Abstract
According to the histone code hypothesis, histone variants and modified histones provide binding sites for proteins that change the chromatin state to either active or repressed. Here, we identify histone variants that regulate the targeting and enzymatic activity of poly(ADP-ribose) polymerase 1 (PARP1), a chromatin regulator in higher eukaryotes. We demonstrate that PARP1 is targeted to chromatin by association with the histone H2A variant (H2Av)—the Drosophila homolog of the mammalian histone H2A variants H2Az/H2Ax—and that subsequent phosphorylation of H2Av leads to PARP1 activation. This two-step mechanism of PARP1 activation controls transcription at specific loci in a signal-dependent manner. Our study establishes the mechanism through which histone variants and changes in the histone modification code control chromatin-directed PARP1 activity and the transcriptional activation of target genes.
Keywords: poly(ADP-ribosyl)ation, poly(ADP-ribose) glycohydrolase, nucleosome, Hsp70
Activation of the poly(ADP-ribose) polymerase 1 (PARP1) protein in chromatin in response to developmental and environmental stress is known to mediate changes in chromatin and stimulate transcription (1–4). Although PARP1 protein is abundant in the nucleus, its localization in chromatin is not uniform and is restricted to promoters of a specific subset of genes (5, 6). The histone modification code hypothesis suggests that nucleosomal code could control positioning in chromatin and the activation of effector proteins such as PARP1 (7, 8). Indeed, PARP1 protein interacts with histones, and the N-terminal domains of histones regulate activation of PARP1 enzymatic reaction (9).
A potential functional relationship between PARP1 protein activation and histone H2A variant H2Ax phosphorylation has been suggested by a substantial body of experimental research (10–13). These observations suggest that interaction between PARP1 and H2Ax may be a general mechanism for PARP1 regulation. In addition to H2Ax, most eukaryotes have another histone H2A variant, H2Az, which is highly homologous to H2Ax (14). Although a direct relationship between PARP1 and H2Az has not yet been demonstrated, these proteins display a strong functional connection. Similar to PARP1 protein (3, 5), H2Az has been implicated in the maintenance of silencing (15, 16), transcriptional activation (17), and maintenance of cell viability (18).
In this study, we examined the relationship between the H2Ax/H2Az pathway and PARP1 regulation using Drosophila as a model. Drosophila has a single H2A variant, H2Av, which combines specific features of the two mammalian homologs, H2Ax (the SQ phosphorylation domain) and H2Az (the amino-terminal tail) (19, 20). The role of H2Av in PARP1 regulation is supported by a considerable body of evidence. For example, both PARP1 (5) and H2Av (16) are involved in heterochromatin formation and maintenance. Both proteins demonstrate a nonuniform pattern of chromatin association, localizing to thousands of discrete euchromatic loci as well as to heterochromatin (3, 19). Recent studies report that the Drosophila histone H2Av binds specifically to active chromatin (21), indicating a likely functional linkage between PARP1 and H2Av. Histone H2Av is enriched in heat shock (19) and in other “open chromatin” loci where the PARP1 protein is responsible for chromatin decondensation and transcriptional induction of numerous genes, including heat-shock protein 70 genes (hsp70) (1, 2). These facts allow us to propose and test the hypothesis that histone H2Av and PARP1 protein interact in chromatin and that phosphorylation of H2Av triggers changes in the local chromatin environment, which, in turn, stimulate PARP1 enzymatic activity and PARP1-dependent transcription.
Results
Histone H2Av Is Required for PARP1 Protein Activation and Localization in Chromatin.
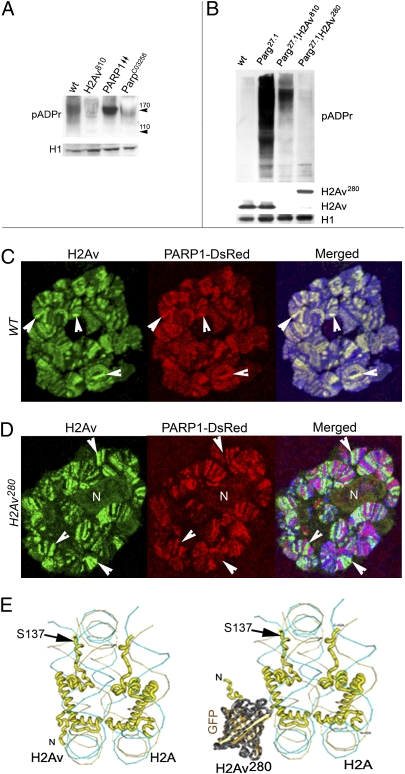
Similar to Parp1C03256 (22) and poly(ADP- ribose) glycohydrolase (Parg27.1) mutants (23), the H2Av-null mutant (16) (H2Av810) has late lethality, with mutant homozygous animals arrested in development in late third-instar larvae stage or as prepupae. The level of poly(ADP ribosyl)ation (pADPr) in the H2Av-mutant Drosophila is reduced significantly compared with the level in WT animals but is similar to that of mutant Parp1C03256 Drosophila (Fig. 1A), supporting the idea that PARP activity may be regulated by H2Av. Precise measurement of pADPr levels in WT cells is complicated by the abundance of PARG protein, which rapidly cleaves pADPr in vivo as well as during protein extract preparation. Therefore, to measure pADPr levels accurately, we performed assays in the absence of endogenous PARG. In this way, total pADPr could be compared precisely among flies of different genotypes (23). As expected, pADPr accumulates in a greater quantity in Parg-mutant animals than in WT animals at the same developmental stage (Fig. 1B). However, in double-mutant animals, which, in addition to the Parg mutation, carry null mutation of H2Av810, accumulation of pADPr is suppressed (Fig. 1B). The dominant mutation, H2AvG00280 (H2Av280) (24), in the H2Av locus, caused by GFP-exon insertion in the N-terminal tail of this histone, also severely reduces the amount of pADPr (Fig. 1B).
Fig. 1.
Histone H2Av is required for PARP1 protein activation and localization in nuclei. (A) Mutating H2Av decreases pADPr in vivo. Equal amounts of total nuclear protein extracts from WT, H2Av810 overexpressing PARP1 (PARP1↑↑), and ParpC03256 prepupae were analyzed after PAGE on Western blot using anti-pADPr antibody. Histone H1 (H1) was used as a loading control and was detected by the mAb antibody. (B) A comparative analysis of PARP1 protein activity of the WT, parg27.1, parg27.1;H2Av810, and parg27.1;H2Av280 mutant third-instar larvae. To detect pADPr and proteins on a Western blot, the following antibodies were used: mAb 10H against pADPr; pAb against H2Av (gift of R. Glaser, Division of Genetic Disorders, Wadsworth Center, Albany, NY), and mAb against histone H1 (loading control). (C and D) PARP1 protein colocalizes with histone H2Av in Drosophila nuclei in WT (C). Mutating H2Av by insertion of GFP protein into the N-terminal tail disrupts PARP1 and H2Av colocalization (D). A single larval salivary gland nucleus is shown for each sample. To detect H2Av (green), pAb antibody staining was used. PARP1-DsRed protein was detected by DsRed autofluorescence (red). DNA was stained with Draq5 dye (blue). Arrowheads indicate regions of protein colocalization (C) and antagonistic localization (D) in H2Av280 mutants. N, nucleolus. (E) The structures of H2Av and H2Av280 are compared in the context of the nucleosomes. To reconstruct the 3D structure of the nucleosome, we used Cn3D 4.1 software (the National Center for Biotechnology Information) and structural information about core histones and nucleosomes from the National Center for Biotechnology Information database. DNA is indicated by a “wire-like” structure. H2Av is shown in yellow. The GFP insertion in H2Av280 is shown in brown. Other histones are hidden in this diagram. The SQ phosphorylation site of the C-terminal domain of H2Av is indicated by the black arrow.
These results show that the presence of intact histone H2Av is crucial for the regulation of enzymatic activity of PARP1 protein in vivo. Therefore we tested whether these two proteins colocalize in chromatin. Indeed, we found that histone H2Av colocalizes with PARP1 in WT nuclei (Fig. 1C and Fig. S1). In contrast, mutating H2Av by inserting the GFP exon disrupts PARP1 and H2Av colocalization (Fig. 1D and Fig. S1). Because the GFP “barrel” in H2Av280 mutants blocks PARP1 and H2Av colocalization, these results suggest that PARP1 localization is mediated by its interaction with H2Av-containing nucleosomes and that H2Av may regulate precise PARP1 localization along specific genes.
Histone H2Av Is Colocalized with PARP1 Protein in the hsp70 Promoter Region.
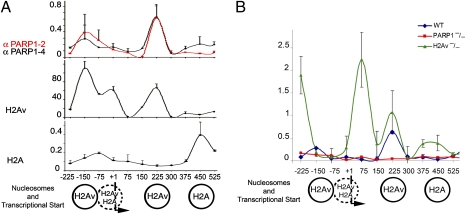
Previously, we found that transcriptional activation of hsp70 genes upon heat shock is totally PARP1 dependent and that it is abolished in Parp mutants or in animals treated with PARP1 inhibitor (1). Therefore, by using a ChIP assay, we determined the localization of PARP1 and H2Av in the chromatin hsp70 locus. Both proteins localize in hsp70 chromatin (1, 19) and have the same dynamics upon heat-shock treatment (Fig. S1). Moreover, we found that PARP1 protein and H2Av, but not histone H2A, colocalize precisely in the promoter region of the hsp70 locus in WT animals (Fig. 2A). Finally, depletion of H2Av in H2Av-null mutants (H2Av810) was shown to disrupt proper localization of PARP1 protein. Furthermore, ChIP analysis detected mislocalization and overaccumulation of PARP1 along the hsp70 promoter (Fig. 2B). The general profile of PARP1 protein distribution was altered. Specifically, in addition to positioning on the −150, + 1, and +225 nucleosomes (Fig. 2), PARP1 protein overaccumulated on the −225, +75, +375, and +450 sites (Fig. 2B). Even though H2Av regulates the precise localization of PARP1 protein in chromatin, this finding demonstrates that the absence of H2Av does not, in general, disrupt PARP1 interaction with chromatin and that mislocalization of PARP1 may not be the only reason for the blockage of PARP1 activation in H2Av-null mutants. Therefore, we propose that alterations in chromatin coupled to H2Av or posttranslational modifications of H2Av mediate PARP1 activation.
Fig. 2.
Histone H2Av is colocalized with PARP1 protein in the hsp70 promoter. (A) ChIP assay reveals PARP1 protein and histone H2Av colocalization in the promoter of Drosophila hsp70 locus. The ChIP assay was performed using anti-Drosophila melanogaster PARP1 (anti-DmPARP1) antibody (α-PARP1/2; α-PARP1/4) (SI Experimental Procedures) in addition to anti-H2Av and anti-H2A histones (gift of R. Glaser). Nucleotide positions are shown according to hsp70 transcriptional start (+1). (B) ChIP assay reveals PARP1 protein mislocalization and overaccumulation in H2Av810 mutants in the promoter of hsp70 locus (green) compared with WT (blue). ParpC03256 mutants were used as a negative control (red). The ChIP assay was performed using anti-DmPARP1 antibody (α-PARP1/2).
Phosphorylation of H2Av Controls PARP1 Activation in Vivo.
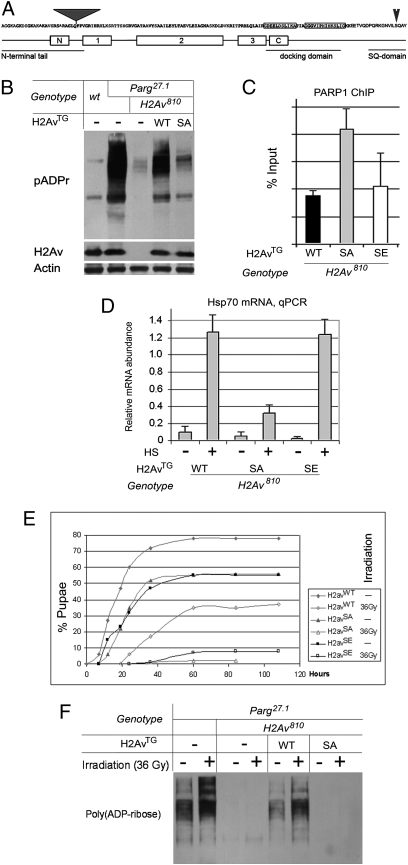
Phosphorylated H2Ax colocalizes with pADPr in the DNA repair pathway and during spermatogenesis (11, 12), raising the possibility that phosphorylated H2Ax may promote PARP activation. Drosophila H2Av contains a phosphorylation domain homologous to the SQ domain of H2Ax (Fig. 3A). It also has been shown that H2Av is phosphorylated in the same enzymatic pathways as mammalian histone H2Ax (25). In Drosophila, phosphorylated histone H2Av localizes to multiple sites, including developmental loci 75A–C (26), where we previously demonstrated a high level of pADPr (1). Therefore, we hypothesized that H2Av phosphorylation may trigger activation of PARP1.
Fig. 3.
Phosphorylation of H2Av controls PARP1 activation in vivo. (A) Protein sequence of Drosophila H2Av protein is shown. Positions of the N-terminal tail, docking domain, phosphorylation SQ domain, and the positions of the GFP insertion in H2Av280 (gray triangle) and the Ser137 residue, which could be phosphorylated, are shown. Boxes identified as “N,” “1,” “2,” “3,” and “C” indicate conserved alpha-spiral regions. (B) Mutating the Ser137 phosphorylation site affects pADPr in vivo. pADPr and proteins were detected using the following antibodies: mAb anti-pADPr, pAb anti-H2Av, and mAb anti-actin. (C) Level of PARP1-ECFP protein binding in the hsp70 gene was compared with animals expressing WT, phospho-mutant (SA), and phospho-mimic (SE) H2Av. (D) Comparison of PARP1-dependent hsp70 gene transcriptional activation in animals expressing WT, phospho-mutant (SA), and phospho-mimic (SE) H2Av before (−) and after (+) 40 min of heat-shock treatment. The level of hsp70 mRNA was detected using quantitative RT-PCR. (E) Phosphorylation of H2Av is critical for viability of the flies after gamma-irradiation. H2Av810-mutant flies were “rescued” by expression of WT, phospho-mimic (SE), and phospho-mutant (SA) H2Av transgenes. Third-instar larvae were irradiated with a semilethal dose (36 Gy). The numbers of pupae were counted for different genotypes. A significant number of animals with WT and SE H2Av genotypes survive to pupal stage, but those with the SA H2Av genotype die at the larval stage. (F) H2Av phosphorylation controls PARP1 activation upon gamma-irradiation. Parg27.1 mutants, Parg27.1; H2Av810 double-mutant animals, and Parg27.1; H2Av810 animals “rescued” by expression of WT or phospho-mutant (SA) H2Av transgenes were irradiated with a semilethal dose (36 Gy) as third-instar larvae. Equal amounts of total protein extracts were analyzed after PAGE on Western blot using anti-pADPr antibody. PARP1 protein activation after gamma-irradiation is clearly detected in Parg27.1 and Parg27.1; H2Av810 animals expressing WT H2Av but is blocked by expression of the H2Av phospho-mutant SA isoform.
To test this hypothesis, we generated three transgenic Drosophila constructs: WT (H2Avwt); a phospho-mutant in which Ser137 is replaced by Ala residue (H2AvSA); and a construct mimicking constant phosphorylation, in which Ser137 is replaced by Glu residue (H2AvSE). Although H2Avwt rescued H2Av810-null mutants, which survived to adulthood, expression of H2AvSA or H2AvSE failed to rescue the H2Av810 mutants fully. Consistent with the observed rescue of H2Av810 mutants, pADPr accumulation was restored by expression of H2Avwt in Parg27.1;H2Av810 double-mutants (Fig. 3B). However, expression of H2AvSA did not restore accumulation of pADPr in the Parg27.1;H2Av810 mutant animals (Fig. 3B). Expression of the H2AvSE transgene was found to be lethal at the early embryonic stage for the Parg27.1;H2Av810 double-mutants, preventing quantification of pADPr levels for this genotype.
These results demonstrate that phosphorylation of H2Av is necessary for activation of PARP1 protein and synthesis of pADPr in vivo. To examine signal-dependent PARP1 activation, we measured PARP1-dependent transcriptional activation of the hsp70 locus in H2Av810 mutants bearing WT or phospho-mutant and phospho-mimic H2Av transgenes. Strikingly, mutating the SQ domain does not disrupt PARP1 protein localization in the promoter of hsp70 genes (Fig. 3C). However, H2AvSA mutants show no significant increase of hsp70 mRNA production upon heat shock and die upon heat-shock treatment, demonstrating a requirement for the SQ domain in PARP1-dependent transcriptional activation of hsp70 locus (Fig. 3D). In contrast, H2Avwt and H2AvSE mutants have the same level of hsp70 mRNA after heat shock (Fig. 3D). These data confirm our hypothesis that histone H2Av is sufficient for proper PARP1 protein localization, whereas the activation of PARP1 in vivo and the start of PARP1-dependent processes in chromatin are triggered by phosphorylation of histone H2Av.
Core H2Av Phosphorylation Is Required for Genotoxic Stress-Dependent PARP1 Activation and for the Recovery After Genotoxic Stress.
The role of PARP1 protein in protecting the genome from genotoxic stress, such as irradiation, has been known for a long time (27). Similarly, the presence of phosphorylated H2Av (γ-H2Ax) has been used for decades as an indicator of damaged genomic DNA (14). Here we found that histone H2Av phosphorylation also is required for the survival of animals after irradiation (Fig. 3E), suggesting that H2Av-mediated PARP1 activation may be a conserved mechanism. To test this possibility directly, we examined H2Av roles in PARP1 regulation during the genotoxic stress response using a PARG-mutant genetic background. As expected, pADPr accumulates in irradiated Parg-mutant animals because of PARP1 protein enzymatic activation (Fig. 3F). However, mutating either histone H2Av or the SQ domain of H2Av alone completely impairs pADPr accumulation after semilethal irradiation (Fig. 3F). Moreover, mutating the H2Av SQ domain leads to 100% larval lethality after gamma-irradiation (Fig. 3E). The results of these experiments indicate that PARP1 function during genotoxic stress response requires H2Av phosphorylation. These findings again indicate that H2Av and, more specifically, H2Av phosphorylation, is required not only for transcription-coupled PARP1 activation but also for regulation of PARP1 protein in the genotoxic stress response pathway.
H2Av280 Mutation Disrupts the PARP1-Dependent Silencing of Retrotransposable Elements.
Although PARP1 protein is known primarily as a regulator of genotoxic stress response and an activator of transcription, it also mediates transcriptional silencing (5). PARP1 and H2Av are both localized in silent chromatin (5, 16, 19). In Drosophila, PARP1 is required for silencing of heterochromatic repeated DNAs, such as the retrotransposable elements copia and gypsy (5, 22). It has been shown that enzymatic activation of PARP1 is not required to establish and maintain PARP1-dependent transcriptional silencing (5). To test if H2Av function is required for PARP1-mediated silencing, we examined the expression of copia and gypsy retrotransposons in H2Av810 mutants, in which PARP1 protein localization with chromatin is not disrupted (Fig. S1), as well as in H2Av280 mutants, in which PARP1 protein is displaced from a number of loci (Fig. S1). As expected, we found that H2Av810 mutants express a low level of retrotransposable mRNA (Fig. S2 A and B), but H2Av280 mutants overproduce mRNA of both retrotransposons (Fig. S2 A and B) and accumulate retroviral particles in the nucleoplasm (Fig. S2 C–E). Thus, we conclude that neither H2Av function nor H2Av-mediated PARP1 activation is required for PARP1-dependent transcriptional silencing, whereas mutating the N-terminal tail by GFP insertion of H2Av in H2Av280 displaces PARP1 from silent chromatin and leads to the desilencing of retrotransposable elements.
Activation of the PARP1 Protein Involves Two Nucleosomes, One Containing H2Av and One Containing H2A.
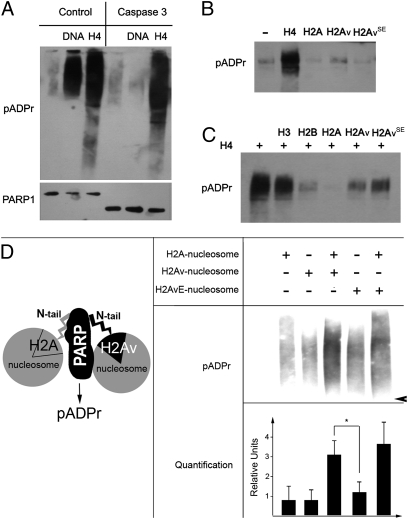
To understand what program of events might lead to H2Av-mediated PARP1 activation, we used an in vitro assay (Fig. 4). To avoid a complication arising from DNA-dependent PARP1 activation, the experiment used a form of PARP1 that lacks a Zn finger-bearing domain. PARP1 lacking Zn fingers can be activated by histones but not by DNA (Fig. 4A and Fig. S3). This characteristic allowed us to measure directly the effects of H2Av and H2Av-bearing nucleosomes on PARP1. Surprisingly, neither purified WT nor phospho-mimic forms of H2Av activated PARP directly (Fig. 4B). Instead, PARP activation was stimulated by histone H4 (H4) (Fig. 4B). Furthermore, because incubation of PARP with H2Av or H2A inhibited H4-mediated PARP activation to different degrees (Fig. 4C), it follows that H2Av is required in PARP activation but not because of direct stimulation of PARP1 activity. Instead, we propose that H2Av phosphorylation modifies nucleosomal structure, exposing the H4 domains for interaction with PARP.
Fig. 4.
Interaction with an H2Av-containing nucleosome complex activates PARP1 in vitro. (A) Interaction with the purified core histone H4 activates PARP1 in a DNA-independent manner. Full-length PARP1 protein (Left) and PARP1 protein cleaved by caspase-3 (Right) were preincubated with randomly broken DNA or core histone H4, followed by mixing with NAD. The products of PARP1 enzymatic activity, poly(ADP ribose), were detected after PAGE on a Western blot using the anti-pADPr antibody. These data clearly demonstrate that the DNA-binding domain of PARP1 (Zn fingers I and II) is not required for histone-dependent PARP1 activation. (B) Interaction with purified histones H2Av and H2AvSE does not activate PARP1 in vitro. (C) The inhibitory effects of H2A, H2Av, and H2AvSE on PARP1 in vitro: Purified core histone H2A completely inhibits H4-dependent PARP1 activation, but H2Av and H2AvSE do not block H4-dependent PARP1 activation. (D) PARP1 enzymatic activity is stimulated by H2Av-containing nucleosomes in the presence of an equimolar amount of the WT nucleosome. PARP1 protein was preincubated with different coregulators, followed by mixing with NAD. The products of PARP1 enzymatic activity, poly(ADP-ribose), were detected after PAGE on a Western blot using the anti-pADPr antibody and were quantified independently using the Image Quant Software Package. The arrowhead indicates the position of the 180-kDa protein.
To test this hypothesis, we used nucleosomes assembled from purified histones containing WT H2Av and phospho-mimic H2AvSE to activate PARP1. Given that H2Av represents only 20% of the total H2A protein in a cell, histones H2A and H2Av must be mixed in chromatin, and both may be required for PARP1 activation to occur. Consistent with the idea that both H2A- and H2Av-containing nucleosomes are important, pure mononucleosomes containing histone H2A, histone H2Av, or H2AvSE recombinant histones did not lead to any significant increase in pADPr (Fig. S4). In contrast, a strong stimulation of pADPr was detected after combining equimolar amounts of PARP1 and H2A- and H2Av-containing nucleosomes (Fig. 4D). These results indicate that activation of PARP1 involves at least two nucleosomes, one containing H2Av and another containing H2A. Mimicking phosphorylation in the H2Av nucleosome (H2AvSE) further stimulates PARP1 enzymatic activation: The level of pADPr increased by ∼26% relative to that detected after H2A/H2Av stimulation (Fig. 4D), suggesting that the phosphorylation of H2Av promotes PARP1 activation in vitro. This result corroborates our prior observation that PARP1 activation is triggered by phosphorylation of H2Av in vivo.
Discussion
The histone code hypothesis has long been accepted in the study of epigenetics, but there never has been a clear demonstration of the direct activation of an effector protein in response to changes in the histone environment. Here, we find that PARP1 activation and PARP1- mediated transcription depend on the regulation of a nucleosome's microenvironment, a mechanism that involves the phosphorylation of a histone variant. This result supports the histone code hypothesis, and the underlying work also reveals a mechanism for PARP1 activation that is functionally important for the regulation of transcription, response to genotoxic stress, and silencing.
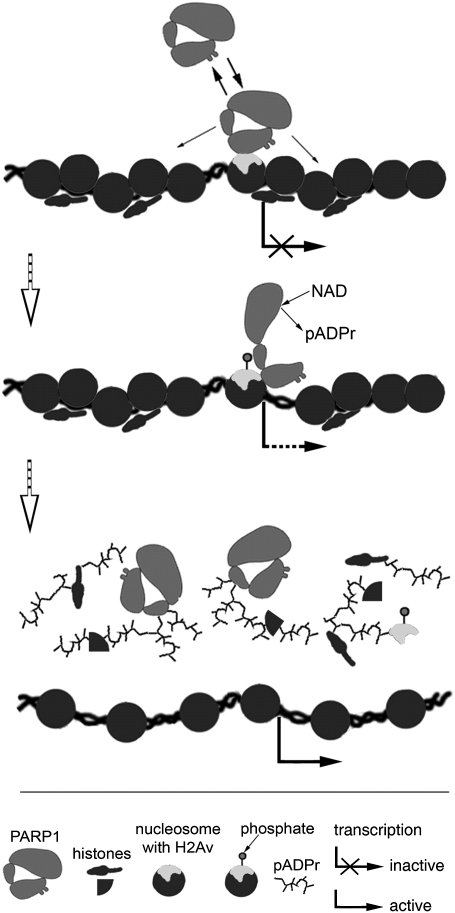
Histone variant H2Av in Drosophila (ref. 21 and this study)—and its homologs in Arabidopsis (28) and Saccharomyces cerevisiae (29)—localize in the promoter region of a subset of genes. Our findings demonstrate that this localization is functionally significant. Specifically, H2Av is involved in the positioning and activation of the PARP1 protein. Nucleosomes containing H2Av form high-affinity sites at which the effector protein PARP1 binds with specific promoters. Thereafter, phosphorylation of H2Av alters the interaction of PARP1 with the nucleosomal histone H4, an event which, in turn, activates PARP1, leading to chromatin opening (1) and facilitating transcription (Fig. 5) (1–3). Therefore, taken as a whole, the results of our study show that, by recruiting PARP1 protein, H2Av controls the chromatin state as well as transcription activation and genotoxic stress response.
Fig. 5.
Model of PARP1 protein regulation by histone H2Av variant and H2Av phosphorylation. (Top) H2Av-containing nucleosome forms a high-affinity binding site for PARP1 protein in chromatin. (Middle) Phosphorylation of histone H2Av in the content of nucleosome changes the conformation of the nucleosome and triggers PARP1 activation. (Bottom) The PARP1 protein modifies itself and surrounding proteins, thereby removing them from DNA and loosening chromatin locally to promote transcription.
Having established that H2Av controls the PARP1 function both in vivo and in vitro, we examined the causal underlying mechanism. Nucleosomes containing H2Av have been reported previously to have a “more open” and stable conformation (30), suggesting that the presence of H2Av may increase access to other core histones, i.e., those hidden in H2A-containing nucleosomes. As we previously demonstrated, among the histones, PARP1 protein preferentially interacts with H3/H4 tetramers (9), possibly explaining the enrichment of PARP1 in the presence of H2Av-containing nucleosomes in vivo, as reported here. In other words, chromatin in “more open” H2Av nucleosomes, with a high level of H3/H4 exposure, has a greater affinity for binding with PARP1 than does unexposed chromatin. Moreover, we found that interaction with the N-terminal tail of the histone H4 triggers PARP1 protein activation (Fig. 4B) (9). The SQ domain of histone H2Av, the phosphorylation of which controls PARP1 activation in vivo—as reported here—is positioned in close proximity to the N-tail of H4 in the nucleosome (Fig. S5). Therefore, we propose that the histone-replacement machinery positions H2Av within the promoter region of specific genes, thereby creating nucleosomes with an “open” configuration (30). Within these nucleosomes, exposed H3/H4 histones bind PARP1 protein and properly determine its localization in promoters. Phosphorylation of the H2Av C terminus then leads to exposure of the H4 histone N-tail, promoting its interaction with PARP1, and the activation of the PARP1 protein.
Although our results establish a direct connection between PARP1, H2Av-containing nucleosomes, H2Av phosphorylation, and pADPr, we cannot exclude a possible role for H2Av phosphorylation itself in regulating the activity of the PARP1 protein on a higher level of chromatin organization. This possibility is suggested by the observed difference between the in vivo and in vitro results. Although phosphorylation of H2Av was required to elicit PARP1 activation in vivo (Figs. 1 and 2), purified nucleosomes containing either H2A or H2Av were able to elicit PARP1 activation in vitro (Fig. 4). Mimicking the phosphorylation of H2Av (as in H2AvSE) resulted in a 26% increase in the activation of PARP1 in vitro (Fig. 4). These observations suggest that although phospho-H2Av may act directly on PARP1, it also may mediate changes in the higher-order chromatin microenvironment (which could not be reproduced in vitro), leading to the disruption of PARP1 interaction with inhibitors and/or the interaction of PARP1 with activating epitopes in the context of the local chromatin. Alternatively, H2Av phosphorylation may be involved in a “system restart” in vivo; i.e., phosphorylation of H2Av has been linked to the replacement of this histone in the local chromatin (13). Consequently, multiple repeated acts of transcriptional initiation, and therefore multiple acts of H2Av phosphorylation, may be required, for example, during heat-shock gene expression (2). Thus, in the absence of H2Av phosphorylation, H2Av replacement will be blocked, and transcriptional restart will be arrested.
H2Av (H2Az/H2Ax) may have other roles in the nucleus in addition to the regulation of PARP1. For instance, although the yeast genome does not encode any obvious PARP1 homolog, yeasts have H2Az (HTZ1) and H2Ax homologs (14). Moreover, both yeast histones play essential roles. Histone H2Ax phosphorylation is involved in genotoxic stress response (31, 32), but HTZ1 regulates chromatin remodeling (33), transcription, and transcriptional silencing in heterochromatin (15, 34). Although PARP1 is a target that performs a critical role in higher-order chromatin, which otherwise cannot be accomplished by yeast, these observations suggest that the function of H2Av may not be restricted to PARP1 activation.
Because PARP1 activation has been shown in this work to be mediated through H2Av phosphorylation, we further asked what signaling pathway and kinases might be responsible for such H2Av phosphorylation. During genotoxic stress response, cell-cycle checkpoint kinases such as ataxia telangiectasia-mutated/ataxia telangiectasia and Rad- related (ATM/ATR) and DNA-PK kinase, are shown to phosphorylate the C-terminal tail of H2Ax (35, 36). Although we cannot exclude the possible roles of the Drosophila homolog of these enzymes in chromatin regulation and transcription, kinases such as Jil-1 kinase, which functions inside the puffs of polytene chromosomes (37), seem to be more promising candidates for performing this function. Therefore, one of the future directions for investigating the mechanism of PARP1 regulation in chromatin is to identify the kinase enzyme responsible for triggering H2Av-mediated PARP1 activation.
The current paradigm for the role of the PARP1 protein has two parts. The first part assigns to PARP1 the role of DNA repair and genotoxic stress response, and the second part assigns to PARP1 functional roles in the regulation of chromatin structure and transcription. In demonstrating that phosphorylation of H2Av (the H2Az/H2Ax homolog) controls the activity of the PARP1 protein in both pathways, we have established a more universal mechanism for PARP1 regulation. Our findings also support the notion that PARP1 is not simply a component of either the DNA-repair or transcriptional complexes but instead is a universal regulator of high-order chromatin, which in eukaryotes needs management during both DNA repair and transcription. The activation of the PARP1 protein by histone H2Av phosphorylation ultimately leads to the loosening of compacted chromatin and opens access for either the DNA repair machinery or the transcriptional apparatus.
Experimental Procedures
Experimental procedures are discussed in detail in SI Experimental Procedures. Sequence information for primers used for ChIP-quantitative PCR is given in Table S1.
Drosophila Strains and Genetics.
The fly stocks were generated by the standard genetic methods or were obtained from the Bloomington Drosophila Stock Center and the Exelixis Collection at the Harvard Medical School, except as indicated. Genetic markers are described in Flybase (38). H2Av810 stock is described in ref. 16. H2Av280 stock was generated by the L. Cooley laboratory (24). The Parpc03265 strains were generated in a single pBac-element mutagenesis screen (39). The Parg27.1 mutation was constructed in ref. 40. pP{w1, UAS::PARP1-DsRed} was described in ref. 5. The transgenic stock with pP{w1, UAS::PARP1-ECFP} was described in ref. 9. The following GAL4 driver strains were used: arm::GAL4 (Bloomington stock no. 1560), da::GAL4 (gift of A. Veraksa, University of Massachusetts, Boston), and 69B-GAL4 (41). Balancer chromosomes carrying Kr::GFP, i.e., TM3, Sb, P{w+, Kr-GFP} and FM7i, P{w1, Kr-GFP} (42) were used to identify heterozygous and homozygous H2Av810, H2Av280, Parpc03265, and Parg27.1.
Construction of Transgenic Drosophila.
To make UAS::H2AvWT, UAS::H2AvSA, and UAS::H2AvSE transgenic Drosophila constructs, full-length H2Av DNA was synthesized by PCR using following oligos:
5′-CAC Cat ggc tgg cgg taa agc agg-3′, direct
5′-GTA GGC CTG CGA CAG AAT GAC GT-3′, reverse, WT
5′-GTA GGC CTG CGC CAG AAT GAC GT-3′, reverse, SA
5′-GTA GGC CTG CTC CAG AAT GAC GT-3′, reverse, SE
The resulting PCR products were cloned through the Drosophila Gateway Vector Cloning System (Carnegie Institution of Washington) into a pTW vector for Drosophila transformation. y,w67c23(2) was used as the host for transformation. Transformation was performed as described in ref. 43.
Supplementary Material
Acknowledgments
We thank Dr. R. Glaser for providing materials. Drs. A. O'Reilly, K. Zaret, J. Lis, and E. Pechenkina provided comments on the manuscript. The research was supported by Grants R01 GM077452 and R01 DK082623 from the National Institutes of Health (to A.V.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019644108/-/DCSupplemental.
References
- 1.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 2.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 7.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of PARP1 protein binding to chromatin and induction of PARP1 enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 11.Lankenau S, Bürkle A, Lankenau DH. Detection of poly(ADP-ribose) synthesis in Drosophila testes upon gamma-irradiation. Chromosoma. 1999;108:44–51. doi: 10.1007/s004120050350. [DOI] [PubMed] [Google Scholar]
- 12.Meyer-Ficca ML, Scherthan H, Bürkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma. 2005;114:67–74. doi: 10.1007/s00412-005-0344-6. [DOI] [PubMed] [Google Scholar]
- 13.Du YC, et al. The dynamic alterations of H2AX complex during DNA repair detected by a proteomic approach reveal the critical roles of Ca(2+)/calmodulin in the ionizing radiation-induced cell cycle arrest. Mol Cell Proteomics. 2006;5:1033–1044. doi: 10.1074/mcp.M500327-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Redon C, et al. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 15.Dhillon N, Kamakaka RT. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell. 2000;6:769–780. doi: 10.1016/s1097-2765(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stargell LA, et al. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993;7(12B):2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Li B, Gorovsky MA. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol Cell Biol. 1996;16:4305–4311. doi: 10.1128/mcb.16.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach TJ, et al. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 20.Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotova E, Jarnik M, Tulin AV. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci USA. 2010;107:6406–6411. doi: 10.1073/pnas.0914152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tulin A, Naumova NM, Menon AK, Spradling AC. Drosophila poly(ADP-ribose) glycohydrolase mediates chromatin structure and SIR2-dependent silencing. Genetics. 2006;172:363–371. doi: 10.1534/genetics.105.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelso RJ, et al. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004;32(Database issue):D418–D420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 26.Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, Zhimulev IF. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res. 2008;16:851–862. doi: 10.1007/s10577-008-1244-4. [DOI] [PubMed] [Google Scholar]
- 27.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Raisner RM, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 31.Kim J-A, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paull TT, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 33.Li B, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 35.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 36.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 37.Deng H, et al. Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development. 2008;135:699–705. doi: 10.1242/dev.015362. [DOI] [PubMed] [Google Scholar]
- 38.FlyBase The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artavanis-Tsakonas S. Accessing the Exelixis collection. Nat Genet. 2004;36:207. doi: 10.1038/ng1316. [DOI] [PubMed] [Google Scholar]
- 40.Hanai S, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manseau L, et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 42.Casso D, Ramírez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- 43.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.