov EO, Shah SV, Wang X, Bogdanov KV, Buzder T, Stewart AG, Basnakian AG. Induction of renal endonuclease G by cisplatin is reduced in DNase I-deficient mice. J Am Soc Nephrol. 2007;18:2544–2553. doi: 10.1681/ASN.2006080896. [DOI] [PubMed] [Google Scholar]
- 37.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]