Abstract
Since the identification of two functional polymorphisms in pre-miRNAs ﹛miR-146a rs2910164 and miR-196a2 rsll614913), a number of studies were published in the past several years to evaluate the associations between the two SNPs and cancer risk. However, the findings remain conflicting rather than conclusive. This meta-analysis included 12 published case-control studies, 5,916 cancer patients and 6,869 control subjects for SNP rs2910164, and 6,574 cases and 7,601 controls for SNP rsll614913. The allele C of rsll614913 was associated with a significantly increased risk of overall cancers (CC vs. TT: OR=1.18; 95%CI: 1.02-1.37; CT vs. TT: OR=1.11; 95% CI: 1.02-1.21; CC/CT vs. TT: OR=1.13; 95%CI: 1.04-1.23). However, we failed to find main effects for rs2910164 on overall cancer risk in different genetic models tested, while significantly increased risk was evident among cancers other than breast (GG/GC vs. CC: OR=1.30, 95%CI: 1.03-1.64; between study heterogeneity test: P = 0.007). The two functional SNPs may represent tissue specific effect on cancer susceptibility; however, additional well-designed large studies are required for the validation of the associations.
Keywords: miRNA, polymorphism, cancer, susceptibility, meta-analysis
Introduction
Lots of efforts have been made in the field of cancer research and enormous genes were found to be involved in carcinogenesis. Recently, the addition of noncoding RNAs to cancer etiopathogenesis has provided stimulus for further researches. MicroRNAs (miRNAs) are about 20-nucleotide-long small noncoding RNAs, which control gene activity and affect the expression of proteins by base pairing with target mRNAs at the 3'-untranslated regions (3'UTR), leading to mRNA cleavage or transla-tional repression [1-3]. It has been proposed that miRNAs are involved in various biological processes [4], and the information obtained to date has suggested that miRNA may play a vital important role in carcinogenesis [5-9]. Genes encoding miRNAs located in chromosomal regions that are amplified in cancers can function as oncogenes, while those in regions deleted in cancers may act as tumor suppressors [9-11]. Newly published evidence has also shown that a global reduction in miRNA processing was cancer prone, and miRNA profiling have been successfully applied to classify tumors [10-13].
It has been reported that SNPs (single nucleo-tide polymorphism) or mutations located in miRNA regions might change the processing of the miRNA as well as alter the target binding affinity and specificity [14]. In 2008, Jazdzewski etal. found that the SNP rs2910164 in pre-miR-146a was associated with mature miR-146a expression and differently influenced target report gene levels [15]. Likewise, Xu etal. showed rs2910164GG conferred a higher expression level of mature miR-146a by in vitro cell model compared with the CC genotype [16]. However, in the contrary, Shen et al. suggested that breast/ovarian cancer patients carrying the C allele of rs2910164 had high levels of mature miR-146 than the G allele [17]. For rsll614913 in miR-196a2, in a genotype-phenotype correlation analysis of lung cancer tissues, homozygote rsll614913CC was associated with a significant increased mature miR-196a expression but without changes in the levels of its precursor [18]. The same study also revealed that rsll614913 could affect the binding of mature miR-196a2-3p to its target mRNA [18]. Furthermore, Hoffman et al. reported similar evidence that the risky miR-196a2-C allele led to more efficient processing of the miRNA precursor to its mature form as well as enhanced capacity to regulate target genes [19].
For the past several years, emerging molecular epidemiological studies studied the associations of the two functional SNPs ﹛miR-146a rs2910164 and miR-196a2 rsll614913) and susceptibility of diverse cancers in different populations. The tumor types in the case populations included lung, breast, prostate, bladder, renal, gastric, glioma, thyroid, hepatocellular and esophageal [15,16,19-29]. However, these published studies presented contradicting findings. Considering the amount of accumulated published data are available, it is necessary for us to carry out a systematic review and meta-analysis to assess the overall tumor risk associated with SNPs rs2910164 and rsll614913 and to quantify the potential between-study heterogeneity.
Materials and methods
Identification and eligibility of relevant studies
We have attempted to include all the case-control studies published to date on cancers with genotyping data for at least one of the two SNPs ﹛miR-146a rs2910164 and miR-196a2 rsll614913). In order to obtain all possible articles we need, we searched the electronic literature PubMed for relevant reports (last search update April 20th, 2010, using the search terms “miRNA or microRNA and cancer and polymorphism”) by two independent investigators (T.T. and Y.X.) and also did hand search from the references of related articles. Overall, we obtained thirteen published articles focused on the relationship between the two SNPs and tumor risk. However, one study was excluded because of the lack of detailed genotyping information [23]. Finally, the data for this analysis were available from twelve case-control studies, including 5,916 cancer cases and 6,869 controls for miR-146a rs2910164 (from 8 studies), and 6,574 cancer cases and 7,601 controls for miR-196a2 rsll614913 (from 9 studies).
Data Extraction
Two investigators independently extracted data and reached consensus on all of the items. Data collected from these articles included the first author's name, year of publication, country of origin,ethnicity, type of cancer, number of cases and controls, genotype frequencies for cases and controls, characteristics of cancer cases and controls, and racial descent.
Statistical-analysis
We used the fixed-effect model and the random-effect model based on the Mantel-Haenszel method and the DerSimonian-Laird method, respectively, to combine values from single study [30]. When the effects were assumed to be homogenous, the fixed -effects model was used; otherwise, the random-effects model was more appropriate. Statistical heterogeneity was assessed with the x2-based Q test, and the heterogeneity was considered significant when P < 0.1 [31]. Subgroup analyses were processed, according to tumor type [categorized as breast cancer and other cancers (only breast cancer has more than two published studies)] and ethnicity (categorized as Asian and Caucasian descents). The inter-study variance (t2) was used to quantify the degree of heterogeneity between studies and the percentage of t2 was used to describe the extent of explained heterogeneity [31]. Egger's test was utilized to provide diagnosis of publication bias (Linear regression analysis [32]). All analyses were done by using the Statistical Analysis System software (v.9.1.3; SAS Institute, Cary, NC, USA) and STATA7.0 (Stata-Corp, College Station, TX, USA). All the P values were two-sided.
Results
Characteristics of the studies
We established a database according to the extracted information from each study (Table 1). There are totally 8 case-control studies concerning SNP rs2910164, and 9 for SNP rsll614913. All studies indicated that the distribution of genotypes in the controls was consistent with Hardy-Weinberg equilibrium, except for one study for rs2910164 [28] and one study for rsll614913 [26]. However, none of the comparisons had the statistical power greater than 80% when we assumed an allelic risk was 1.2 forthetwoSNPs.
Table 1.
Characteristics of literatures included in the meta-analysis
| Author | Year | Origin | Ethnicity | Tumor type | Sample size (case/control) | HWE | MAFin controls | Powerb |
|---|---|---|---|---|---|---|---|---|
| Horikawa | 2008 | American | Caucasian | Renal cell cancer | 276/277(261/235) | 0.024(0.648) | 0.576(0.736) | 0.182(0.139) |
| Jazdzewski | 2008aa | Finland | Caucasian | Papillary thyroid cancer | (206/274) | (0.915) | (0.739) | (0.133) |
| Jazdzewski | 2008ba | Poland | Caucasian | Papillary thyroid cancer | (201/475) | (0.661) | (0.774) | (0.141) |
| Jazdzewski | 2008ca | American | Caucasian | Papillary thyroid cancer | (201/152) | (0.508) | (0.763) | (0.104) |
| Xu | 2008 | Chinese | Asian | Hepatocellular cancer | (479/504) | (0.119) | (0.362) | (0.284) |
| Yang | 2008 | American | Caucasian | Bladder cancer | 736/731(691/674) | 0.329(0.137) | 0.586(0.763) | 0.399(0.287) |
| Hoffman | 2009 | American | Caucasian | Breast cancer | 426/466 | 0.583 | 0.602 | 0.260 |
| Hu | 2009 | Chinese | Asian | Breast cancer | 1009/1093(1009/1093) | 0.207(0.221) | 0.436(0.417) | 0.548(0.545) |
| Peng | 2009 | Chinese | Asian | Gastric cancer | 213/213 | 0.936 | 0.514 | 0.153 |
| Tian | 2009 | Chinese | Asian | Lung cancer | 1058/1035(1058/1035) | 0.700(0.853) | 0.453(0.406) | 0.549(0.541) |
| Catucci | 2010aa | German | Caucasian | Breast cancer | 1101/1496(805/904) | 0.711(0.753) | 0.623(0.769) | 0.594(0.339) |
| Catucci | 2010ba | Italian | Caucasian | Breast cancer | 751/1243(754/1243) | 0.315(0.019) | 0.649(0.732) | 0.458(0.402) |
| Dou | 2010 | Chinese | Asian | Glioma | 643/656 | 0.119 | 0.451 | 0.374 |
| Qi | 2010 | Chinese | Asian | Hepatocellular cancer | 361/391 | 0.869 | 0.487 | 0.238 |
| Xu | 2010 | Chinese | Asian | Prostate cancer | (251/280) | (0.191) | (0.461) | (0.181) |
Different populations from one study.
Power was calculated by the DSTPLAN4.2 software
NOTE: The data given in parenthesis were related to rs2910164, others were related to rs11614913.
Quantitative synthesis
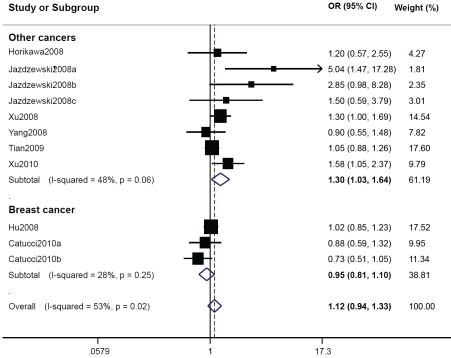
MiR-146a rs2910164. There was a distinct variation in the allele frequency of rs2910164G across different ethnicities, ranging from 0.362 in an Asian population [16] to 0.774 in a Caucasian population [15]. The mean frequency of rs2910164G was 0.408 for Asians and 0.753 for Caucasians. In the meta-analysis, we estimated the risks of genotypes GG and GC, compared with CC, respectively, and then evaluated the risk of GG/GC versus CC. However, we failed to find significant main effects for rs2910164 on cancer risk in all genetic models tested (Table 2). In the stratified analyses, significantly increased risk was found among Asians by comparing GG/GC with CC (OR=1.12; 95% CI: 1.00-1.25; P = 0.15 for heterogeneity test) and in cancer patients other than breast cancer in all tested genetic models (GG vs. CC: OR=1.44; 95% CI: 1.10-1.89; P = 0.09 for heterogeneity test; GC vs. CC: OR=1.31; 95% CI: 1.00-1.72; P = 0.02 for heterogeneity test; and GG/GC vs. CC: OR=1.30; 95% CI: 1.03-1.64; P = 0.06 for heterogeneity test) (Table 2, Figure 1).
Table 2.
Associations of the two polymorphisms and cancer risk
| mir-146a rs2910164 | mir-196a2 rs11614913 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | OR(95%CI) | ||||||||||||||
| No. of Comparisons | GG VS. CC | P | GCVS. CC | P | GG/GCVS. CC | P | No. of Comparisons | CCVS.TT | P | CT VS. TT | P | CC/CT VS. TT | P | ||
| Total | F | 11 | 1.14(1.00-1.29) | 0.048 | 1.07(0.97-1.19) | 0.184 | 1.09(0.99-1.20) | 0.082 | 10 | 1.17(1.06-1.29) | 0.002 | 1.11(1.02-1.21) | 0.018 | 1.13(1.04-1.23) | 0.003 |
| R | 11 | 1.19(0.96-1.48) | 0.109 | 1.12(0.92-1.35) | 0.256 | 1.12(0.94-1.33) | 0.193 | 10 | 1.18(1.02-1.37) | 0.027 | 1.11(1.02-1.22) | 0.021 | 1.13(1.03-1.25) | 0.010 | |
| Ethnicity | |||||||||||||||
| Asian | F | 4 | 1.22(1.04-1.43) | 0.013 | 1.08(0.96-1.22) | 0.174 | 1.12(1.00-1.25) | 0.053 | 5 | 1.18(1.03-1.35) | 0.018 | 1.10(0.98-1.23) | 0.096 | 1.13(1.01-1.25) | 0.027 |
| R | 4 | 1.33(0.99-1.77) | 0.056 | 1.08(0.97-1.22) | 0.176 | 1.15(0.98-1.34) | 0.088 | 5 | 1.16(0.94-1.43) | 0.175 | 1.10(0.98-1.23) | 0.096 | 1.13(1.01-1.25) | 0.027 | |
| Caucasian | 7 | 1.00(0.81-1.23) | 0.966 | 1.03(0.83-1.28) | 0.776 | 1.01(0.82-1.24) | 0.934 | 5 | 1.15(1.00-1.33) | 0.047 | 1.12(0.98-1.29) | 0.087 | 1.14(1.00-1.30) | 0.051 | |
| R | 7 | 1.08(0.78-1.50) | 0.643 | 1.30(0.83-2.05) | 0.256 | 1.17(0.80-1.70) | 0.418 | 5 | 1.20(0.96-1.51) | 0.102 | 1.16(0.96-1.40) | 0.116 | 1.19(0.97-1.46) | 0.095 | |
| Tumor type | |||||||||||||||
| Breast | F | 3 | 0.91(0.75-1.10) | 0.311 | 0.95(0.81-1.11) | 0.493 | 0.95(0.81-1.10) | 0.471 | 4 | 1.25(1.09-1.43) | 0.002 | 1.15(1.01-1.31) | 0.033 | 1.19(1.05-1.34) | 0.005 |
| R | 3 | 0.91(0.75-1.09) | 0.307 | 0.90(0.70-1.16) | 0.424 | 0.92(0.75-1.12) | 0.399 | 4 | 1.30(1.01-1.68) | 0.041 | 1.17(0.98-1.40) | 0.086 | 1.22(1.00-1.50) | 0.053 | |
| Othera | F | 8 | 1.36(1.15-.161) | 0.000 | 1.17(1.03-1.34) | 0.020 | 1.21(1.06-1.37) | 0.004 | 6 | 1.09(0.95-1.26) | 0.204 | 1.08(0.96-1.22) | 0.206 | 1.09(0.97-1.21) | 0.151 |
| R | 8 | 1.44(1.10-1.89) | 0.008 | 1.31(1.00-1.72) | 0.047 | 1.30(1.03-1.64) | 0.024 | 6 | 1.09(0.92-1.31) | 0.324 | 1.08(0.96-1.22) | 0.208 | 1.09(0.97-1.21) | 0.152 | |
F: Fixed-effects model, R: Random-effects model;
Including cancers of renal, thyroid, bladder, lung, hepatocellular, and prostate for SNP miR-146a rs2910164; and cancers of glioma, bladder, renal, gastric, hepatocellular, and lung for SNP miR-196a2 rs11614913
Figure 1.
ORs (log scale) of different cancers associated with rs2910164 for the GG/GC genotypes compared with the CC genotype.
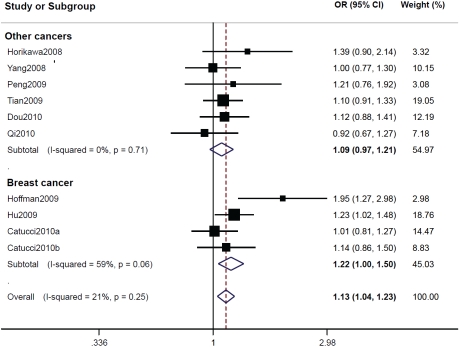
MiR-196a2 rsll614913. Similarly, there was a distinct variation in the allele frequency of rsll614913C across different ethnicities, ranging from 0.436 in an Asian population [20] to 0.649 in a Caucasian population [28]. The mean frequency of rsll614913C was 0.619 for Caucasians and 0.455 for Asians. When all the eligible studies were pooled, the genotypes including rsll614913C allele were associated with increased tumor risk in all genetic models tested (CC vs. TT: OR=1.18; 95% CI: 1.02-1.37; P = 0.03 for heterogeneity test; CT vs. TT: OR=1.11; 95% CI: 1.02-1.21; P = 0.41 for heterogeneity test; and CC/CT vs. TT: OR=1.13; 95% CI, 1.04-1.23; P = 0.25 for heterogeneity test; Table 2). In the subgroup analyses, we reached significantly elevated risks associated with the rsll614913C harboring genotypes for breast cancer in all models tested (CC vs. TT: OR=1.30; 95% CI: 1.01-1.68; P = 0.03 for heterogeneity test; CT vs. TT: OR=1.15; 95% CI: 1.01-1.31; P = 0.16 for heterogeneity test; and CC/CT vs. TT: OR=1.22; 95% CI: 1.00-1.50; P = 0.06 for heterogeneity test), while significantly increased risk was found among Asians by comparing CC/CT with TT (OR = 1.13; 95% CI, 1.01-1.25; P = 0.66 for heterogeneity test) (Table 2, Figure 2).
Figure 2.
ORs (log scale) of different cancers associated with rs11614913 for the CC/CT genotypes compared with the TT genotype.
Test of between subgroup heterogeneity and source of between study heterogeneity
We also tested between subgroup heterogeneity and evaluated the source of between study heterogeneity for rs2910164 (GG versus CC) and rsll614913 (CC versus TT) by tumor type and ethnicity. We found that tumor type (x2 = 7.36, df = 1, P = 0.007) do contribute to substantial altered between subgroup heterogeneity, but not the ethnicity (x2 = 0.87, df = 1, P = 0.351) for SNP rs2910164. Furthermore, meta-regression analyses revealed that tumor type can explain 65.5% (P = 0.035) of the t2, whereas ethnicity cannot explain significant between study heterogeneity (P = 0.397). For rsll614913, neither tumor type (x2 = 1.41, df = 1, P = 0.235) nor ethnicity (x2 = 0.05, df = 1, P = 0.830) contributed substantially to the between subgroup heterogeneity. Meta-regression analyses also revealed that none of these two factors could explain significant between study heterogeneity (tumor type: P = 0.342; ethnicity: P = 0.786). Similar results were not shown for other genetic models tested (GC VS.CC and GG/ GC VS.CC for rs2910164 and CT VS.TT and CC/ CT VS. TT for rsll614913).
Publication bias
We used Egger's test to access the publication bias of literatures. The result of Egger's test did not show any statistically significant evidence for publication bias for the two SNPs (rs2910164: t = 1.58, P = 0.15 for GG/GC vs. CC and rsll614913: t = 1.13, P = 0.29 for CC/ CT vs.TT).
Discussion
Several studies have reported that the sequence variations in pre-miRNA may affect the maturation process of miRNAs and binding activity to their target mRNAs, including rs2910164 and rsll614913 [15-19, 29]. Jaz-dzewski et al. proposed the allele C of rs2910164 decreased pri-miR-146a nuclear processing efficiency, reduced production of mature miRNA and resulted in less efficient inhibition of the target genes including TRAF6, IRAKI and PTC1 [15]. Other studies also showed the SNP rs2910164C allele was related to decreased production of mature miR-146a 16, 29, but Shen et al. got a contrary result for breast/ovarian cancer patients [17]. Interestingly, our meta-analysis found a significant association for rs2910164 and cancer risk other than breast and at least an apparently controversial risk estimation between the two subgroups (P for between subgroup heterogeneity: 0.007). Therefore, we think rs2910164 may affect tumor susceptibility in a tissue specific way with yet unidentified mechanism.
The allele C of rsll614913 also influenced endogenous processing of the miRNA precursors to its mature form and affected the binding of mature miRNA to its target genes [18, 19]. Furthermore, a significant association of SNP rsll614913 with G2 cell cycle delay has been found, which was functionally verified that the SNP play a important role in tumorigenesis [19]. In our meta-analysis, we found consistently increased breast cancer risk for the rsll614913C allele. However, the associations with other cancers warrant further studies. In stratified analyses by race, the associations between the two SNPs and cancer risk were significant in Asian but not in Caucasians and the between subgroup heterogeneity was not evident. Interestingly, the risk alleles of the two SNPs were rare in Asians, but common in Caucasians. Therefore, these two SNPs may truly causative SNPs instead of tagging ones.
An increasing number of studies have also reported that miR-146a and miR-196a were associated with multiple kinds of malignant tumors. He et al. reported that miR-146a was up-regulated in patients with papillary thyroid carcinoma (PTC) [33]. Overexpression of miR-146a in PTC was also proved by one another study [15]. In addition, it has also been reported that miR-146a was highly expressed in pediatric acute leukemia [34]. Moreover, Pacifico et al. found that miR-146a was strongly up-regulated in human thyroid carcinoma FRO cells and the inhibition of miR-146a expression in FRO cells could decrease their oncogenic potential [35]. For miR-196a, which was up-regulated in breast cancer compared with normal breast [13], suggesting it might potentially act as oncogene in breast cancer. Highly elevated levels of miR-196a were also detected in other cancers, such as colorectal cancer [36] and esophageal ade-nocarcinoma [37]. Additionally, cell culture experiments supported high miR-196a levels could supress the activities of various cancer related genes, such as ANXA1 (annexin A1) [38], suppression of which was well documented in various cancers [39, 40].
In conclusion, considering the population evidence provided by this meta-analysis together with the functional evaluations on the two SNPs, we think it is clearer that the two SNPs ﹛miR-146a rs2910164 and miR-196a2 rs 11614913) play a crucial role in cancer susceptibility. The functional relevant of the SNPs and their related miRNAs may represent tissue specific effect, especially miR-146a rs2910164.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (30972541 and 30800946).
References
- 1.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl AcadSci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13ql4 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory Rl, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–12. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microR-NAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, lorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl AcadSci US A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.lorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 14.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 15.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 17.Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–6. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. MicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 21.Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, Ma H, Chen Y, Shen H. A functional genetic variant in microRNA- 196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA -related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–7. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 23.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–9. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi P, Dou TH, Geng L, Zhou FG, Gu X, Wang H, Gao CF. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010 doi: 10.1016/j.humimm.2010.02.017. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of MicroRNA-196a-2 Gene Polymorphism with Gastric Cancer Risk in a Chinese Population. Dig Dis Sci. 2009 doi: 10.1007/s10620-009-1007-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–62. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou T, Wu Q, Chen X, Ribas J, Ni X, Tang C, Huang F, Zhou L, Lu D. A polymorphism of mi-croRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010 doi: 10.1007/s00432-010-0844-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K, Sutter C, Wappen-schmidt B, Dick M, Arnold N, Bugert P, Niederacher D, Meindl A, Schmutzler RK, Bartram CC, Ficarazzi F, Tizzoni L, Zaffaroni D, Manoukian S, Barile M, Pierotti MA, Radice P, Burwinkel B, Peterlongo P. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance al-leles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31:1052–7. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 29.Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang ZD, Tong N, Wang JF, Song NH, Zhang W, Hua LX, Wu HF. A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate. 2010;70:467–72. doi: 10.1002/pros.21080. [DOI] [PubMed] [Google Scholar]
- 30.Petitti DB. New York: Oxford University Press; 1994. Meta-analysis, decision analysis, and cost-effectiveness analysis. [Google Scholar]
- 31.Lau J, loannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–6. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 32.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacifico F, Crescenzi E, Mellone S, lannetti A, Porrino N, Liguoro D, Moscato F, Grieco M, Formisano S, Leonardi A. Nuclear factor-{kappa} B contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab. 2010;95:1421–30. doi: 10.1210/jc.2009-1128. [DOI] [PubMed] [Google Scholar]
- 36.Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–96. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A, Luthra R. MicroRNA-196a is a potential marker of progression during Barrett's meta-plasia-dysplasia- invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregula-tion in cancers. Oncogene. 2008;27:6667–78. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 39.Kang JS, Calvo BF, Maygarden SJ, Caskey LS, Mohler JL, Ornstein DK. Dysregulation of annexin I protein expression in high-grade prostatic intra-epithelial neoplasia and prostate cancer. Clin Cancer Res. 2002;8:117–23. [PubMed] [Google Scholar]
- 40.Shen D, Nooraie F, Elshimali Y, Lonsberry V, He J, Bose S, Chia D, Seligson D, Chang HR, Goodglick L. Decreased expression of annexin A1 is correlated with breast cancer development and progression as determined by a tissue microar-ray analysis. Hum Pathol. 2006;37:1583–91. doi: 10.1016/j.humpath.2006.06.001. [DOI] [PubMed] [Google Scholar]