Abstract
A major objective in the treatment of Alzheimer's disease is amyloid plaque reduction. Transgenic mouse models of Alzheimer's disease provide a controlled and consistent environment for studying amyloid plaque deposition in Alzheimer's disease. Magnetic resonance imaging is an attractive tool for longitudinal studies because it offers non-invasive monitoring of amyloid plaques. Recent studies have demonstrated the ability of magnetic resonance imaging to detect individual plaques in living mice. This review discusses the mouse models, MR pulse sequences, and parameters that have been used to image plaques and how they can be optimized for future studies.
Keywords: MR microscopy, Alzheimer's disease, magnetic resonance imaging, magnetic resonance micro imaging, transgenic mice
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disease characterized by the loss of cognitive function. Because the prevalence of AD increases exponentially after 65 years of age, AD is a leading public health concern as the population ages [1]. The cardinal pathologies of AD are amyloid plaque deposition, cerebral atrophy, and neurofibrillary tangles; however, there are no currently accepted diagnostic tests for AD short of autopsy. It is widely believed that abnormal amyloid metabolism is an early step in the progression to clinical AD; therefore, reducing or preventing amyloid plaque accumulation is a major therapeutic objective in AD research [2].
Transgenic mouse models of AD have been created by inserting one or more of the mutated genes known to cause familial AD in humans [3,4]. The mutations are associated with presenilin 1 (PS1), presenilin 2 (PS2), and amyloid precursor protein (APP). These mouse models allow controlled study of plaque deposition and testing of interventions. Currently the only well-established method of detecting individual amyloid plaques is through post-mortem histological analysis. A non-invasive tool to detect and quantify individual plaques in mice would be an invaluable tool, particularly in longitudinal studies where the number of mice in the study can be a limiting factor. Positron emission tomography (PET) ligands such as Pittsburgh Compund B detect fibrillar amyloid β (Aβ) in plaques, but the resolution of PET is too low to identify individual plaques or to quantify plaque density [5-8].
Magnetic Resonance Imaging (MRI) provides the spatial resolution necessary to visualize and quantify plaques with the potential to track individual plaques over time. Amyloid plaques are very small (< 100 μm) compared to typical MRI resolution, so they are difficult to image. Reliable imaging requires very long scan times, so even small improvements in imaging efficiency can significantly increase the feasibility of studies involving large numbers of mice, multiple imaging time points, or both. Despite several reports of successful plaque imaging, the variety of transgenic mouse models and imaging techniques have made it difficult to compare the studies to determine an optimal imaging strategy. Furthermore, imaging plaques in humans is currently far from feasible, but it may be possible after refining imaging techniques on transgenic mice.
Ex Vivo Imaging
The high resolution needed to image amyloid plaques requires long scan times that make some techniques impractical in vivo. In 2004 Zhang et al. reported ex vivo imaging of amyloid plaques in the APP/PS1 mouse model with a spin-echo pulse sequence [9]. They were able to see individual cortical and hippocampal plaques in the fixed ex vivo brain of 15.5 month old mice. However, the long scan times (8-24 hr) made in vivo imaging with that technique impossible. Also in 2004, Lee et al. reported visualizing plaques in the ex vivo brain of APP/PS1 mice [10]. This work used a fast-spin-echo (FSE) technique to image cortical and hippocampal plaques, but the imaging time of 10-11 hr was also too long to be translated to in vivo imaging.
Faber et al. reported bright contrast detection of plaques using the CRAZED sequence [11]. They were able to generate bright contrast from the thalamic plaques of an APP × ADAM10-dn mouse in less than 3 min. However, they determined that the low signal intensity in the image would make CRAZED imaging impractical for routine in vivo plaque imaging.
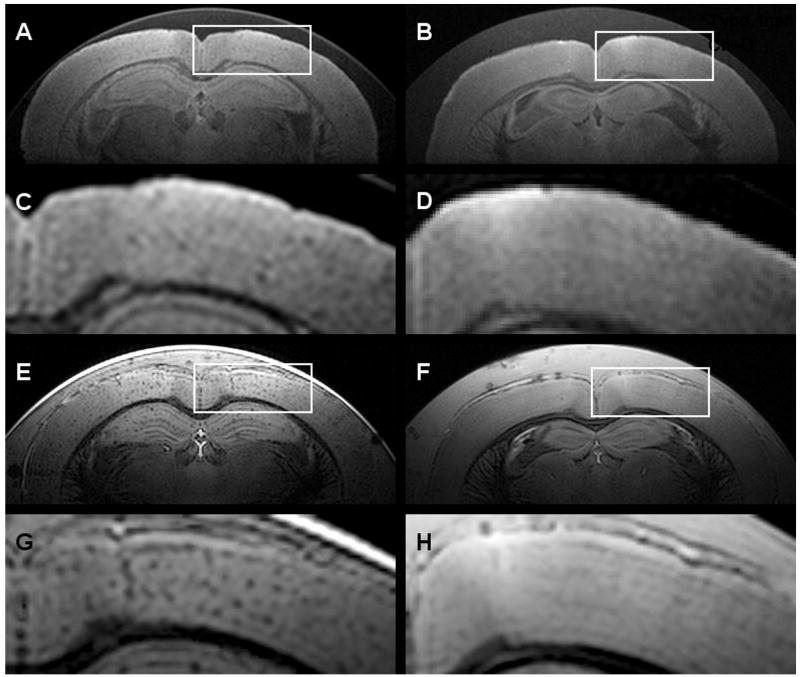
Our group reported the advantage of using susceptibility-weighted imaging (SWI) [12] for ex vivo plaque imaging [13]. The SWI method provided high plaque contrast in a relatively short scan time (1hr 30 min). Fig. (1) shows spin-echo and SWI images of ex vivo brain of a 9-month old APP/PS1 mouse brain and an ex vivo brain of a wild-type (WT) mouse. The plaque contrast in the SWI image is clearly greater, but the SWI method was found to be impractical in vivo due the susceptibility artifacts from the air-to-skull interface and the surface vessels.
Fig. (1).
Comparison of spin-echo and SWI images of ex vivo APP/PS1 and WT mouse brains. (A) and (C) Spin-echo image of an APP/PS1 brain, with (C) representing the magnified box in (A). (B) and (D) Spin-echo image of a WT brain. (E) and (G) SWI image of an APP/PS1 brain. (F) and (H) SWI image of a WT brain.
In Vivo Imaging
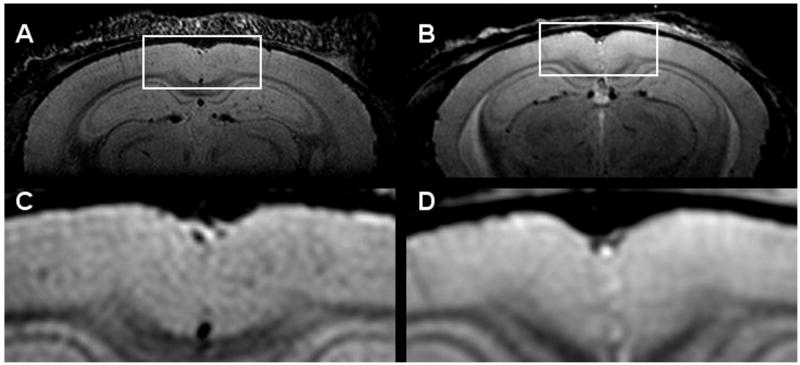
In vivo imaging of plaques has proven more difficult than ex vivo imaging due to restricted scan times and motion artifacts from the mouse's cardiorespiratory pattern. In vivo imaging of amyloid plaques was first reported in 2004 by Jack et al [14]. In this work, individual cortical plaques in APP/PS1 mice were visible with an adiabatic spin-echo sequence in a nominal 1 hr 40 min scan. Fig. (2) shows a spin-echo image from a 13 month old APP/PS1 mouse and a wild-type mouse. One can see the lack of hypointensities in the wild-type mouse. The scan was triggered off of the cardiorespiratory pattern, so the actual scan time was slightly longer and varied from mouse to mouse. This work was also the first to compare multiple techniques and parameter sets to optimize the imaging. First, it was subjectively determined that a spin-echo sequence was superior to a gradient-echo sequence, then it was subjectively determined that the optimal through-plane resolution was 120 μm given and in-plane resolution of 60 μm × 60 μm. Using this technique it was possible to register in vivo MR images, ex vivo MR images and histology images to match plaques across the three modalities. Plaques were matched between MRI and histology to determine that the smallest plaque that could be resolved in vivo had a diameter of 35 μm [15,16]. It was also demonstrated that the earliest age at which plaques were visible in the APP/PS1 mouse using the adiabatic spin-echo sequence was 9 months. In 2006, Braakman et al. imaged the cortical plaques of APP mice using a FSE sequence [17]. The multi-slice FSE sequence allowed them to visualized plaques in a relatively short scan time (25 minutes). This study is currently the only one to report longitudinal imaging of the same mice. Individual plaques were matched between images of the same mouse at 12, 14, 16, and 18 months of age. Borthakur et al. were also able to visualize the cortical and hippocampal plaques of APP/PS1 mice in vivo using T1ρ-weighting [18]. The T1ρ-weighting generates sufficient plaque contrast, but the scan time was very long (3 hours) compared to the other techniques, and only a single slice was acquired.
Fig. (2).
In vivo images using a multi-asymmetric spin-echo sequence. (A) and (C) Images of a 13 month old APP/PS1 mouse. (B) and (D) Images of a 4 month old WT mouse.
Thalamic plaques have also been imaged by several groups. Vanhoutte et al. demonstrated in vivo thalamic plaque imaging in APP mice using a gradient-echo pulse sequence [19]. The thalamic plaques of APP/PS1 were also imaged using a gradient-echo sequence by Dhenain et al [20]. Faber et al. imaged the thalamic plaques of APPxADAM10-dn mice in vivo using a fast, low-resolution gradient-echo sequence [11]. Thalamic plaques in all of these mouse models are relatively large and have a high iron concentration; therefore, they can be successfully imaged with a lower resolution than that needed to image cortical plaques [21,22]. These techniques would be unable to see cortical or hippocampal plaques.
Optimizing Plaque Imaging
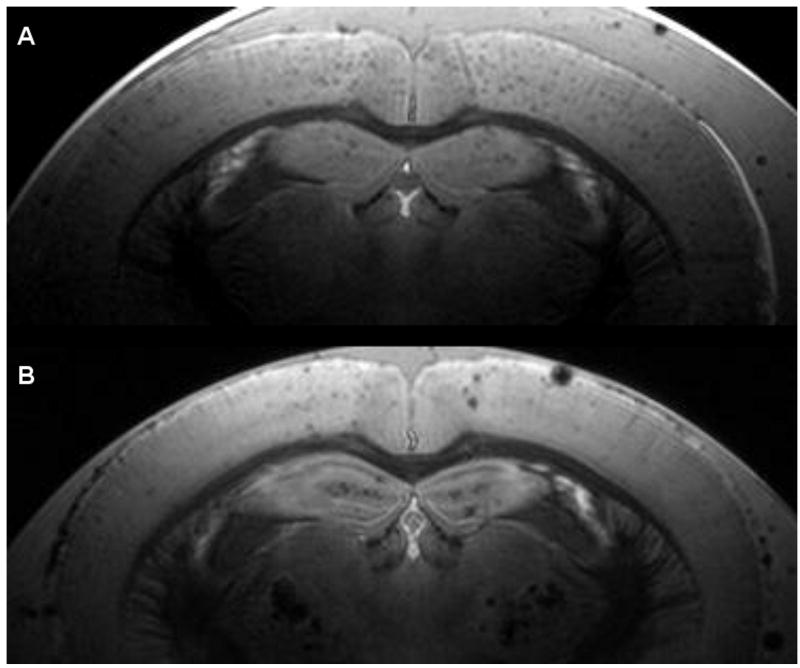
Our group has chosen the cortical plaques of the APP/PS1 transgenic mouse model as the preferred target for imaging and therapeutic studies. The APP/PS1 mouse model is a well established model with plaque formation beginning at three months of age [22]. Compared to the APP mouse model, the plaques in the APP/PS1 model are generally smaller, more numerous, and also develop more rapidly beginning at a younger age (Fig. (3)). This makes them more difficult to image, but more attractive for therapeutic and longitudinal studies, due to the shorter time and costs required for housing the mice. The cortical, as well as hippocampal, plaques are preferred to the thalamic plaques in the APP and APP/PS1 transgenic mice for MR imaging because they more closely mimic the morphology and distribution of human amyloid plaques. Thalamic plaques are very large and contain high concentrations of iron. This makes them easy to image, but they are not closely related to human amyloid plaques. While both cortical and thalamic plaques are thioflavine S/Congo red positive and are immunopositive for Aβ peptides, cortical and hippocampal plaques in both humans and transgenic mice contain much lower levels of iron that are only detectable by DAB-enhanced Perl's staining [23]. Thalamic plaques are highly visible by standard Perl's (Prussian blue) staining [24]. With regard to anatomical distribution, just like the cortical and hippocampal plaques in APP/PS1 mice, amyloid plaques in humans are also found predominantly in the gray matter of the cerebral cortex and hippocampus. The morphology of thalamic plaques is also different from cortical plaques in humans and transgenic mice. Thalamic plaques have a compact and highly spherical morphology with well-defined, regular margins (unpublished observations). Cortical plaques in both transgenic mice and humans generally have a more irregular shape, displaying a dense core of intense thioflavine S/Congo red staining with irregular fibrils radiating outward. These are generally referred to as neuritic, dense-core, or senile plaques. Diffuse plaques are less common in transgenic mice than humans, but in our experience only thioflavine S-positive plaques are detectable by MRI. It is our opinion that if successful MR techniques for imaging individual amyloid plaques in human AD patients are to be developed using AD transgenic mouse models, then only the pathological features that most closely replicate the human disease should be targeted.
Fig. (3).
Comparison of a 9 month old APP/PS1 brain (A) and a 22 month old APP brain (B). Even at less than half the age of the APP mouse, the APP/PS1 mouse has more numerous but smaller plaques.
Ex vivo imaging can be used as a tool to empirically compare and optimize in vivo imaging techniques, since performing more than two or three scans in a single session is impossible in vivo. Recently two reports have focused on using ex vivo imaging to analyze and optimize amyloid plaque imaging [13,25].
Meadowcroft et al. used a novel histological coil to image 60 μm thick slices of brain tissue [25]. The slices were then stained for histologic analysis. This technique allows perfect registration between the 2D MRI and the histologic images. This method was performed on both human AD tissue and APP/PS1 tissue. The MR images were compared to thio-S stains and iron stains to determine the properties of plaques that were MR visible. It was concluded that the contrast generated by plaques in APP/PS1 mice was most likely via decreased T2 due to dense fibrillar Aβ deposits, not via decreased T2* due to high iron concentration. Our group corroborated this theory by mapping T2, T2*, T1 and proton density in ex vivo samples [13]. We found that T1 and proton density did not contribute to plaque contrast, and that the T2 and T2* values were very similar in plaques (32.1 ms and 31.1 ms, respectively). This indicates that the dominant mechanism of plaque contrast is due to increased transverse relaxation rate, probably due to dense Aβ aggregates. If the iron concentration were high inside the plaque, one would expect that T2* would be significantly different than T2.
Although there have been many reports of successful plaque imaging, no two groups have used the same pulse sequence or parameters. Our group used ex vivo imaging to compare the plaque contrast generated by a variety of pulse sequences in order to determine which sequences provide greater plaque visibility [13]. We found that fast-spin-echo techniques are inferior to techniques that average multiple echoes for a given total scan time. This is because the T2 decay between echoes causes blurring in the final image [26]. Since plaques are typically smaller than a single voxel, any blurring will significantly degrade the plaque visibility. We also determined that the greatest plaque contrast can be generated ex vivo by a multi-gradient-echo based SWI sequence. Unfortunately, we found that this sequence would be impractical in vivo due to the very long echo time required to get optimal T2* and phase contrast. Therefore, we concluded that the optimal in vivo pulse sequence was a multi-asymmetric-spin-echo sequence with less than 8 ms of spin-echo asymmetry.
Summary
MR imaging of amyloid plaques in mouse models of AD has proven possible and reliable, but there are still no reports of it being used as a tool in a therapeutic study. Recent studies have gone beyond proof of principle to optimization and analysis of plaque contrast [13,25], but there is still room for improvement. Previous studies have based the image resolution on qualitative image sharpness and plaque contrast. A quantitative analysis of the imaging efficiency for a range of image resolutions could significantly improve plaque imaging efficiency. Also, a more detailed analysis like that done by Meadowcroft could illuminate the differences between plaques that are MR visible and those that are MR invisible in order to better design experiments. Even without these further improvements, we believe that plaque imaging can be valuable tool for AD research in mice.
With enough optimization and understanding of plaque imaging in mouse models, it is possible that human plaque imaging will be realized. The difficulties in moving from mouse imaging to human imaging are many. First, the movement artifacts will be much larger since a human patient will not be immobilized and anesthetized as a mouse is. Second, the imaging time will need to be reduced to a more reasonable length – less than 10 minutes would be ideal. Third, the human brain is much larger than the mouse brain, so the FOV used for typical human clinical MRI will need to be severely reduced to a restricted area of interest within the human brain. The most attractive targets for reduced FOV imaging would be the lateral temporal, prefrontal, or parietal cortex because these areas have high plaque loads but are also close to the surface, so a surface coil could be used to achieve maximal SNR. Despite the difficulties, there are a couple ways in which human plaque imaging may be easier. First, the location of interest in humans will be further from the sources of susceptibility interfaces than in mice; therefore, SWI may be feasible. This is advantageous because we found that SWI can provide significantly greater plaque contrast than other techniques. Second, human plaques contain more iron than APP/PS1 cortical plaques [25], so the SWI contrast will be significantly higher. Because these advantages of human plaques, it is our opinion that the most significant hurdle in human plaque imaging is patient motion. If motion can be adequately accounted for, human plaque imaging should be possible.
Acknowledgments
This work was supported in part by funds from NIH P41 RR008079, NIH P30 NS057091, the Keck Foundation, and the Minnesota Partnership for Biotechnology and Medical Genomics.
References
- 1.Evans D, Funkenstein H, Albert M. Pevalence of Alzheimer's disease in a community population of older persons. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe D. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Holcomb L, Gordon M, McGowan E, et al. Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med. 1998;4:97–100. doi: 10.1038/nm0198-097. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao K, Chapman P, Nilsen S, et al. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 5.Klunk W, Engler H, Nordberg A, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55(3):306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 6.Klunk W, Wang Y, Huang G, et al. The binding of 2-(4′-methylaminophenyl)benzothiazole to postmortem brain homogenates is dominated by the amyloid component. J Neurosci. 2003;23(6):2086–2092. doi: 10.1523/JNEUROSCI.23-06-02086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sair H, Doraiswamy P, Petrella J. In vivo amyloid imaging in Alzheimer's disease. Neuroradiology. 2004;46:93–104. doi: 10.1007/s00234-003-1034-9. [DOI] [PubMed] [Google Scholar]
- 8.Shoghi-Jadid K, Small G, Agdeppa E, et al. Localization of neurofibrillary tangles and beta-amyloid plaques in the brains of living patients with Alzheimer disease. Am J Geriatr Psychiatry. 2002;10(1):24–35. [PubMed] [Google Scholar]
- 9.Zhang J, Yarowsky P, Gordon MN, et al. Detection of amyloid plaques in mouse models of Alzheimer's disease by magnetic resonance imaging. Magn Reson Med. 2004;51:452–457. doi: 10.1002/mrm.10730. [DOI] [PubMed] [Google Scholar]
- 10.Lee SP, Falangola MF, Nixon RA, Duff K, Helpern JA. Visualization of β-amyloid plaques in a transgenic mouse model of Alzheimer's disease using MR microscopy without contrast reagents. Magn Reson Med. 2004;52:538–544. doi: 10.1002/mrm.20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faber C, Zahneisen B, Tippmann F, Schroeder A, Fahrenholz F. Gradient-echo and CRAZED imaging for minute detection of Alzheimer plaques in an APPV717I × ADAM10-dn mouse model. Magn Reson Med. 2007;57:696–703. doi: 10.1002/mrm.21201. [DOI] [PubMed] [Google Scholar]
- 12.Haacke E, Xu Y, Cheng YCN, Reichenbach JR. Susceptibility Weighted Imaging (SWI) Magn Reson Med. 2004;52:612–618. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain R, Reyes DA, Curran GL, et al. Comparison of Amyloid Plaque Contrast Generated by T2-Weighted, T2*-Weighted, and Susceptibility-Weighted Imaging Methods in Transgenic Mouse Models of Alzheimer's Disease. Magn Reson Med. 2009;61:1158–1164. doi: 10.1002/mrm.21951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Garwood M, Wengenack TM, et al. In vivo visualization of Alzheimer's amyloid plaques by magnetic resonance imaging in transgenic mice without a contrast agent. Magn Reson Med. 2004;52:1263–1271. doi: 10.1002/mrm.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR, Marjanska M, Wengenack TM, et al. Magnetic resonance imaging of Alzheimer's pathology in the brains of living transgenic mice: A new tool in Alzheimer's disease research. Neuroscientist. 2007;13(1):38–48. doi: 10.1177/1073858406295610. [DOI] [PubMed] [Google Scholar]
- 16.Jack CR, Wengenack TM, Reyes DA, et al. In vivo magnetic resonance microimaging of individual amyloid plaques in Alzheimer's transgenic mice. J Neurosci. 2005;25(43):10041–10048. doi: 10.1523/JNEUROSCI.2588-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braakman N, Matysik J, van Duinen SG, et al. Longitudinal assessment of Alzheimer's β-amyloid plaque development in transgenic mice monitored by in vivo magnetic resonance microimaging. J Magn Reson Imaging. 2006;24:530–536. doi: 10.1002/jmri.20675. [DOI] [PubMed] [Google Scholar]
- 18.Borthakur A, Gur T, Wheaton AJ, et al. In vivo measurement of plaque burden in a mouse model of Alzheimer's disease. J Magn Reson Imaging. 2006;24:1011–1017. doi: 10.1002/jmri.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanhoutte G, Dewachter I, Borghgraef P, Van Leuven F, Van der Linden A. Noninvasive in vivo MRI detection of neuritic plaques associated with iron in APP[V717I] transgenic mice, a model for Alzheimer's disease. Magn Reson Med. 2005;53:607–613. doi: 10.1002/mrm.20385. [DOI] [PubMed] [Google Scholar]
- 20.Dhenain M, El Tannir El Tayara N, Wu TD, et al. Characterization of in vivo MRI detectable thalamic amyloid plaques from APP/PS1 mice. Neurobiol Aging. 2009;30:41–53. doi: 10.1016/j.neurobiolaging.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Wengenack TM, Curran GL, Poduslo JF. Targeting Alzhiemer amyloid plaques in vivo. Nat Biotechnol. 2000;18:868–872. doi: 10.1038/78482. [DOI] [PubMed] [Google Scholar]
- 22.Wengenack TM, Whelan S, Curran GL, Duff KE, Poduslo JF. Quantitative histological analysis of amyloid deposition in Alzheimer's double transgenic mouse brain. Neuroscience. 2000;101:939–944. doi: 10.1016/s0306-4522(00)00388-2. [DOI] [PubMed] [Google Scholar]
- 23.LeVine SM. Iron deposits in multiple sclerosis and Alzheimer's disease brains. Brain Res. 1997;760:298–303. doi: 10.1016/s0006-8993(97)00470-8. [DOI] [PubMed] [Google Scholar]
- 24.Vanhoutte G, Dewachter I, Borghgraef P, Van Leuven F, Van der Linden A. Noninvasive in vivo MRI detection of neuritic plaques associated with iron in APP[V717I] transgenic mice, a model for Alzheimer's disease. Mag Reson Med. 2005;53:607–13. doi: 10.1002/mrm.20385. [DOI] [PubMed] [Google Scholar]
- 25.Meadowcroft MD, Connor JR, Smith MB, Yang QX. MRI and histological analysis of beta-amyloid plaques in both human Alzheimer's disease and APP/PS1 transgenic mice. J Magn Reson Imaging. 2009;229:997–1007. doi: 10.1002/jmri.21731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Constable RT, Gore JC. The loss of small objects in variable TE imaging: Implications for FSE, RARE, and EPI. Magn Reson Med. 1992;28:9–24. doi: 10.1002/mrm.1910280103. [DOI] [PubMed] [Google Scholar]