Abstract
Protein arginine methylation is a common posttranslational modification catalyzed by a family of the protein arginine methyltransferases (PRMTs). We have previously reported that PRMT1 methylates Forkhead box O transcription factors at two arginine residues within an Akt consensus phosphorylation motif (RxRxxS/T), and that this methylation blocks Akt-mediated phosphorylation of the transcription factors. These findings led us to hypothesize that the functional crosstalk between arginine methylation and phosphorylation could be extended to other Akt target proteins as well as Forkhead box O proteins. Here we identify BCL-2 antagonist of cell death (BAD) as an additional substrate for PRMT1 among several Akt target proteins. We show that PRMT1 specifically binds and methylates BAD at Arg-94 and Arg-96, both of which comprise the Akt consensus phosphorylation motif. Consistent with the hypothesis, PRMT1-mediated methylation of these two arginine residues inhibits Akt-mediated phosphorylation of BAD at Ser-99 in vitro and in vivo. We also demonstrate that the complex formation of BAD with 14-3-3 proteins, which occurs subsequent to Akt-mediated phosphorylation, is negatively regulated by PRMT1. Furthermore, PRMT1 knockdown prevents mitochondrial localization of BAD and its binding to the antiapoptotic BCL-XL protein. BAD overexpression causes an increase in apoptosis with concomitant activation of caspase-3, whereas PRMT1 knockdown significantly suppresses these apoptotic processes. Taken together, our results add a new dimension to the complexity of posttranslational BAD regulation and provide evidence that arginine methylation within an Akt consensus phosphorylation motif functions as an inhibitory modification against Akt-dependent survival signaling.
A complex interplay between pro- and antiapoptotic members of the B-cell lymphoma 2 (BCL-2) family of proteins regulates apoptosis by governing mitochondrial outer membrane permeabilization and subsequent caspase activation (1). BCL-2 antagonist of cell death (BAD) is a BCL-2 homology domain 3 (BH3)-only proapoptotic BCL-2 family member inactivated by phosphorylation through survival kinases, including Akt/protein kinase B, protein kinase A (PKA), and p90 ribosomal S6 kinase (RSK) (2). In the dephosphorylated state, BAD binds and inhibits antiapoptotic BCL-XL/BCL-2, thereby derepressing proapoptotic BCL-2 antagonist killer (BAK)/BCL-2-associated X protein (BAX), which in turn triggers apoptosis by facilitating the release of mitochondrial cytochrome c into the cytoplasm, apoptosome assembly, and activation of caspases executing cell apoptosis (2, 3). In contrast, Akt-mediated phosphorylation at Ser-99 has been shown to repress BAD function by causing it to dissociate from mitochondria and bind to 14-3-3 proteins in the cytoplasm (4–6). Alternatively, PKA and RSK are known to phosphorylate BAD at Ser-75 and Ser-118, promote the 14-3-3 binding, and disrupt the BCL-XL/BCL-2 interaction (7–11). Several phosphatases, including protein phosphatase 1α (PP1α), PP2A, and calcineurin, have been shown to dephosphorylate BAD and enhance its proapoptotic activity (12–14). Thus, although phosphorylation has been established as a central regulatory mechanism of BAD function, whether BAD might undergo other posttranslational modifications, which fine tune the BAD-mediated apoptotic program, remains unclear.
Protein arginine methyltransferases (PRMTs) are enzymes that catalyze the transfer of a methyl group from donor S-adenosylmethionine (SAM) to the guanidino nitrogen atom in target arginine residues (15). To date, 11 PRMTs have been reported in mammals, and these PRMTs were classified into the following two groups: type 1 consisting of PRMT1, 2, 3, 4, 6, and 8 that catalyze asymmetric dimethylation, and type 2 consisting of PRMT5, 7, and 9 that catalyze symmetric dimethylation (16).
Recently, accumulating evidence has revealed that arginine methylation plays an important role in many aspects of biological processes, such as signal transduction, transcriptional regulation, RNA processing, and DNA damage response (17). In a previous report, we demonstrated that Forkhead box O (FOXO) transcription factors downstream of the PI3-kinase/Akt-signaling pathway are substrates for PRMT1-mediated methylation (18). Importantly, this methylation occurs at arginine residues within an Akt consensus phosphorylation motif (RxRxxS/T) of FOXO proteins, thereby blocking their phosphorylation by Akt (18). Given that an RxR motif often exists in substrates for PRMT1 (19, 20), our finding led us to hypothesize that other Akt target proteins containing the RxRxxS/T motif could be methylated by PRMT1, and that this methylation also counteracts Akt-mediated phosphorylation of the target proteins.
In this study, we identify BAD as a unique substrate for PRMT1. We show that PRMT1 specifically methylates BAD at Arg-94 and Arg-96 within the Akt consensus phosphorylation motif. As expected, this methylation inhibits Akt-mediated BAD phosphorylation at Ser-99 in vitro and in vivo. We also demonstrate that PRMT1 knockdown increases complex formation of BAD with 14-3-3β, and conversely, represses mitochondrial localization of BAD and its binding to BCL-XL. Furthermore, BAD-mediated apoptosis observed in HEK293T and C2C12 cells is significantly suppressed by PRMT1 knockdown. Collectively, our results suggest that PRMT1 functions as a proapoptotic mediator that methylates BAD, and this methylation counteracts Akt-dependent survival signaling.
Results
BAD is a Unique Substrate for PRMT1.
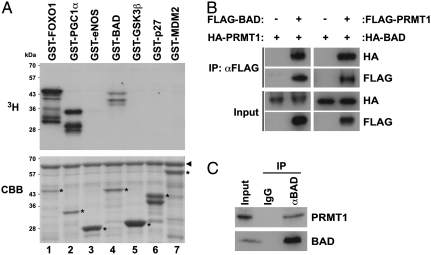
We have previously demonstrated that PRMT1 methylates the transcription factor FOXO1 at two arginine residues within an Akt consensus phosphorylation motif (RxRxxS/T), and this methylation directly blocks Akt-mediated phosphorylation of FOXO1 (18). This finding prompted us to hypothesize that functional crosstalk between arginine methylation and phosphorylation could be extended to other Akt target proteins that include the RxRxxS/T motif. To assess this hypothesis, we screened for unique substrates for PRMT1 from among a number of Akt target proteins, including FOXO1, peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α), eNOS, BAD, glycogen synthase kinase 3β (GSK3β), p27, and murine double minute 2 (MDM2), by in vitro methylation assays (21, 22). These proteins were purified as GST-fused fragments containing the Akt consensus phosphorylation motif and were then incubated with GST-PRMT1 in the presence of 3H-labeled SAM. Consistent with our previous study (18), both FOXO1 and PGC-1α were methylated by PRMT1 (Fig. 1A, lanes 1 and 2). Notably, we found that PRMT1 also specifically methylated the proapoptotic protein BAD from among other Akt target proteins in vitro (Fig. 1A, lane 4).
Fig. 1.
BAD is a unique substrate for PRMT1. (A) Several Akt target proteins were prepared as GST-fusion proteins and incubated with GST-PRMT1 in the presence of [3H]SAM. Reaction products were analyzed by autoradiography (Upper) and Coomassie brilliant blue staining (Lower). The arrowhead and asterisks indicate total amounts of GST-PRMT1 and GST-fused Akt target proteins, respectively. (B) HEK293T cells were transfected with expression vectors as indicated. Whole-cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblotting with anti-HA or anti-FLAG antibodies. (C) Whole cell lysates from HEK293T cells were immunoprecipitated with normal IgG or anti-BAD antibody, followed by immunoblotting with anti-PRMT1 or anti-BAD antibodies.
To assess the possibility that BAD is indeed targeted by PRMT1, we examined whether PRMT1 binds to BAD in vivo. HEK293T cells were transfected with either or both of BAD and PRMT1 expression vectors, and then whole cell lysates were immunoprecipitated with anti-FLAG antibody. As shown in Fig. 1B, a reciprocal interaction between two proteins was observed. In addition, their interaction at endogenous levels was also confirmed by coimmunoprecipitation assay with anti-BAD and anti-PRMT1 antibodies (Fig. 1C). Together, these results suggest that BAD appears to be a bona fide substrate for PRMT1.
PRMT1 Methylates BAD at Arg-94 and Arg-96 in Vitro and in Vivo.
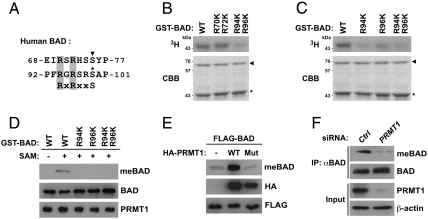
We next determined whether PRMT1-dependent methylation occurs at arginine residues within the Akt consensus phosphorylation motif in the BAD protein. Because BAD has two RxRxxS/T motifs, of which one is targeted by both PKA and RSK (70-RSRHSS-75) and the other is targeted by Akt (94-RGRSRS-99; Fig. 2A) (5–8), we constructed GST-BAD mutants in which two arginines in each RxRxxS/T motif were substituted with lysine (R70K/R72K and R94K/R96K). In vitro methylation assays demonstrated that PRMT1 methylated the R70K/R72K mutant to the same extent as wild-type BAD, whereas the methylation was markedly reduced in the R94K/R96K mutant (Fig. 2B). To further determine whether either or both Arg-94 and Arg-96 are responsible for PRMT1-mediated methylation of BAD, BAD mutants in which Arg-94 and Arg-96 were substituted with lysine, individually or in combination, were subjected to in vitro methylation assays. As shown in Fig. 2C, the R96K single mutant showed significantly decreased BAD methylation, and the R94K and R94K/R96K double mutants exhibited marked decrease in overall methylation. These data indicate that PRMT1 methylates BAD at Arg-94 and Arg-96 within the Akt consensus phosphorylation motif in vitro.
Fig. 2.
PRMT1 methylates BAD at Arg-94 and Arg-96 in vitro and in vivo. (A) Alignment of the human BAD sequence with the Akt consensus phosphorylation motif (RxRxxS). The corresponding arginine residues are shaded in gray. Two known phosphorylation sites of PKA/RSK and Akt are shown by the arrowhead and asterisk, respectively. (B and C) A series of GST-BAD point mutants in which one or two arginine residues were substituted with lysine were incubated with GST-PRMT1 in the presence of [3H]SAM. Reaction products were analyzed by autoradiography (Upper) and Coomassie brilliant blue staining (Lower). The arrowhead and asterisk indicate total amounts of GST-PRMT1 and GST-BAD, respectively. (D) A series of GST-BAD point mutants in which one or two arginine residues were substituted with lysine were incubated with GST-PRMT1 in the presence or absence of SAM. Reaction products were analyzed by immunoblotting with anti-MeBAD, anti-BAD, or anti-PRMT1 antibodies. (E) HEK293T cells were transfected with FLAG-BAD with or without wild-type or G98R mutant of HA-PRMT1. Whole-cell lysates were analyzed by immunoblotting with anti-MeBAD, anti-HA, or anti-FLAG antibodies. (F) Whole-cell lysates from HEK293T cells transfected with control (Ctrl) or PRMT1-specific siRNA were immunoprecipitated with anti-BAD antibody, followed by immunoblotting with anti-MeBAD or anti-BAD antibodies.
To investigate whether the two arginine residues in BAD can be methylated in vivo, we generated an anti-MeBAD antibody that specifically recognized asymmetric dimethylation at both Arg-94 and Arg-96 in BAD. The specificity of the antibody was tested by dot blot analysis using BAD peptides unmodified or methylated at Arg-94 and/or Arg-96 (Fig. S1A), and was further validated by the finding that this antibody recognized wild-type but not the RK mutant BAD after in vitro methylation reaction with GST-PRMT1 (Fig. 2D). Then, by using the anti-MeBAD antibody, we demonstrated that overexpression of PRMT1 in cells led to a marked increase in BAD methylation at Arg-94 and Arg-96, whereas the G98R catalytically inactive mutant PRMT1 failed to methylate BAD (Fig. 2E and Fig. S1B). Moreover, to assess whether endogenous PRMT1 is responsible for basal methylation of BAD, lysates from HEK293T cells transfected with siRNAs against GFP (control) or PRMT1 were immunoprecipitated with anti-BAD antibody and subsequently immunoblotted with the anti-MeBAD antibody. As shown in Fig. 2F, the extent of endogenous BAD methylation was diminished by PRMT1 knockdown. Collectively, these results suggest that BAD methylation at Arg-94 and Arg-96 is almost entirely dependent on PRMT1 in vivo.
Arginine Methylation of BAD Inhibits Its Phosphorylation by Akt.
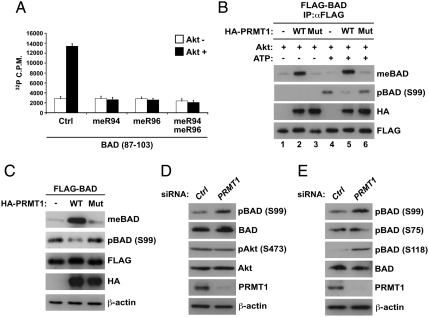
To determine whether arginine methylation within the RxRxxS motif of BAD inhibits Akt-mediated phosphorylation, we performed in vitro Akt kinase assays using two BAD peptides (amino acids 87–103) that included an Akt phosphorylation site at Ser-99. They were unmodified (control) or had asymmetric dimethylarginines at Arg-94 and/or Arg-96 for the assays. Recombinant Akt readily phosphorylated the unmodified peptides at Ser-99, whereas phosphorylation was completely blocked when either or both of the arginine residues were methylated (Fig. 3A). To further prove the inhibition of Akt-mediated phosphorylation of BAD by arginine methylation, a sequential in vitro methylation/phosphorylation assay was conducted using immunoprecipitated FLAG-BAD proteins from HEK293T cells transfected with wild-type or mutant PRMT1. After immunoprecipitation, basal phosphorylation of FLAG-BAD was preliminary removed by treatment with alkaline phosphatase and then they were incubated with recombinant Akt. As shown in Fig. 3B, Akt-dependent phosphorylation of BAD at Ser-99 was observed in the presence of ATP, whereas the extent of phosphorylation was substantially reduced by prior methylation of BAD (Fig. 3B, lanes 4–6).
Fig. 3.
Arginine methylation of BAD inhibits its phosphorylation by Akt. (A) BAD peptides (amino acids 87–103) that were unmodified control (Ctrl) or methylated at Arg-94 and/or Arg-96 (meR94, meR96, and meR94/meR96) were incubated with or without recombinant Akt1 in the presence of [32P]ATP. The extent of BAD phosphorylation was measured using a scintillation counter. Values are means ± SD of three independent experiments. (B) HEK293T cells were transfected with FLAG-BAD together with or without wild-type or G98R mutant of HA-PRMT1. Whole-cell lysates were immunoprecipitated with anti-FLAG antibody, and then the immunopurified FLAG-BAD was treated with bacterial alkaline phosphatase, followed by incubation with recombinant Akt1 in the presence or absence of ATP. Reaction products were analyzed by immunoblotting as indicated. (C) HEK293T cells were transfected with FLAG-BAD with or without the wild-type or G98R mutant of HA-PRMT1. Whole-cell lysates were analyzed by immunoblotting as indicated. (D and E) Whole-cell lysates from HEK293T cells transfected with control (Ctrl) or PRMT1-specific siRNA were analyzed by immunoblotting as indicated.
We next investigated whether the mutual antagonism of arginine methylation and phosphorylation occurs in BAD in vivo. Overexpression of PRMT1 enhanced BAD methylation in an enzyme activity-dependent manner, whereas the levels of BAD phosphorylation at Ser-99 were inversely correlated with an increase in methylation (Fig. 3C). In addition, silencing of PRMT1 resulted in a significant increase in BAD phosphorylation at Ser-99 (Fig. 3D). To exclude the possibility that siRNA transfection could influence the insulin signaling pathway upstream of Akt, we confirmed that levels of Akt phosphorylation at Ser-473 remained unchanged after PRMT1 knockdown (Fig. 3D).
PKA and RSK have been shown to phosphorylate BAD at both Ser-75 and Ser-118, and notably, BAD phosphorylation at Ser-118 is accompanied by a prior Akt-mediated phosphorylation at Ser-99 (9). We thus examined the effect of arginine methylation on these two phosphorylation sites. As shown in Fig. 3E, PRMT1 knockdown had no effect on BAD phosphorylation at Ser-75, whereas a marked increase in phosphorylation was observed at Ser-118, probably due to the up-regulation of this phosphorylation at Ser-99. It is also possible that PRMT1 might directly interfere with the activity of a kinase to phosphorylate BAD at Ser-118. Next, we asked whether phosphorylation of BAD could oppositely affect its methylation in vivo and in vitro. Overexpression of a constitutively active form (Myr-p110α) or a dominant negative form (Δp85) of PI3K, upstream of Akt, caused no alteration in BAD methylation (Fig. S2). Moreover, in vitro phosphorylation/methylation assays demonstrated that a prior phosphorylation of BAD by Akt or PKA had insignificant effects on the PRMT1-dependent methylation of BAD (Fig. S3 A and B). Taken together, these data indicate that PRMT1 blocks Akt-mediated phosphorylation of BAD by methylating arginine residues within the Akt consensus phosphorylation motif, but not vice versa.
PRMT1 Is Involved in Binding of BAD to 14-3-3 and BCL-XL.
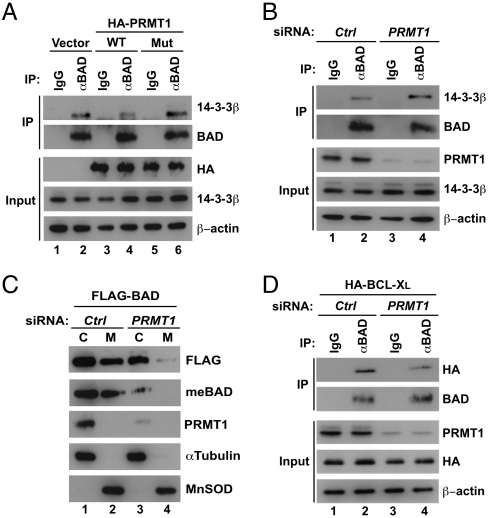
BAD phosphorylation at Ser-99 has been shown to result in binding of BAD to 14-3-3 proteins, which in turn sequesters BAD as an inert complex in the cytoplasm (4). We thus explored the role of PRMT1 in regulating the BAD/14-3-3 interaction. As shown in Fig. 4A, overexpression of wild-type but not mutant PRMT1 attenuated the endogenous BAD/14-3-3β interaction when compared with vector control. We also found that PRMT1 knockdown led to a significant increase in the endogenous BAD/14-3-3 interaction (Fig. 4B). Because BAD phosphorylation and subsequent 14-3-3 binding facilitate the dissociation of BAD from mitochondria (4, 23), PRMT1 knockdown was predicted to inhibit mitochondrial localization of BAD. To assess this hypothesis, HEK293T cells transfected with siRNAs and FLAG-BAD were fractionated into cytoplasmic and mitochondrial fractions, and then the abundance of BAD in each fraction was analyzed by Western blotting. As shown in Fig. 4C, total and methylated BAD were observed in both fractions, whereas knockdown of PRMT1 substantially abrogated the mitochondrial localization of BAD. In mitochondria, dephosphorylated BAD is known to bind and inhibit antiapoptotic BCL-XL and to derepress proapoptotic BAK/BAX (2, 3). Thus, we next determined whether PRMT1-mediated methylation of BAD is involved in complex formation with BCL-XL. Coimmunoprecipitation assays revealed that PRMT1 knockdown weakened the BAD/BCL-XL interaction (Fig. 4D). Taken together, these findings support our hypothesis that PRMT1-mediated methylation of BAD inhibits phosphorylation by Akt and thereby promotes a subsequent series of processes, such as mitochondrial localization and binding to BCL-XL.
Fig. 4.
PRMT1 regulates binding of BAD to 14-3-3 and BCL-XL. (A) HEK293T cells were transfected with or without wild-type or G98R mutant of HA-PRMT1. Whole-cell lysates were immunoprecipitated with normal IgG or anti-BAD antibody, followed by immunoblotting with anti-14-3-3β or anti-BAD antibodies. (B) Whole-cell lysates from HEK293T cells transfected with control or PRMT1-specific siRNA were immunoprecipitated with anti-BAD antibody, followed by immunoblotting with anti-14-3-3β or anti-BAD antibodies. (C) HEK293T cells were transfected with control or PRMT1-specific siRNA together with empty or FLAG-BAD expression vectors. Whole-cell lysates were separated into cytoplasmic (C) and mitochondrial (M) fractions. Equal loading and verification of fractionation was confirmed by immunoblotting with anti-α-tubulin (cytoplasm) and anti-MnSOD (mitochondria) antibodies. (D) HEK293T cells were transfected with control or PRMT1-specific siRNA together with the HA-BCL-XL expression vector. Whole-cell lysates were immunoprecipitated with anti-BAD antibody, followed by immunoblotting as indicated.
PRMT1 Knockdown Suppresses BAD-Mediated Apoptosis.
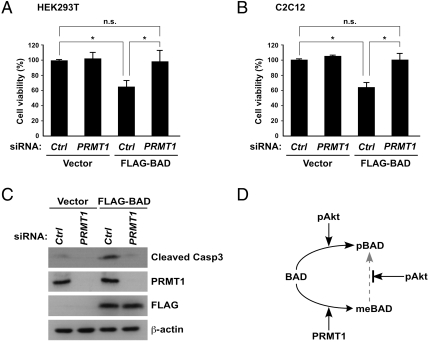
Finally, to examine the biological significance of BAD methylation, we investigated the role of endogenous PRMT1 in BAD-mediated apoptosis. As shown in Fig. 5A, FLAG-BAD overexpression in HEK293T cells resulted in a 40% reduction in cell viability (column 1 vs. 3), whereas PRMT1 knockdown prior to FLAG-BAD transfection abrogated BAD-mediated cell death (column 3 vs. 4). Importantly, similar results were obtained in C2C12 myoblasts and MCF7 breast cancer cells (Fig. 5B and Fig. S4). To further confirm that PRMT1 indeed affects an apoptotic program cascade through BAD signaling, we measured the level of activated caspase-3 by detecting the appearance of cleaved caspase-3 fragments. Consistent with the cell viability data, caspase-3 activation triggered by BAD overexpression was abolished when PRMT1 was simultaneously depleted by siRNA (Fig. 5C). Thus, these results indicate that PRMT1 plays a critical role in BAD-mediated apoptosis.
Fig. 5.
PRMT1 knockdown suppresses BAD-mediated cell death. (A and B) HEK293T (A) and C2C12 (B) cells were transfected with control or PRMT1-specific siRNA together with empty or FLAG-BAD expression vectors. Cell viability was measured using the WST-8 assay. Data are represented as relative cell viability compared with cells with control siRNA. Values are means ± SD of three independent experiments (P < ∗0.05; n.s, not significant). (C) Whole-cell lysates from C2C12 cells transfected with control or PRMT1-specific siRNA together with empty or FLAG-BAD expression vectors were analyzed by immunoblotting as indicated. (D) Proposed working model.
Discussion
In this study, we demonstrated an opposite interplay between arginine methylation and phosphorylation of the proapoptotic protein BAD (Fig. 5D). We show that PRMT1 binds and methylates BAD at two arginine residues within the Akt consensus phosphorylation motif. This methylation prevents Akt-mediated phosphorylation of BAD, thereby precluding 14-3-3β binding. Consequently, PRMT1-mediated methylation of BAD causes its mitochondrial localization and complex formation with BCL-XL, which in turn results in apoptosis. Thus, our findings add another dimension to the complexity of BAD regulation as per which PRMT1-mediated methylation of BAD at arginine residues directs cell fate toward apoptosis by counteracting Akt-dependent survival signaling. In addition, strong conservation of the methylated arginine residues of BAD orthologs in vertebrates suggests the biological significance of a negative crosstalk between arginine methylation and phosphorylation in BAD.
Our initial hypothesis was that PRMT1 could methylate Akt target proteins, which have potential RxR methylation motifs within the Akt consensus phosphorylation sequence. However, we identified only one target protein during screening with five known Akt target proteins, including eNOS, BAD, GSK3β, p27, and MDM2. This result indicates that Akt target proteins are not necessarily substrates for PRMT1-mediated methylation and instead raises the question as to what determines the substrate preference of PRMT1. At least, two plausible explanations exist. First, the recognition properties depend on the binding affinity of PRMT1 for substrates. Supporting this idea, FOXO1 and BAD tightly bind to PRMT1 in vitro (Fig. S5). Second, substrate specificity is attributed to the surrounding sequence of the RxR motif, in which some additional code may be embedded. An extensive comparison of the BAD sequence with those of FOXO transcription factors, however, revealed no common feature between them. Further identification of a unique PRMT1 substrate whose methylation sites overlap an Akt consensus phosphorylation motif may provide more information on modes of substrate recognition in both Akt-mediated phosphorylation and PRMT1-mediated methylation.
Here we identified BAD as a second substrate for PRMT1 among several Akt target proteins. A common feature between BAD and that previously identified FOXO1 is that both function as proapoptotic genes downstream of the cell-survival signaling cascade, and their inactivation confers a proliferative and survival advantage to cancer cells. Indeed, bad-deficient mice exhibited lymphoma (24), and conditional deletion of three principal foxo genes, (foxo1, foxo3a, and foxo4) induces thymic lymphomas and hemangiomas (25). Given that PRMT1-mediated methylation of BAD and FOXO1 enhances their activity by inhibiting Akt signal transduction, PRMT1 likely plays a critical role in the apoptotic program and serves as a tumor suppressor. However, because PRMT1 knockout embryos fail to develop beyond day E6.5 (26), the physiological significance of PRMT1 in tumorigenesis remains unclear. Further studies using conditional knockout mice will be necessary to address the potential role of PRMT1 in BAD- and FOXO-mediated tumor suppression.
Although the presence of basal BAD methylation was observed in cells under normal culture conditions, this methylation level was insufficient to elicit apoptosis, indicating that a higher proportion of methylated BAD is required to trigger apoptosis. Thus, the situations in which BAD methylation levels increase needs to be clarified. Unlike in FOXO1 (18), oxidative stress by treatment with hydrogen peroxide did not influence the BAD methylation in both cytoplasmic and mitochondrial fractions (Fig. S6A). Alternatively, we also found that proapoptotic stimulation with etoposide or thapsigargin, which are known to be activators of BAD-mediated apoptosis (8, 24), resulted in no alteration in BAD methylation (Fig. S6 B and C). Meanwhile, the mechanism underlying the regulation of PRMT1 activity is largely unknown, and also the reversibility of arginine methylation remains controversial (27, 28). Further studies are required to reveal the link between extracellular stimuli and BAD methylation.
Is the crosstalk between arginine methylation and phosphorylation restricted to Akt? Interestingly, because the consensus sequences for SGK and Pim kinase are known to contain an RxR motif (29, 30), phosphorylation appears to be blocked if either or both of two arginine residues are methylated. On the other hand, several kinases such as PKA and AuroraB/C have consensus sequences harboring arginine residues, but not an overlapping glycine and arginine-rich (GAR) motif that is the canonical target for PRMTs (RRxS/T; PKA). However, given the recent evidence that arginine methylation occurs frequently beyond a GAR motif (31), PRMTs may counteract PKA- or AuroraB/C-mediated signaling through arginine methylation. Importantly, this notion may be further extended to the crosstalk with not only phosphorylation but also other posttranslational modifications, such as lysine methylation, acetylation, ubiquitination, and poly(ADP-ribosyl)ation. Thus, our findings provide further insight into the functional significance of arginine methylation in diverse biological processes.
Materials and Methods
Plasmids and Antibodies.
Full-length cDNAs encoding human BAD, BCL-XL, PRMT1, and PRMT1 (G98R) mutant (32) were inserted into pcDNA3-FLAG, pcDNA3-HA, and pGEX-6P vectors. A series of BAD (Arg to Lys) mutations were generated by site-directed mutagenesis. pGEX-FOXO1 (amino acids 208–409) and PGC-1α (amino acids 511–640) were described previously (18). Deletion mutants of eNOS (amino acids 1151–1202), p27 (amino acids 101–197), and MDM2 (amino acids 1–236) were inserted into pGEX-6P vectors. Deletion mutant of GSK3β (amino acids 1–49) was generated from human GSK3β cDNA (gift from A. Kikuchi, Osaka University, Osaka, Japan) and inserted into pGEX-6P vector. A rabbit polyclonal antibody against methylated Arg-94 and Arg-96 of BAD was raised by using the methylated peptide GEEPSPFRGRSRSAPPN (R, asymmetric dimethylarginine) and purified over a peptide-affinity column. The following antibodies were purchased: anti-BAD (#9239), anti-phospho-BAD (Ser112, #9296), (Ser136, #9295 and #5286), (Ser155, #9297), anti-panAkt (#2920), anti-phospho-Akt (Ser473, #9271), anticleaved caspase3 (#9664) from Cell Signaling Technology; anti-BAD (C-7) and agarose-conjugated anti-BAD (C-7) from Santa Cruz Biotechnology; anti-FLAG (M2), anti-αTubulin (B-5-1-2), anti-β-actin (AC-74) from Sigma-Aldrich; anti-HA (3F10, Roche), anti-PRMT1 (#07-404, Millipore), and anti-MnSOD (SOD-110, Stressgen).
Cell Culture and Transfection.
Details are described in SI Materials and Methods. siRNA against human PRMT1 (5′-GGACAUGACAUCCAAAGAUTT-3′) (18) and mouse PRMT1 (5′-GACAUGACAUCCAAAGACUTT-3′) (33) were synthesized by Nippon EGT. Control siRNA against GFP (5′-CUACAACAGCCACAACGUC-3′) was purchased from B-Bridge.
Coimmunoprecipitation Assay.
Coimmunoprecipitation assay was carried out as described previously (34).
In Vitro Methylation Assay.
GST-BAD or GST-fused Akt substrate proteins were incubated with GST-PRMT1 in the presence or absence of S-adenosyl-L-[methyl-3H]methionine (55 Ci/μmol) at 37 °C for 6 h. After washing the beads, the reaction products were analyzed by fluorography and Coomassie brilliant blue staining.
In Vitro Phosphorylation Assay.
One hundred micromolar of the indicated peptides were incubated in phosphorylation buffer [20 mM MOPS (pH 7.2), 5 mM EGTA, 1 mM dithiothreitol, phosphatase inhibitor cocktail], followed by the addition of recombinant Akt1 (Upstate) and [γ-32P]ATP solution (100 μCi). After incubation for 10 min at 30 °C, the aliquot was transferred to P81 phosphocellulose paper. The squares were washed three times with 0.75% phosphoric acid and once with acetone. Relative activities were measured by a scintillation counter. The following peptides were synthesized by Anygene: control, GEEPSPFRGRSRSAPPN; meR94, GEEPSPFRGRSRSAPPN; meR96, GEEPSPFRGRSRSAPPN; meR94/meR96, GEEPSPFRGRSRSAPPN (R, asymmetric dimethylarginine). The authenticity of the peptides was verified using mass spectroscopy. In vitro sequential methylation and phosphorylation assay is described in SI Materials and Methods.
Subcellular Fractionation.
Subcellular fractionation was performed as described in ref. 35 with some modifications. Details are described in SI Materials and Methods.
Cell Viability Assay.
Relative cell viability was measured by 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt (WST-8) assay using Cell Counting kit-8 (Dojindo). Briefly, 36 h after siRNA transfection, HEK293T and C2C12 cells were further transfected with empty or FLAG-BAD expression vectors and cultured for 36 h. The cells were collected and 1/10 vol (100 μL) of the cell suspension was incubated with 10 μL of WST-8 solution in a 96-well plate for 2 h at 37 °C. Absorbance of the samples at 450-nm wavelength was measured by an ARVO SX1420 multilabel counter. An average of three independent counts for each sample point was performed.
Supplementary Material
Acknowledgments.
We thank Dr. Akira Kikuchi (Osaka University) for providing the human GSK3β cDNA. We thank Dr. Shinya Yamanaka (Kyoto University) for providing pCAG-Myr-p110-IH plasmid. We also thank members of the Fukamizu laboratory for helpful discussions. This work was supported by research grants from the Novartis Foundation (Japan) for the Promotion of Science (H.D.), Grants-in-Aid for Scientific Research on Priority Areas (17054004 to A.F.), Grants-in-Aid for Young Scientists (20780237 and 22688029 to H.D.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Grants-in-Aid for Japan Society for the Promotion of Science Fellows (10J00366 to J.-i.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015328108/-/DCSupplemental.
References
- 1.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 2.Danial NN. BAD: Undertaker by night, candyman by day. Oncogene. 2008;27(Suppl 1):S53–70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- 3.Yang E, et al. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 4.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 5.Datta SR, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 6.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 7.Harada H, et al. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 8.Bonni A, et al. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 9.Datta SR, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 10.Tan Y, Demeter MR, Ruan H, Comb MJ. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 11.Virdee K, Parone PA, Tolkovsky AM. Phosphorylation of the pro-apoptotic protein BAD on serine 155, a novel site, contributes to cell survival. Curr Biol. 2000;10:1151–1154. doi: 10.1016/s0960-9822(00)00702-8. [DOI] [PubMed] [Google Scholar]
- 12.Ayllon V, Martinez AC, Garcia A, Cayla X, Rebollo A. Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 2000;19:2237–2246. doi: 10.1093/emboj/19.10.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang CW, et al. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin- 3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood. 2001;97:1289–1297. doi: 10.1182/blood.v97.5.1289. [DOI] [PubMed] [Google Scholar]
- 14.Wang HG, et al. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 15.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res. 2009;60:466–474. doi: 10.1016/j.phrs.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Smith JJ, et al. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 20.Wada K, Inoue K, Hagiwara M. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim Biophys Acta. 2002;1591:1–10. doi: 10.1016/s0167-4889(02)00202-1. [DOI] [PubMed] [Google Scholar]
- 21.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 23.Hekman M, et al. Reversible membrane interaction of BAD requires two C-terminal lipid binding domains in conjunction with 14-3-3 protein binding. J Biol Chem. 2006;281:17321–17336. doi: 10.1074/jbc.M600292200. [DOI] [PubMed] [Google Scholar]
- 24.Ranger AM, et al. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2003;100:9324–9329. doi: 10.1073/pnas.1533446100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 28.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 29.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 30.Friedmann M, Nissen MS, Hoover DS, Reeves R, Magnuson NS. Characterization of the proto-oncogene pim-1: Kinase activity and substrate recognition sequence. Arch Biochem Biophys. 1992;298:594–601. doi: 10.1016/0003-9861(92)90454-5. [DOI] [PubMed] [Google Scholar]
- 31.Wooderchak WL, et al. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry. 2008;47:9456–9466. doi: 10.1021/bi800984s. [DOI] [PubMed] [Google Scholar]
- 32.Kwak YT, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 33.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 34.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djouder N, et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.