Abstract
Background
Potential epigenetic mechanisms underlying fetal alcohol syndrome (FAS) include alcohol-induced alterations of methyl metabolism, resulting in aberrant patterns of DNA methylation and gene expression during development. Having previously demonstrated an essential role for epigenetics in neural stem cell (NSC) development and that inhibiting DNA methylation prevents NSC differentiation, here we investigated the effect of alcohol exposure on genome-wide DNA methylation patterns and NSC differentiation.
Methods
NSCs in culture were treated with or without a 6-hr 88mM (“binge-like”) alcohol exposure and examined at 48 hrs, for migration, growth, and genome-wide DNA methylation. The DNA methylation was examined using DNA-methylation immunoprecipitation (MeDIP) followed by microarray analysis. Further validation was performed using Independent Sequenom analysis.
Results
NSC differentiated in 24 to 48 hrs with migration, neuronal expression, and morphological transformation. Alcohol exposure retarded the migration, neuronal formation, and growth processes of NSC, similar to treatment with the methylation inhibitor 5-aza-cytidine. When NSC departed from the quiescent state, a genome-wide diversification of DNA methylation was observed—that is, many moderately methylated genes altered methylation levels and became hyper- and hypomethylated. Alcohol prevented many genes from such diversification, including genes related to neural development, neuronal receptors, and olfaction, while retarding differentiation. Validation of specific genes by Sequenom analysis demonstrated that alcohol exposure prevented methylation of specific genes associated with neural development [cutl2 (cut-like 2), Igf1 (insulin-like growth factor 1), Efemp1 (epidermal growth factor-containing fibulin-like extracellular matrix protein 1), and Sox 7 (SRY-box containing gene 7)]; eye development, Lim 2 (lens intrinsic membrane protein 2); the epigenetic mark Smarca2 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2); and developmental disorder [Dgcr2 (DiGeorge syndrome critical region gene 2)]. Specific sites altered by DNA methylation also correlated with transcription factor binding sites known to be critical for regulating neural development.
Conclusion
The data indicate that alcohol prevents normal DNA methylation programming of key neural stem cell genes and retards NSC differentiation. Thus, the role of DNA methylation in FAS warrants further investigation.
Keywords: Epigenetics, Epigenomics, MeDIP-Chip, Neural development, Fetal alcohol syndrome
INTRODUCTION
Multifactorial neurodevelopmental deficit, such as fetal alcohol spectrum disorder (FASD), is likely to occur through gene-environment interactions that alter the fate of early developing precursor cells. Neural stem cells (NSC) are capable of self-renewal and differentiate along neuronal and glial lineages. These processes are defined by the dynamic interplay between extracellular cues, and intracellular transcriptional signaling programs. Recently, epigenetic mechanisms—including DNA methylation, histone modifications, and non-coding RNA expression--have been shown to be closely associated with the fate specification of NSCs. These epigenetic alterations could provide coordinated systems for regulating gene expression at each step of neural cell differentiation. Epigenetic modifications regulate key developmental events, including germ cell imprinting (Bartolomei, 2003), stem cell maintenance (Cheng et al., 2005; Kondo, 2006; Zhang et al., 2006; Meshorer, 2007; Surani et al., 2007; Tang and Zhu, 2007), cell fate, and tissue patterning (Kiefer, 2007). Aberrant epigenetic alterations are known to disrupt key developmental events, particuarly in the nervous system, leading to conditions such as Rett's syndrome (Shahbazian and Zoghbi, 2002), immunodeficiency centromeric instability and facial syndrome (ICF) (Hansen et al., 1999, Tao et al., 2002, Ueda et al., 2006), and Prader-Willi/Angelman syndrome (Lalande et al., 1999; Xin et al., 2003; Lalande and Calciano, 2007). Environmental input also has a significant influence on development and can alter epigenetic programming. Recently, we and others have reported that alcohol exposure alters DNA methylation, resulting in genetic and phenotypic changes (Qiang et al., 2010; Ouko et al., 2009; Haycock, 2009; Pandey et al., 2008; Moonat et al., 2010; Shukla et al., 2008; Oberlander et al., 2008; Miranda et al., 2010; Liu et al., 2009; Kaminen-Ahola et al., 2010).
Alcohol has multiple effects on methyl donors (Mason and Choi, 2005) and appears to interfere with the folate-methyl metabolic pathway for methyl donors by inhibiting methionine synthase and methionine adenosyltransferase. Inhibition of methionine synthase also creates a “methylfolate trap,” analogous to what occurs in vitamin B12 deficiency (Cravo and Camilo, 2000; Mason and Choi, 2005). Alcohol ingestion in animals has been shown to inhibit folate-mediated methionine synthesis, thereby interrupting critical methylation processes mediated through s-adenosyl methionine, the activated form of methionine and substrate for biologic methylation. In addition, some evidence indicates that alcohol may redirect the utilization of folate toward serine synthesis and thereby interfere with a critical function of methylene tetrahydrofolate and thymidine synthesis (LaBaume et al., 1987; Cravo and Camilo, 2000). Thus, alcohol exposure, by resulting in downstream epigenetic modifications, could bridge the gene-environment interaction, providing a new causal paradigm for the neurodevelopmental deficit.
Alcohol exposure, thus far, has been associated with the development of decreasing DNA methylation transferase (DNMT) mRNA levels in the sperm of male rats exposed to alcohol (Bonsch et al., 2006), and substantial evidence indicates that DNA methylation plays a significant role in neural cell lineage differentiation and early brain development (Takizawa et al., 2001; Setoguchi et al., 2006; Feng et al., 2007; MacDonald and Roskams, 2009). We recently demonstrated that alcohol exposure during early neurulation altered neural tube development and induced changes in DNA methylation, resulting in functional consequences on gene expression, cell cycle regulation, and neural development (Liu et al., 2009). The possibility of alcohol-induced alteration of stem cell DNA methylation changes is not clear, and evidence for alcohol causing aberrant stem cell development through altered DNA or histone methylation has not been examined.
In the current study, we used an established dorsal root ganglial neural stem cells, in which multipotency and a differential phenotype profile has been characterized (Singh et al., 2009a). Epigenetic changes at the cellular level have also been demonstrated in this model system (Singh et al., 2009a). By using MeDIP-chip (anti-5-methylcytidine antibody immunoprecipitation followed by microarray analysis) a prominent effect of alcohol on DNA methylation profiles in neural stem cells was discovered. We report here that the normal DNA methylation program during differentiation is disrupted by alcohol, including those genes mediating early embryonic development.
RESULTS
Alcohol Effects on Neural Stem Cell Differentiation and Cellular DNA methylation
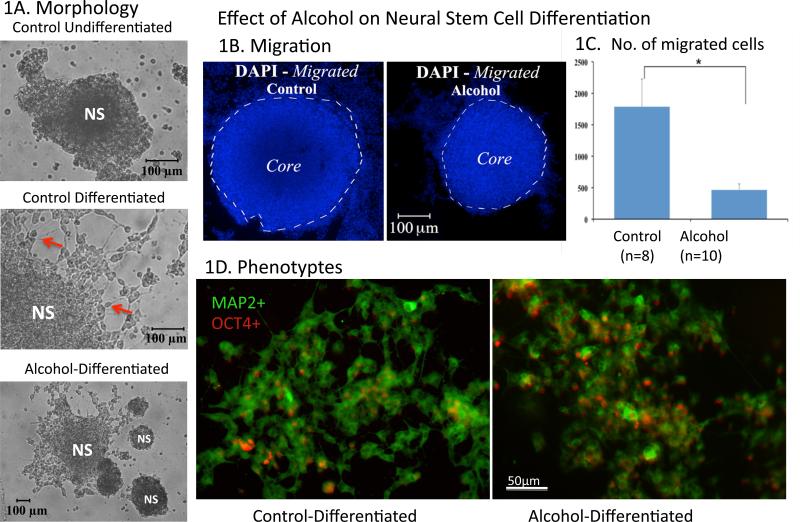
As shown in Figure 1, undifferentiated NSCs in neurospheres were characteristically round and small with a distinct nucleus in the center of the small cytoplasm, as reported previously (Singh et al., 2009a). After a two-day culture on substrate-coated chamber slides, the neurospheres were flattened in appearance and cells in the outer layer of the neurosphere increased in size (Fig 1A). On the fourth day in culture (DIC4), noticeable zoning for a differential differentiating state was evident, as reported previously (Singh et al., 2009a). In brief, in the core of the neurosphere, the NSCs remained round and small (Fig 1A). However, in the periphery of the neurosphere, much like the undifferentiated groups, many NSCs remained round, but a subpopulation of the cells formed angular cytoplasmic expansions, some with fiber extensions. In the migrated zone (out of neurosphere), more differentiated cells grew processes, MAP2-immunostained (im), and acquired neuronal morphology with varicosities (Figure 1A); on the other hand, the OCT4-im was reduced in the peripheral area. The alcohol-treated NSC neurospheres, on the fourth day, displayed decreased migration, i.e. migrated cells traveled in smaller scales (Figure 1B, outside the core ) and fewer NSCs left the neurosphere (Figure 1C). The MAP2-im / OCT4-im cell ratio was reduced in comparison with that of Controls in the DIC4.
Figure 1. Alcohol treatment on Differentiation and Migration of neural stem cells.
[1A] Brightfield Microscopy shows morphology of DRG neurospheres (NS) under three treatment groups: undifferentiated, differentiated, and alcohol-differentiated. Cells migrated out of neurosphere and grew processes as going through differentiation (Arrows). Alcohol reduced migration and fiber growth. DAPI staining (1B) shows less migration of differentiating cells in the alcohol-differentiated group (right, 1B and 1C) as compared with the control-differentiated group by student's t-test (left, IB and 1C) (*=p <0.05). 1D shows two phenotypes of the NSCs during differentiation. The MAP2-postive (MAP2+, green fluorescence, indicating neurons) cells some with fibers prevailed in the control differentiation (1D, left); the extend of MAP+ staining and fiber extension is apparently reduced when treated with alcohol (1D, right). Contrary to the MAP2+, the OCT4-positive (OCT4+, red fluorescence, indicating stem cells here) cells reduced through normal differentiation (1D, left), was held up when treated with alcohol (1D, right). Scale bars: 1A,100μm; 1B, 100μm; 1D, 50μm.
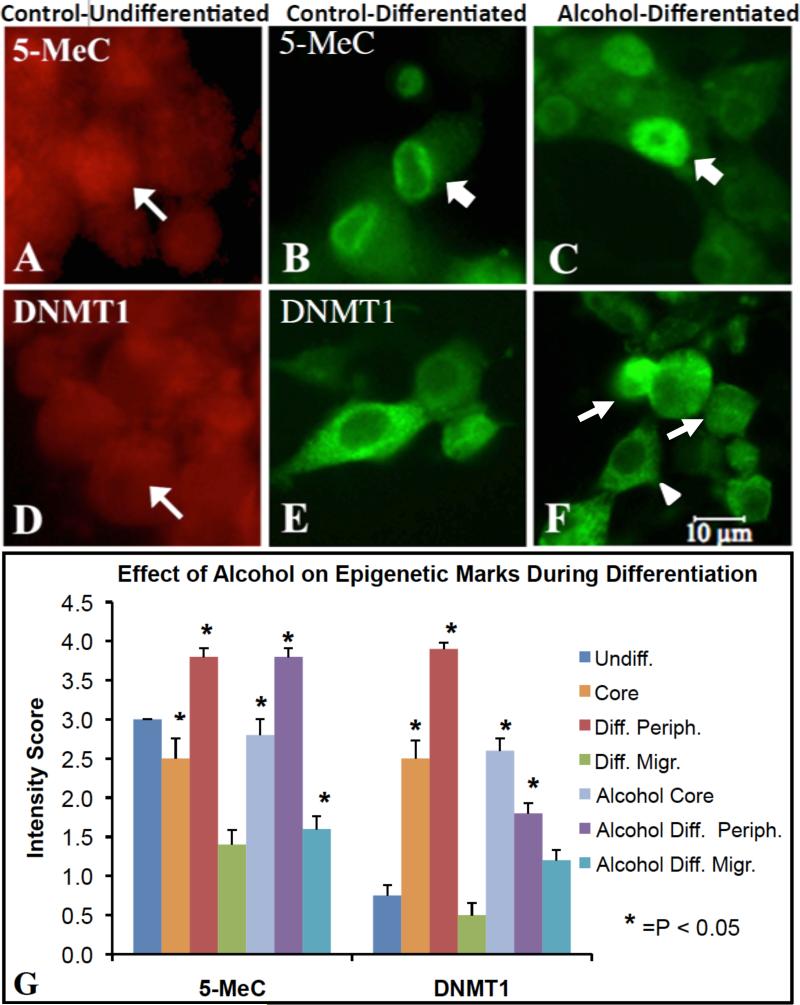
In parallel with analysis of the genome-wide DNA methylation, it was next of interest to examine the effect of alcohol on the cellular distribution of DNA methylation marks, including DNA methylation mark 5-methyl cytidine (5-MeC) and DNA methyltransferase (DNMT). In undifferentiated NSC, 5-MeC immunostaining (im) was moderately intense and distributed throughout the nucleus. However, in the control-undifferentiated group, increased 5-MeC-im intensity was seen in the nucleus, but preferentially distributed near the nuclear membrane into differentiation. A redistribution of 5-MeC-im throughout most of the nucleus was observed in alcohol-treated NSC during the differentiation stage (Figure 2A-C). A redistribution of DNMT1-im was also evident in the three treatment groups. The DNMT1 was distributed similarly to 5-MeC-im--low and throughout the nucleus--but translocated into cytoplasm in differentiated NSC. Alcohol exposure prevented this redistribution in a subpopulation of the NSC during differentiation (Figure 2D-E). The significance of these translocations may be pertinent to euchromatic vs heterochromatic distribution of epigenetic marks associated genes.
Figure 2. Immunostaining for 5-Methyl-cytidine (5-MeC) and DNMT1 epigenetic markers in neural stem cells during differentiation upon alcohol treatment.
A-F shows translocation of epigenetic marks during differentiation. The 5-MeC immunoreactivity distributed throughout the nucleus (A, arrow) in undifferentiated NSCs, but relocated to the perinucleus (B, arrow) upon differentiation. Following alcohol treatment, 5-MeC was found not only in the perinucleus (C, arrow) but also regained nuclear-wide distribution (C, arrowhead). In undifferentiated NSCs, DNMT1 was expressed in the nucleus and cytoplasm (D, arrow); however, following differentiation, DNMT1 expression became exclusively cytoplasmic around nucleus (E). With alcohol treatment, DNMT1 was distributed in both the cytoplasm (F, arrowhead), and nucleus (F, arrows) which is normally seen in undifferentiated NSCs. Scale bars: A-F= 10um. A, D: Alexa633 (red fluorescence), and B, C, E, F: Alexa488 (Gree fluorescence). G, shows, through semiquantitative measurement, that 5-MeC and DNMT1 were dynamically changed from the undifferentiated control neurosphere (undiff.) to the differentiated Control and Alcohol groups in core and periphery (Periph.) of neurospheres and in migrated (Migr.) cells (n=10-12, *=p<0.05). The methods and statistics are described in the Methods.
A semi-quantitative measurement of staining indicated that 5-MeC and DNMT1 were dynamically changed from the undifferentiated control neurosphere to the differentiated Control and Alcohol groups in core and periphery of neurospheres and in migrated (Figure 2G).
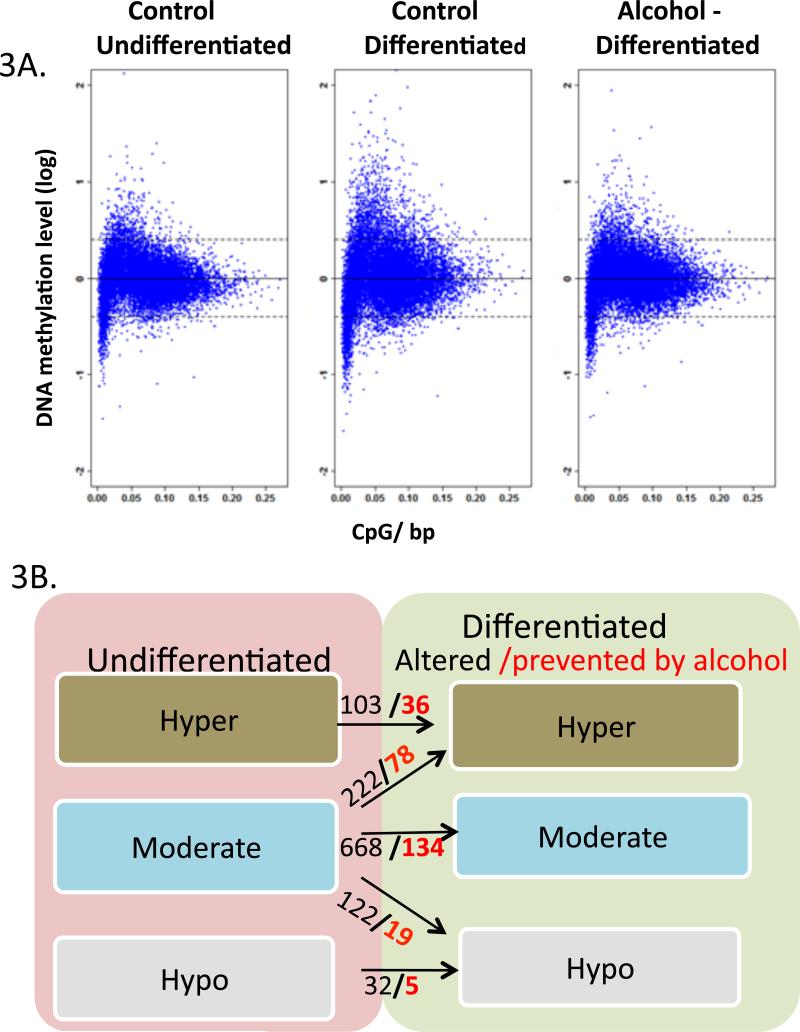
The distribution of multiple DNA methylation profiles changed during differentiation by presenting an increased diversity with more hypermethylated and hypomethylated genes, and a modified landscape of gene methylation. During the undifferentiated/renewal state, the vast majority of genes (93%) were moderately methylated (between 0.4 and -0.4 log mean methylation, see definition in Method) (Figure 3A). During differentiation, a total of 1143 genes changed the methylation status, and, out of those, alcohol prevented the change of 272 genes (p <0.05). Among the 344 genes that were changed from moderate methylation during differentiation, 222 became hypermethylated and 122 hypomethylated. When the DRG stem cells were treated with alcohol during differentiation, 97 genes out of those 344 changes were prevented or reversed; 78 genes were prevented from hypermethylation, and 19 genes changed from hypomethylation back to moderate methylation status (Figure 3B).
Figure 3. Genome-wide Distribution of NSC DNA methylation during differentiation and upon alcohol treatment.
(3A) Scatter-plotting of promoter methylation levels against CpG content in NSC of three treatment groups demonstrated the pattern of DNA methylation. X-axis denotes the observed-to-expected CpG ratio, and Y-axis represents the average log2 transformation of the methylation signal level through all the probes in -1,300 to +500-bp from transcription start site. The dash lines at y=0.4 and -0.4 are defined as cutoff for hypermethylation (above upper line) and hypomethylation (below lower line), which represents DNA-methylation levels greater than 1.3 folds of the genome-wide median methylation level. The genes with DNA methylation levels between the two lines are defined as moderately methylated. (3B) shows the numbers of genes with DNA methylation changed during differentiation (Diff). The numbers in genes with DNA methylation changed during differentiation is indicated in black in three DNA methylation categories, and the number of genes prevented from change by alcohol during differentiation is shown in red and bold (p <0.05). For example, there were 103 hypermethylated genes that become more hypermethylated during differentiation, out of which 36 genes were prevented by alcohol; 222 moderate-methylated genes became hypermethylated, of which 78 were prevented (remained a moderate) by alcohol during differentiation.
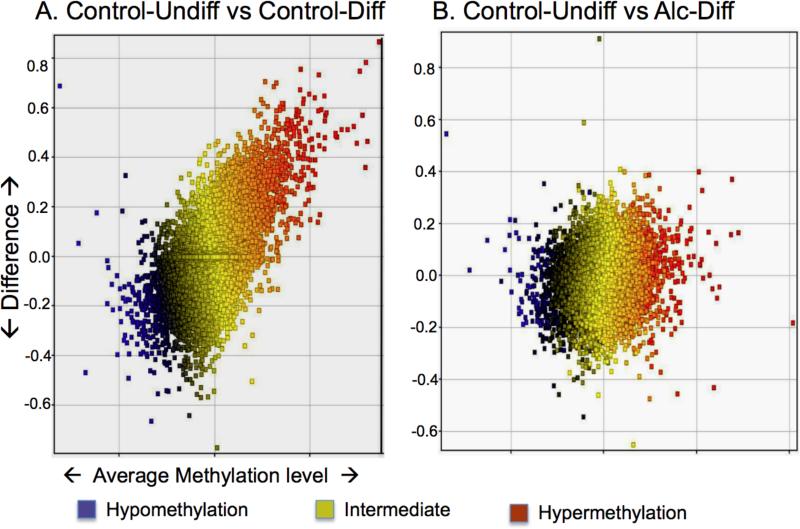
The genome-wide MvA plot displayed difference of DNA methylation levels between two groups of all the genes according to their average DNA methylation levels e.g. control-undifferentiated vs. control-differentiated, control-differentiated vs. alcohol-differentiated. There was strong diversification of methylation during differentiation (comparing the undifferentiated with the differentiated). Such differentiation-related diversification was greatly reduced when treated with alcohol (comparing the undifferentiated with the differentiated + alcohol) (Figure 4), indicating alcohol prevented the diversification of DNA methylation
Figure 4. Genome-wide MvA plot compares DNA methylation difference between two groups.
The difference of methylation between two groups is plotted on the Y-axis against the methylation level (X-axis, also indicated by color, see color legend). In (A) Control-Undifferentiated (Control-Undiff) vs. Control-Differentiated (Control-Diff), a large number of genes differ in methylation levels (Y-axis), and the degree of DNA methylation change is wide (X-axis). In contrast, in (B) Control-Undifferentiated (Control-Undiff) vs. Alcohol-Differentiated (Alc-Diff), the number of genes with methylation difference is reduced (Y-axis), and the degree of changes on DNA methylation level is smaller (X-axis).
Functional profile of Hyper- and Hypomethylated genes altered by Alcohol
The 78 genes that were prevented or reversed from hypermethylation by alcohol from moderate methylation category (Supplement Table 1) are involved in multiple key functions. The most noticeable group of genes are related to neuronal receptors, including glutamate transmitter receptors AMPA3 (Gria3) and glutamate receptor interacting protein 1 (Grip1), cholinergic receptor muscarinic 1 (Chrm1), Adrenergic receptor a1 (Adra1a), and water channels proteins Aqp8 and Aqp9. Other genes include neural development (Dlx3, Clcf1), Cell Cycle (Adra1a, Tnf, Pik3r1, Sh3bp2), path finding of retinal ganglion cells and extension of axons (Pou4f2, Pou4f3), synaptic transmission (Chrm1, Gria3, Tnf, Kcna1, Ptprc), and stem cell (Edg6, Pik3r1, Wnt16). An alcohol metabolism enzyme, alcohol dehydrogenase 4 (Adh4), was also affected by alcohol. Of the 19 genes in moderate methylation category prevented from hypomethylation by alcohol, many were noticeably associated with a group of sensory genes, including the olfactory receptor (Olr214, Olr304, Olr408, Olr611, Olr1646, Olr1139), and the taste receptor genes (Tas2r123, Tas2r7, T2r140, T2r18) (Supplement Table 2).
Two-consecutive probe analysis
Since the effect of DNA methylation can be localized (e.g. within 200bp), probe-wise analyses were performed. We adopted a two-consecutive probe analysis (in which two adjacent probes detected the same direction of DNA methylation changes), in parallel with the average DNA methylation analysis, to verify regional changes in DNA methylation. Based on the linear mixed model, 1054 (FDR <30%) two-consecutive probe regions showed a significant difference in DNA methylation between the control and alcohol-treated NSCs during differentiation. Among those genes, alcohol decreased methylation of 681 and increased methylation of 373. These regional changes were also used as the basis for validating regional DNA methylation changes of specific genes using Sequenom.
Genes validated by Sequenom
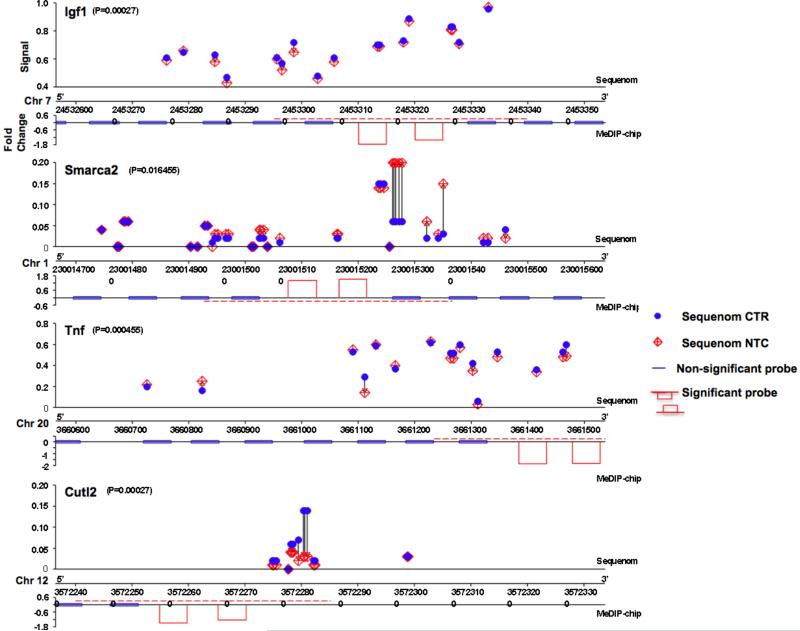
A set of genes with promoter DNA methylation changes during differentiation as well as by alcohol, i.e. genes altered during differentiation and affected by alcohol (changed both in control differentiated vs. control undifferentiated, and in control differentiated vs. alcohol differentiated) and related to the nervous system, were validated using Sequenom MASSarray technology. Overall, based on the two criteria in the Methods, we validated 12 genes out of 26 tested from the MeDIP-microarray results. Their methylation alteration site is shown in Table 1. Examples of comparison of MeDIP-ChIP and Sequenom is shown in Figure 5.
Table 1.
Function and Biology of genes with their DNA methylation altered by alcohol during differentiation in adult rat DRG stem cells validated by Sequenom.
| Gene Symbol | Description | Methylation altered by Alcohol | Function | Biology |
|---|---|---|---|---|
| Cutl2 (Cux2) | cut-like 2 (Drosophila) (predicted) | Down | Transcription factor | Regulates cell cycle progression in neural progenitor cells during spinal cord neurogenesis, dorsal interneuron formation. |
| Igf1 | insulin-like growth factor 1 | Down | Growth factor | Mediates neuron like differentiation of adipose derived stem cells, progenitor cell proliferation, inhibits cell death and promotes recovery of differentiated neurons. |
| Tnf | tumor necrosis factor (TNF superfamily, member 2) | Down | Cytokine | Mediates downregulation of GLT1 and neuronal death in brainstem and cervical motor neurons, proinflammation in adult neural stem/progenitor cells |
| Pou4f3 | POU domain, class 4, transcription factor 3 (predicted) | Down | Transcription factor | Retinal neuron specification, differentiation and survival, autoregulation of inner ear and CNS sensory neurons. |
| Lim2 | lens intrinsic membrane protein 2 | Down | Receptor | Lens development |
| Sox7 | SRY-box containing gene 7 (predicted) | Down | Transcription factor | Activator of FGF-3 transcription in embryonal stem cells |
| Smarca2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Up | Chromatin remodeling complex | Essential for differentiation, cell growth control, cell growth inhibitory activity cyclin D3 through C/EBP alpha and Brm complex |
| Efemp1 | epidermal growth factor-containing fibulin-like extracellular matrix protein 1 | Down | Extracellular matrix protein with EGF domain | Tumor growth |
| Hal | histidine ammonia lyase | Down | Cytosolic enzyme | Histidine catabolism |
| Dgcr2 | DiGeorge syndrome critical region gene 2 | Up | Adhesion receptor protein | Developmental deficits |
| Pard6a | par-6 (partitioning defective 6,) homolog alpha (C. elegans) | Up | Asymmetrical cell division and cell polarization | Migration of immature granule neurons, neuroblast cell polarity |
Figure 5. Confirmation of alcohol-altered DNA methylation during differentiation by Sequenom analysis.
Four genes, Igf1, Smarca2, Tnf, Cutl2 are shown as examples (in each gene, the Sequenom analysis is shown in upper, and MeDIP-Chip, lower). A blue dot indicates the CpG in control undifferentiated; a red diamond indicates CpG in Alcohol-treated. The red dashed lines indicate the significant region of change in methylation by MeDIP-chip.
DISCUSSION
Mammalian development is associated with considerable changes in global DNA methylation levels during genomic reprogramming. The current genome-wide methylation study, showing widespread DNA methylation changes in specific genes of NSCs, supports our previous observation that DNA methylation is an active component and an intrinsic program during differentiation of NSCs (Singh et al., 2009a). In particular, we observed hyper- and hypo-methylation of moderately methylated genes during the reprogramming of quiescent NSCs into differentiation. There are two major questions addressed in this study: first, whether altered DNA methylation in the process of neural stem cell differentiation can be modified by the environmental input, i.e. whether alcohol, which is known to change methyl donor, can alter the intrinsic DNA methylation program during differentiation. Second, whether the altered DNA methylation (by alcohol) has a functional consequence on neural stem cell development. Two categories of alcohol effect are reported here. First, alcohol prevented programmed diversification of DNA methylation during differentiation, i.e. it prevented moderately methylated genes from increased or decreased methylation, or further, becoming significantly hyper- or hypo-methylated. As promoter DNA methylation represses transcription and recruits other repressive chromatin-modifying activities to the chromatin (Bird, 2002), the prevention of programmed hyper- and hypo-methylation of a set number of genes can disrupt chromatin remodeling, and, thus, differentiation reprogramming of quiescent NSCs might be incomplete. The second effect of alcohol is the methylation change of the genes that were not supposed to alter in the normal program at this stage of normal differentiation. The untimely methylation change of these genes also have an opportunity to affect the gene transcription and deviate differentiation.
A number of genes verified by Sequenom MASSarray are known to closely participate in the neural stem cell differentiation program (Table 1). The Igf2 (insulin growth factor 2, an imprinting gene key in development) and Sox 7 (an activator of fibroblast growth factor 3 transcription) are involved in neural stem cell growth and patterning. The Lim2 and Cutl2 (Cux2, Cux2 cut-like homeobox 2), a homeodomain transcription factor, regulates neural stem cell proliferation and differentiation in the developing cortical ventricular zone and in the spinal cord (Cubelos et al., 2008, Iulianella et al., 2008). Cutl2 loss-of-function mouse mutants exhibit smaller spinal cords with deficits in neural progenitor development (Iulianella et al., 2008). Smarca2 (Brm; SWI/SNF related, matrix associated, actin dependent regulator of chromatin) with Brg1 are subunits of SWI/SNF complex essential for the transition from neural stem/progenitors to postmitotic neurons (Lessard et al., 2007). The function of DNA methylation may regulate the recruitment of histone modification enzymes (e.g. histone deactylase or histone methyl transferase) or transcription factor binding. The sites of altered DNA methylation of these genes notably coincide with important transcription factors known for neural specification and neuronal development (Table 2). Multiple binding motifs displayed altered DNA methylation in both Smarca2 and Cutl2. Sp1 has been shown to increase the transcription of Mash1 and promote the RA-induced neuronal differentiation of neural progenitor cells. E2F/p107, another transcription factor complex when bound to Smarca2, mediates cell cycles and proliferation of neural precursor cells (Muchardt and Yaniv, 1999; Vanderluit et al., 2004; McClellan et al., 2009). ER-alpha (Estrogen receptor alpha) is involved in neuronal differentiation when bound to Smarca2 (Merot et al., 2005).
Table 2.
List of predicted transcription factors binding to Smarca2 region with DNA methylation increased by alcohol and Cutl2 region with DNA methylation decreased by alcohol
| Factor | Description | Begin | Sns | Length | Sequence | Function |
|---|---|---|---|---|---|---|
| List of predicted transcription factors binding to Smarca2 region with DNA methylation increased by alcohol | ||||||
| Sp1 | Sp/KLF family transcription factor | 89, 16 | N, R | 6 | AGGCGG, CCGCCT | Transcriptional activator, involved in BMP induced glial expression, MASH1 induction by ASK1 and ATRA in adult neural progenitors. |
| E2F+ p107 | Elongation factor/retinolastoma family member p107 | 22 | R | 4 | TCGC | Cell cycle progression, Neural stem cell expansion |
| ER-alpha | Estrogen receptor alpha | 90 | N | 6 | GGCGGG | Promotes Neuronal differentiation in PC12 cells |
| ZF5 | Zinc finger protein transcription factor | 12 | R | 6 | CGCGCC | Mouse homolog has growth inhibiting activity |
| List of predicted transcription factors binding to Cutl2 region with DNA methylation decreased by alcohol | ||||||
| GCF | GC rich sequence DNA binding factor/chromosome 2open reading frame 3(C2orf3) | 55, 73 | N | 8, 7 | GCGGGGCG, SCGSSSC | Transcriptional repressor which supresses EGF receptor transcription |
| Sp1 | Sp/KLF family transcription factor | 57,70, 43, 34 | R, N | 10 | GGGGCGGAGC, TGCGC GCGCA | Transcriptional activator, involved in BMP induced glial expression, MASH1 induction by ASK1 and ATRA in adult neural progenitors. |
| ETF, CP1 | Nuclear factor/ CCAAT binding factor | 58 | N | 6 | GGGCGG | Transcriptional activator of genes with TATA less promoter |
Among the hypermethylated genes prevented by alcohol, Crabp1 is involved in retinol metabolism, and Wnt16 is involved in Wnt pathways. These genes are key to neural differentiation and neural patterning. Alcohol also affected programmed DNA methylation of a number of genes related to neural phenotype expression. Interestingly, many are related to transmitter receptors, sensory receptors, and an ion channel. The glutamate receptor AMPA (Gria3) and its interacting protein (Grip1), adrenergic receptor alpha 1a (Adra1a), cholinergic receptor muscarinic 1 (Chrm1), and potassium voltage-gated channel (Kcna1) are major receptors/channels key in neuronal transmission, communication, and signal transduction. In addition, of the 19 hypomethylated genes during differentiation prevented by alcohol, it is remarkable that > 50% them are sensory receptor genes specific to olfaction (Olr214, Olr304, Olr408, Olr611, Olr1646, Olr1139) and taste (Tas2r123, Tas2r7, T2r140, T2r18). It was remarkably, since the NSCs in this study are sensory in origin and may share common propensity, , that alcohol targeted the programmed DNA methylation of many sensory receptors. Our previous study in embryo development also revealed that the DNA methylation of a large number of (>100) olfactory genes was altered by alcohol exposure in a similar dose (Liu et al., 2009). These two studies point to a general effect of alcohol on DNA methylation in sensory reception genes, particularly olfaction. It is interesting that the findings also indicated that after prenatal alcohol exposure, a major defect was found in olfactory bulb development (Kirstein et al., 1997, Maier and West, 2001, Abate et al., 2002). The major alteration of the DNA methylation program may provide a mechanism for such neurodevelopmental deficit (e.g. sensory deficits) in FASD. Although the consequence of the methylation on DRG gene expression is yet to be verified, the function of the DNA methylation change was demonstrated by us and others by treating NSCs with 5-azacytidine (5-AZA), which inhibits DNA methyl transferase, and the new DNA-methylation at the time of differentiation (Matsuda, 1990, Singh et al., 2009b). Retardation of migration, process growth, and key neural phenotype expression were evident. The necessity of methylation programming during embryonic development was also demonstrated by 5-AZA treatment and methionine suppression (methyl donor) resulting in growth retardation and neural tube defect (Dunlevy et al., 2006). Alcohol, known to affect methyl donors, also interferes with the DNA methylation program at early NSC differentiation. In conclusion, using neural stem cells for high-throughput analysis of genome-wide methylation, we demonstrated, for the first time, that alcohol prevented moderately methylated genes from hyper- as well as hypomethylation and affected migration and differentiation of neural stem cells. These results indicate that alcohol alters epigenetic programming while affecting stem-cell-ness and its differentiation. The temporal effect of the alcohol-modified DNA methylation on gene transcription and its causal effect on dysregulation of stem cell development remains under investigation.
METHODS
Neural Stem Cell Culture and Alcohol / AZA Treatments
This study utilized adult DRG-derived neural stem cells (NSCs) that were previously established in our laboratory (Singh et al., 2009). The line of DRG-NSC, among others, have been screened for sustained longevity and stability using non-passaged method (Zhou & Chiang, 1998). The multipotency and stability has been tested in these screened neural stem cells (Zhou et al., 2000; Singh et al., 2009a), and the epigenetic profile has been characterized (Singh et al., 2009b). We have raised both adult and embryonic DRG NSCs. After characterization, we found they are extremely similar in multipotency, renewability, capability of migration, and phenotype expression (Singh et al., 2009a). The adult DRG-NSCs were maintained in Dulbecco's Modified Eagle Medium / F-12 Nutrient Mix (D-MEM/F-12) media containing N2 supplement (12μL/mL, Invitrogen, Carlsbad, CA), and penicillin-streptomycin (12μL/mL, Sigma, St. Louis, MO) and grown in a humidified incubator at 37°C and 5% CO2. Media was supplemented with 10ng/mL epidermal growth factor (EGF, Harlan Bioproducts for Science, Indianapolis, IN) and basic fibroblast growth factor twice per week (bFGF, PeproTech, Rocky Hill, NJ) for maintenance of NSCs in neurosphere form. During the medium changes throughout the years, no passaging (trypsin digestion and cell transferring, except dividing into multiple flask) was performed. All neurospheres for analyses were screened by size <1mm shape (round neurosphere) for their robust growth.
The undifferentiated and differentiated neurospheres were defined previously (Singh et al., 2009). In brief, the undifferentiated neurospheres were obtained from the above culture medium minus EGF+bFGF for two days. The neurospheres allowed to differentiate were withdrawn from the EGF+bFGF supplementation and equilibrated for 5-10 minutes in Neurobasal media (no supplements) before being plated into a 10 mm plastic Petri dish (for MeDIP analysis) or in 16-well chamber slides (Nunc, Rochester, NY; for immunocytochemistry), both coated with poly-D-lysine (50μg/mL, Sigma, St. Louis, MO) and laminin (50μg/mL, Sigma). Neurospheres were allowed to differentiate for 2 (for MeDIP or morphological analysis) or 4 days (for morphological analysis only) in culture (2 or 4 DIC) with differentiation media consisting of Neurobasal media supplemented with 10% fetal bovine serum, 1.2% B27, and 1.2% penicillin-streptomycin.
The differentiating neurospheres with the paradigm described above were divided into two groups 18 hrs after initial plating (to allow attachment) — receiving no additions (Differentiating Control group) or alcohol (ALC, 400mg/dL for 6 hrs; Differentiating ALC group). A study with 5-aza-cytidine (AZA, DNA-methylation inhibitor, 50ng/mL, Sigma, St. Louis, MO; Differentiating AZA group) treatment was done previously (Singh et al., 2009b). At the end of the differentiation period, the undifferentiated and differentiating cells were (a) collected at 2 DIC by scraping all the cells from the Petri dish for MeDIP analysis, or (b) washed with 0.1M PBS at 2 or 4 DIC and fixed with the 4% formaldehyde fixative in a chamber slide for immunocytochemical analysis of phenotypes.
Immunocytochemistry and Cellular Markers
Double-staining was done to confirm the phenotype and degree of neuronal differentiation. The pluripotent stem cell marker, OCT4 (goat, 1:400, Santa Cruz Biotechnology), was stained to assess retention of pluripotency. Neurons were identified by monoclonal antibody against microtubule associated protein a, b, and c (MAP2, Sigma, 1:500; detecting immature and mature neurons). DNA methylation was investigated using antibodies against 5-methyl-cytosine (5-MeC, goat-anti-5-MeC, 1:250, GeneTex, San Antonio, TX). Changes in expression of the enzyme responsible for DNA methylation, DNA methyltransferase (DNMT), were immunostained using polyclonal antibodies against DNMT1 (de novo DNA methylation; made in goat, 1:200, Santa Cruz Biotechnology, Santa Cruz, CA). Endogenous peroxide was quenched with 3% H2O2 and 1% Triton X-100 was applied for 30 minutes to permeabilize the cell membranes. For 5-MeC staining, cells were incubated with 2N HCl. Non-specific binding was blocked using 4% normal serum from the species used to make the secondary antibody, plus 0.1% Tx-100 in PBS. Primary antibodies (detailed above) were incubated overnight at room temperature in the blocking buffer corresponding to the species of the secondary antibody. For immunofluorescent staining, the NSCs were washed with PBS and incubated with an Alexa 488 or a 635 fluorophor conjugated secondary antibody and counterstained with 4', 6-diamidino-2-phenylindole (DAPI- A-T-specific DNA stain at 350nm wavelength, Invitrogen). Staining of undifferentiated NSCs was done in a 1.5mL Eppendorf centrifuge tube following the procedures above.
Analysis of Immunostaining Intensity
A semi-quantitative measurement of immunostaining for 5-MeC and DNMT was performed according to the methods described previously (Singh et al., 2009b).
Since the sizes of neurospheres were not all identical, the migration density of cells (i.e. number of differentiated cells per neurosphere) would be affected by this variable. We thus did not analyze the epigenetic marks by cell number but rather the neurosphere was divided into three regions: core, corresponding to the densely packed inner mass of tightly clustered cells; periphery, a zone stretching out approximately 100μm from the edge of the core, consisting of early differentiating cells that were less densely packed, starting to leave the neurosphere, changing morphologically from round neuroepithelial cells to tear-drop shaped neural-like cells, and some at the outer edge of the periphery with extended neurites; and the migrated zone, consisting of all cells beyond the periphery, which have varying degrees of differentiation. Furthermore, since all cells in the neurosphere were not differentiated equally, the intensity of the epigenetic marks and phenotype expression were individually different, which are hypothesized to correlate with their differentiation status. Thus, it is reasonable that we analyze the population of cell intensity, particularly along the axis of their migration path.
For imaging, all pictures were taken using a Leitz microscope interfaced with a Spot RT color camera. Fluorescent intensity observations were determined independently by consensus of two analyzers based on the following criteria. To verify the intra-cellular and intra-nuclear distribution of the epigenetic marks, we have examined the cells under both fluorescent- and phase-contrast bright-field microscopy, alternatively or simultaneously (with dimmed bright-field). Positive stained neurospheres were stained with 5-MeC (Normal, N=12; Alcohol, N=10) or DMNT1 (Normal, N=12; Alcohol N=10), with fluorescent intensity compared to the dark background. Based on the rank values, Mann-Whitney U was used for nonparametric statistical analysis (StatView, Cary, NC) of ranked fluorescent intensity to comparing each neurosphere compartment to the undifferentiated neurospheres.
Cell Migration Assay
Quantitation of the effect of alcohol and 5-AZA on migration of differentiated NSCs was conducted in comparison to Controls. Fluorescent or bright-field images taken at 10x magnification were overlayed with a 1×1-inch grid using Adobe Photoshop software (Adobe Systems, Boston, MA). The neurosphere was divided into three regions: core, corresponding to the densely packed inner mass of tightly clustered cells; periphery, a zone stretching out approximately 100μm from the edge of the core, consisting of early differentiating cells that were less densely packed, starting to leave the neurosphere, changing morphologically from round neuroepithelial cells to angular shaped neural-like cells, and some at the outer edge of the periphery with extended neurites; and the migrated zone, consisting of all cells beyond the periphery, which have varying degrees of differentiation. The cells in the migrated zone were counted in 8 control-differentiated and 10 alcohol-treated-differentiated neurospheres. Positive cells were stained with 4', DAPI or had a clearly distinguishable cytoplasm in brightfield from their nearest neighbor. Tightly clustered or multi-layered areas were excluded from analysis. Statistical analyses, T-tests, were done using StatView (SAS, Carey, NC).
DNA Methylation Immunoprecipitation (MeDIP) Assay
A total of 9 samples (Undifferentiated cells without treatment, differentiated cells without treatment, and differentiated cells with alcohol treatment, n=3 for each) were used for MeDIP analysis. Genomic DNA was extracted from the fresh cells immediately after the culture by using a DNeasy blood and tissue kit (Qiagen, Fremont, CA). Briefly, approximately 5×106 cells from each sample were centrifuged to a cell pellet, resuspended in PBS and lysed with Proteinase K. After lysis, DNA was precipitated with 100% ethanol, washed with buffers and eluted in an elution buffer according to the manufacturer's instructions. The DNA quality and quantity was assessed using a Nanodrop spectrophotometer with A260 ratio >1.7 and A230>1.6 considered to be a criteria for quality control. Approximately, 1.5 μg of genomic DNA in 150μl of the buffer from each sample was sonicated using a Branson sonifier to obtain fragment sizes between 200-1000 bp (verified on 2% agarose gel). A 25 μl of sonicated DNA from each sample was kept as Input DNA and the rest of the 100μl was used for Immunoprecipitation with 5-methylcytosine antibody using a Methyl capture kit (Epigentek, Brooklyn, NY). The 10ng of input DNA and immunoprecipitated DNA was amplified using a Sigma WGA2 Genome Plex kit (Saint Louis, MO) and purified with Qiaquick PCR purification kit (Qiagen, Fremont, CA). All samples were further subjected to quality control analyses by the Nimblegen core facility (Reykjavik, Iceland) and then labeled with Cy3 (input DNA) and Cy5 (immunoprecipitated DNA) dyes. Labeled input and immunoprecipitated samples were both hybridized to the same RN34 Promoter plus CpG island microarray (described below) as a two-color experiment and scanned using Nimblescan. The signal intensities for input DNA and immunoprecipitated DNA were used to calculate the ratio of IP/Input DNA.
Microarray
NimbleGen (080212 RN34 CpG island Pro, Madison, WI) promoter plus CpG island tiling array has a total density of 385,000 probes that represent 19,530 rat promoters and CpG islands per slide. In this array platform, each probe contains ~50 oligonucleotides with ~50 bp gaps within the promoter and CpG island regions. These promoters and CpG islands largely overlap with transcription starting sites of RefSeq genes, covering, by average, 1.3-kb upstream and 0.5-kb downstream of the transcription starting site of 21632 transcripts. In order to avoid the biases due to false assigned promoters in intergenic regions, our analysis mainly focused on these well-annotated promoters.
BioInformatics Analysis
Both average-level and probe-level analyses were performed. In the average-level analysis, we focused on the regulatory regions near the transcription start sites (from 1,300-bp upstream and 500bp downstream of the transcription start sites). This approach avoided noise caused by distal oligonucleotides residing in the upstream intergenic regions and provided a more focused analysis of the region where most transcription factors bind, while allowing our results to be compared with other studies focused on human promoters (Weber et al., 2007).
The probe-level analysis was extended to the entire gene-related promoter and CpG island regions covered by the NimbleGen array, from 1,300 bp upstream to 500 bp downstream of the transcription start site, which ensured characterization of methylation patterns in distal regulatory regions without diluting the signals in the core promoter with increased noise levels. Since sheared DNA fragments ranged from 200-1,000 bp, and each probe covers approximately 50 bp, our conservative analysis used two consecutive probes on the NimbleGen CpG island array (covering 150-200 bp). Using a linear mixed model, we identified DNA regions with increased/decreased methylation levels based on the signal intensities of two consecutive probes between control and alcohol-treated samples. Depending on the NimbleGen probe selection, each promoter or CpG Island contained more than one consecutive probe pair. On average, there were 17 probe pairs for each gene. We identified genes that contained no less than one DNA region (two consecutive probes) with increased/decreased methylation levels within 1,300 bp upstream and 500 bp downstream of transcription start sites.
The effect of CpG methylation is likely to extend beyond the core of promoter regions (Mikkelsen et al., 2007, Weber et al., 2007), and core promoter regions from 500 bp upstream and 200 bp downstream of the transcription start site were also analyzed. A summarized methylation level was calculated for each gene based on the average signal intensity of all the probes within the gene's regulatory region, from -1,300 bp to +500 bp (Average level analysis). We considered promoters with DNA methylation levels higher than 1.3-fold of the genome-wide median methylation level as hypermethylated promoters. This threshold is similar to that used in the MeDIP-chip analysis of human fibroblast. Promoters with DNA methylation levels lower than 1.3-fold of the genome-wide median methylation level were considered hypomethylated (Weber et al., 2007).
Ingenuity Pathway Analysis was used to identify the common function and pathways among the genes that showed significant change in DNA methylation by alcohol. Ingenuity uses a curated database to predict the functions, canonical pathways, and networks from the Ingenuity Knowledge base (IPKB). Fischer's exact test was used to identify the significant functions and pathways relevant to the genes.
Sequenom Mass ARRAY EpiTYPER Methylation Detection
Selected genes with both average difference and two-consecutive differences were further validated with Sequenom Mass ARRAY. Epityper DNA methylation analysis is based on bisulfite conversion of DNA, PCR amplification, followed by in vitro transcription and analysis of cleaved products by Mass Spectrometry (MALDI-TOF). The primers were designed using MethPrimer (www.urogene.org/methprimer) for 26 gene promoter regions with 48 amplicons (103-625bp with median target length of 400bp) covering, on average, 9 CpGs per amplicon. In brief, DNA was isolated as described above for MeDIP analysis from differentiated DRG cells with or without alcohol treatment (n=1 each). The methylation detection was carried out in the Sequenom facility in San Diego, CA. Approximately 1 μg of DNA from each sample was treated with sodium bisulfite using the EZ DNA methylation kit (ZymoResearch, CA) according to the manufacturer's protocol, and amplification was done using the primers with the T7 promoter tag on the reverse primer. The in vitro transcription was done, followed by site-specific cleavage using MassCLEAVE biochemistry. MassARRAY compact MALDI-TOF mass was used to acquire spectral peaks and converted into methylation ratios using EpiTYPER software72 (Ehrich et al., 2005). Two statistical criteria were used to identify genes validated by Sequenom; one with genes showing a methylation difference of greater than 10% in at least one CpG unit; furthermore, another criteria was the methylation change in the same direction of all CpG units tested for that gene amplicon (Liu et al. 2009).
Supplementary Material
SUPPLEMENTARY MATERIALS
Supplemental Table 1. Table showing genes with hypermethylation prevented by alcohol from moderate methylation during differentiation.
Supplemental Table 2. Table showing genes with hypomethylation prevented by alcohol from moderate methylation during differentiation.
ACKNOWLEDGEMENTS
We thank Dr. Bruce Anthony for his assistance in part of the stem cell culture and Yuanyuan Chen for her assistance in preparing figure images. This study is supported by NIH grant AA016698 to FCZ, with YL and KPN as co-investigators.
REFERENCES
- Abate P, Varlinskaya EI, Cheslock SJ, Spear NE, Molina JC. Neonatal activation of alcohol-related prenatal memories: impact on the first suckling response. Alcohol Clin Exp Res. 2002;26:1512–1522. doi: 10.1097/01.ALC.0000034668.93601.8F. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS. Epigenetics: role of germ cell imprinting. Advances in Experimental Medicine and Biology. 2003;518:239–245. doi: 10.1007/978-1-4419-9190-4_21. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Tavazoie M, Doetsch F. Stem cells: from epigenetics to microRNAs. Neuron. 2005;46:363–367. doi: 10.1016/j.neuron.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Cravo ML, Camilo ME. Hyperhomocysteinemia in chronic alcoholism: relations to folic acid and vitamins B (6) and B (12) status. Nutrition (Burbank, Los Angeles County, Calif. 2000;16:296–302. doi: 10.1016/s0899-9007(99)00297-x. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Kim S, Redondo JM, Walsh C, Nieto M. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Dev Neurobiol. 2008;68:917–925. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlevy LP, Burren KA, Chitty LS, Copp AJ, Greene ND. Excess methionine suppresses the methylation cycle and inhibits neural tube closure in mouse embryos. FEBS Lett. 2006;580:2803–2807. doi: 10.1016/j.febslet.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric Research. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC. Fetal Alcohol Spectrum Disorders: The Epigenetic Perspective. Biol Reprod. 2009 doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminen-Ahola N, Ahola A, Maga M, Mallitt KA, Fahey P, Cox TC, Whitelaw E, Chong S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genetics. 2010;6:e1000811. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JC. Epigenetics in development. Dev Dyn. 2007;236:1144–1156. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- Kondo T. Epigenetic alchemy for cell fate conversion. Curr Opin Genet Dev. 2006;16:502–507. doi: 10.1016/j.gde.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kirstein CL, Philpot RM, Dark T. Fetal alcohol syndrome: early olfactory learning as a model system to study neurobehavioral deficits. Int J Neurosci. 1997;89:119–132. doi: 10.3109/00207459708988467. [DOI] [PubMed] [Google Scholar]
- LaBaume LB, Merrill DK, Clary GL, Guynn RW. Effect of acute ethanol on serine biosynthesis in liver. Archives of Biochemistry and Biophysics. 1987;256:569–577. doi: 10.1016/0003-9861(87)90614-x. [DOI] [PubMed] [Google Scholar]
- Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell Mol Life Sci. 2007;64:947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalande M, Minassian BA, DeLorey TM, Olsen RW. Parental imprinting and Angelman syndrome. Adv Neurol. 1999;79:421–429. [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4 doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Progress in Neurobiology. 2009;88:170–183. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Maier SE, West JR. Regional differences in cell loss associated with binge-like alcohol exposure during the first two trimesters equivalent in the rat. Alcohol. 2001;23:49–57. doi: 10.1016/s0741-8329(00)00133-6. [DOI] [PubMed] [Google Scholar]
- Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol. 2005;35:235–241. doi: 10.1016/j.alcohol.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Matsuda M. Comparison of the incidence of 5-azacytidine-induced exencephaly between MT/HokIdr and Slc:ICR mice. Teratology. 1990;41:147–154. doi: 10.1002/tera.1420410204. [DOI] [PubMed] [Google Scholar]
- McClellan KA, Vanderluit JL, Julian LM, Andrusiak MG, Dugal-Tessier D, Park DS, Slack RS. The p107/E2F pathway regulates fibroblast growth factor 2 responsiveness in neural precursor cells. Mol Cell Biol. 2009;29:4701–4713. doi: 10.1128/MCB.01767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merot Y, Ferriere F, Debroas E, Flouriot G, Duval D, Saligaut C. Estrogen receptor alpha mediates neuronal differentiation and neuroprotection in PC12 cells: critical role of the A/B domain of the receptor. J Mol Endocrinol. 2005;35:257–267. doi: 10.1677/jme.1.01826. [DOI] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histology and Histopathology. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D, MicroRNAs Master Regulators of Ethanol Abuse and Toxicity. Alcohol Clin Exp Res. 2010 Jan 26; doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010 Jan;67(1):73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. Semin Cell Dev Biol. 1999;10:189–195. doi: 10.1006/scdb.1999.0300. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008 Mar-Apr;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009 Sep;33(9):1615–27. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008 Apr 2;28(14):3729–37. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny A, Chen J, Ticku MK, Yan B, Henderson G. The site specific demethylation in the 5'-regulatory area of NMDA receptor 2B subunit gene associated with CIE-induced up-regulation of transcription. PLoS One. 2010 Jan 20;5(1):e8798. doi: 10.1371/journal.pone.0008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi H, Namihira M, Kohyama J, Asano H, Sanosaka T, Nakashima K. Methyl-CpG binding proteins are involved in restricting differentiation plasticity in neurons. J Neurosci Res. 2006;84:969–979. doi: 10.1002/jnr.21001. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zoghbi HY. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am J Hum Genet. 2002;71:1259–1272. doi: 10.1086/345360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008 Sep;32(9):1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Singh RP, Cheng YH, Nelson P, Zhou FC. Retentive multipotency of adult dorsal root ganglia stem cells. Cell Transplantation. 2009a;18:55–68. doi: 10.3727/096368909788237177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Shiue K, Schomberg D, Zhou FC. Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplantation. 2009b;18:1197–211. doi: 10.3727/096368909X12483162197204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Developmental Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tang HL, Zhu JH. Epigenetics and neural stem cell commitment. Neuroscience bulletin. 2007;23:241–248. doi: 10.1007/s12264-007-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Huang H, Geiman TM, Lim CY, Fu L, Qiu GH, Robertson KD. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet. 2002;11:2091–2102. doi: 10.1093/hmg/11.18.2091. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- Vanderluit JL, Ferguson KL, Nikoletopoulou V, Parker M, Ruzhynsky V, Alexson T, McNamara SM, Park DS, Rudnicki M, Slack RS. p107 regulates neural precursor cells in the mammalian brain. The Journal of Cell Biology. 2004;166:853–863. doi: 10.1083/jcb.200403156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Xin Z, Tachibana M, Guggiari M, Heard E, Shinkai Y, Wagstaff J. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J Biol Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Anderson TA. MicroRNA: a new player in stem cells. J Cell Physiol. 2006;209:266–269. doi: 10.1002/jcp.20713. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Chiang YH. Long-term nonpassaged EGF-responsive neural precursor cells are stem cells. Wound Repair & Regeneration. 1998;6:337–348. doi: 10.1046/j.1524-475x.1998.60409.x. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Kelley MR, Chiang YH, Young P. Three to four-year old nonpassaged EGF-responsive neural progenitor cells: proliferation, apoptosis and DNA-repair. Exp Neurol. 2000;164:200–208. doi: 10.1006/exnr.2000.7425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIALS
Supplemental Table 1. Table showing genes with hypermethylation prevented by alcohol from moderate methylation during differentiation.
Supplemental Table 2. Table showing genes with hypomethylation prevented by alcohol from moderate methylation during differentiation.