Abstract
The proportion of the human gut bacterial community that is recalcitrant to culture remains poorly defined. In this report, we combine high-throughput anaerobic culturing techniques with gnotobiotic animal husbandry and metagenomics to show that the human fecal microbiota consists largely of taxa and predicted functions that are represented in its readily cultured members. When transplanted into gnotobiotic mice, complete and cultured communities exhibit similar colonization dynamics, biogeographical distribution, and responses to dietary perturbations. Moreover, gnotobiotic mice can be used to shape these personalized culture collections to enrich for taxa suited to specific diets. We also demonstrate that thousands of isolates from a single donor can be clonally archived and taxonomically mapped in multiwell format to create personalized microbiota collections. Retrieving components of a microbiota that have coexisted in single donors who have physiologic or disease phenotypes of interest and reuniting them in various combinations in gnotobiotic mice should facilitate preclinical studies designed to determine the degree to which tractable bacterial taxa are able to transmit donor traits or influence host biology.
Keywords: gut bacterial diversity, nutrient–microbe interactions, translational medicine pipeline for human microbiome
Efforts to dissect the functional interactions between microbial communities and their habitats are complicated by the long-standing observation that, for many of these communities, the great majority of organisms have not been cultured in the laboratory (1). Methodological differences between culture-independent and culture-based approaches have contributed to the challenge of deriving a realistic appreciation of exactly how much discrepancy exists between the culturable components of a microbial ecosystem and total community diversity. Table S1 gives examples of these methodological differences.
The largest microbial community in the human body resides in the gut: Its microbiome contains at least two orders of magnitude more genes than are found in our Homo sapiens genome (2). Culture-independent metagenomic studies of the human gut microbiota are identifying microbial taxa and genes correlated with host phenotypes, but mechanistic and experimentally demonstrated links between key community members and specific aspects of host biology are difficult to establish with these methods alone. The goals of the present study were (i) to evaluate the representation of readily cultured phylotypes in the human gut microbiota; (ii) to profile the dynamics of these cultured communities in a mammalian gut ecosystem; and (iii) to determine whether a clonally arrayed, personalized strain collection could be constructed to serve as a foundation for reassembling varying elements of a human's gut microbiota in vitro or in vivo.
Results
To estimate the abundance of readily cultured bacterial phylotypes in the distal human gut, primers were used to amplify variable region 2 (V2) of bacterial 16S ribosomal RNA (rRNA) genes present in eight freshly discarded fecal samples obtained from two healthy, unrelated anonymous donors living in the United States (n = 1 complete sample per donor at t = 1, 2, 3, and 148 d). Amplicons were subjected to multiplex pyrosequencing, and the results were compared with those generated from DNA prepared from ∼30,000 colonies cultured from each sample under strict anaerobic conditions for 7 d at 37 °C on a rich gut microbiota medium (GMM) composed of commercially available ingredients (“cultured” samples; details of the culturing technique are given in SI Materials and Methods, and a description of GMM is given in Table S2). The resulting 16S rRNA datasets were de-noised to minimize sequencing errors (3, 4), reads were grouped into operational taxonomic units (OTUs) of ≥97% nucleotide sequence identity (ID), and chimeric sequences were removed (SI Materials and Methods).
In total, 632 distinct 97%ID OTUs were observed in the complete samples, and 316 were identified in the cultured samples. The average abundance of cultured OTUs in the complete samples was 0.4%, but the average abundance of uncultured OTUs (i.e., those observed in the complete but not the cultured samples) was significantly lower (0.06%; P < 10−6 by an unpaired, two-tailed Student's t test, not assuming equal variances) (5).
To evaluate the representation of readily cultured taxa in the human gut microbiota at varying phylogenetic levels, we assigned taxonomic designations to each 97%ID OTU (SI Materials and Methods). Each 16S rRNA read from the complete fecal sample was scored as “cultured” if it had a taxonomic assignment that also was identified in the corresponding cultured population. If a 97%ID OTU in the complete sample could not be placed in any known taxonomic group, it was scored as “cultured” only if the same 97%ID OTU was observed in the cultured sample. This analysis indicated that 99% of the 16S rRNA reads derived from the complete fecal samples from either donor belong to phylum-, class- and order-level taxa that are also present in the corresponding cultured sample; 89 ± 4% of the reads are derived from readily cultured family-level taxa, and 70 ± 5% and 56 ± 4% belong to readily cultured genus- and species-level taxa, respectively (Fig. 1A Upper). Two alternate taxonomic binning methods, the Ribosomal Database Project (RDP) Bayesian classifier v2.0 and an arbitrary %ID cutoff, produced similar results (Fig. S1 A–F). Control experiments described in SI Materials and Methods indicate that at least 98% of the reads generated from 30,000 pooled colonies are not derived from nongrowing or lysed bacteria (the percentage of reads from the original fecal samples that are derived from dead cells is unknown).
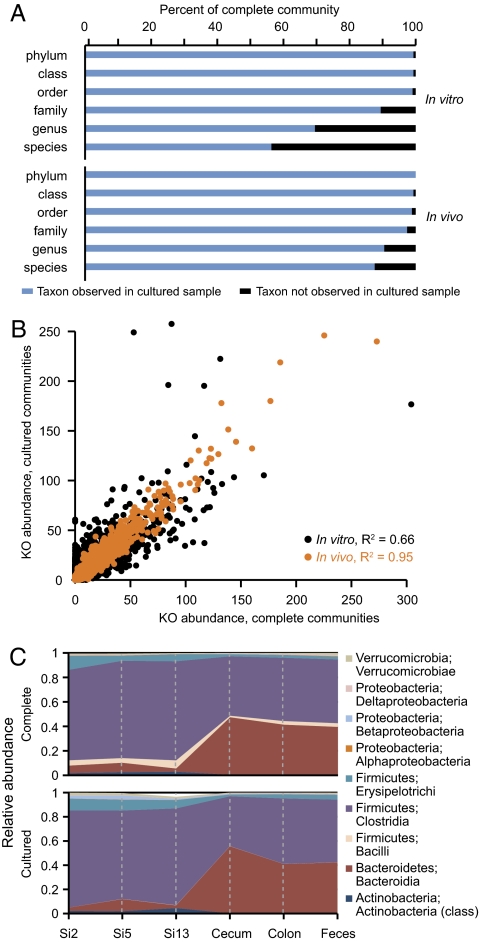
Fig. 1.
Comparison of the taxonomic representation of bacterial species and gene content in complete versus cultured human fecal microbial communities before and after their introduction into gnotobiotic mice. (A) 16S rRNA sequences from complete microbiota were compared with those identified from microbial communities cultured from the same human donors. At each taxonomic level, the proportion of reads in the complete community belonging to a taxonomic group observed in the cultured sample is shown in blue; the proportion of reads belonging to a taxonomic group not observed in the cultured sample (or lacking taxonomic assignment) is shown in black. Data shown are the average of two unrelated human donors. In vitro samples refer to comparisons between human fecal samples and plated material. In vivo samples refer to comparisons between gnotobiotic mice colonized with a complete human fecal microbiota and mice colonized with the readily cultured microbes from the same human fecal sample. (B) Annotated functions identified in the microbiomes of complete and cultured human gut communities. Each point represents a KO designation plotted by relative abundance (average across two donors, per 100,000 sequencing reads). Black points represent KO comparisons between the in vitro samples; orange points represent comparisons between in vivo samples. (C) The distribution of taxa and their relative abundance along the length of the intestine are similar in gnotobiotic mice colonized with complete or cultured human gut communities. Relative abundances of class-level taxa at six locations are shown; data represent the average of mice colonized from two unrelated donors. Si, small intestine divided into 16 equal-size segments and sampled at Si-2 (proximal), Si-5 (middle), and Si-13 (distal). PCoA suggests that gut biogeography, rather than donor or culturing, explains the majority (58%) of variance between samples (Fig. S3 A–C).
Unsupervised hierarchical clustering of the complete and cultured microbial communities, across the two donors and four time points, revealed that cultured samples cluster separately from those that had not been cultured. Both phylogenetic and nonphylogenetic metrics segregate cultured samples by donor, suggesting that the distinctiveness of each donor's microbiota is preserved in their collections of readily cultured representatives (Fig. S1 G and H).
We performed shotgun DNA pyrosequencing to determine the degree to which predicted functions contained in the composite genomes of the complete human fecal microbial communities were represented in the corresponding collection of cultured microbes [n = 4 samples (one complete and one cultured from each of two donors); 119,842 ± 43,086 high-quality shotgun reads per microbiome; average read length, 366 nt]. On average, 90% of the 2,302 distinct KEGG Orthology (KO) annotations identified in the two uncultured samples also were observed in the cultured communities (Fig. 1B, Fig. S2 A and B, and Table S3). This high percentage of functional representation also was observed when the microbiomes were subjected to alternate annotation schemes. On average, 94% of 929 enzyme commission (EC) assignments and 95% of 216 level 2 KEGG pathways associated with the complete fecal samples also were detected in the cultured communities (Fig. S2 C–F and Tables S4 and S5).
To compare the functions represented in the complete and cultured microbiota independent of annotation, we captured antibiotic-resistance genes from their microbiomes in Escherichia coli expression vectors. Each E. coli library contained ∼1 GB of 1.5- to 4-kB fragments of microbiome DNA and was screened against a panel of 15 antibiotics and clinically relevant antibiotic combinations (Table S6). Genes encoding resistance to the same 14 antibiotics were captured in libraries prepared from complete and cultured fecal samples (Fig. S2G and Table S7). In one example, a screen for DNA fragments that confer resistance to the aminoglycoside amikacin produced candidate genes from the microbiomes of both complete and cultured microbial communities from Donor 1 but not from Donor 2. Two genes conferring amikacin resistance (either the 16S rRNA methylase rmtD or the aminoglycoside phosphotransferase aphA-3) were identified in 70% of the DNA fragments captured in selections for this phenotype. Direct culturing of the original fecal communities in the presence of amikacin confirmed that this resistance function is significantly enriched in the readily cultured microbiota of Donor 1 compared with Donor 2 (P < 0.005 based on triplicate samples; unpaired, two-tailed Student's t test assuming equal variances) (Fig. S2H). PCR analysis showed that many of the amikacin-resistant fecal strains harbor rmtD or aphA-3. Sequencing the 16S rRNA genes of a subset of these isolates indicated that rmtD is present in strains of Bacteroides uniformis, B. caccae, and B. thetaiotaomicron in this donor (although, notably, not in the sequenced type strains of these species) and that aphA-3 is contained in the genome of a member of the genus Desulfotomaculum (order Clostridiales).
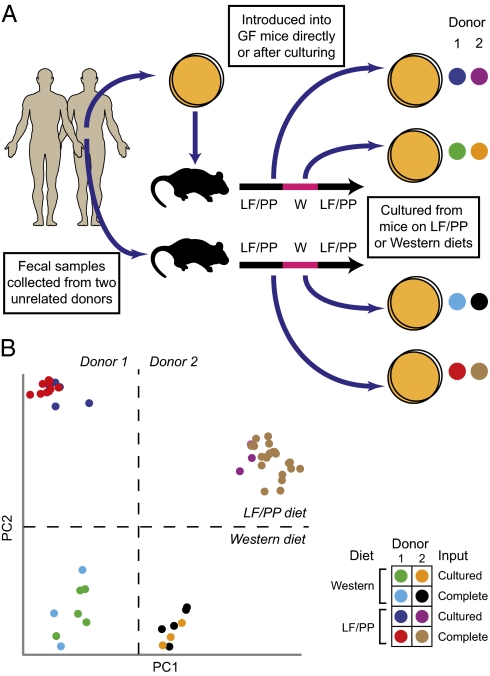
To determine whether a community composed of an individual's readily cultured bacteria exhibits behavior in vivo mirroring that of the individual's complete microbial community, 9-wk-old C57Bl6/J germfree mice were colonized with a complete or cultured microbiota from each of the two human donors (n = 5 recipient mice per sample type). A fecal sample from each donor was divided after collection, and one aliquot was gavaged directly into one group of recipient mice; the other aliquot was cultured on GMM plates for 7 d, as above, harvested, and introduced into a second group of recipient animals. Mice were maintained on a standard autoclaved low-fat, plant polysaccharide-rich (LF/PP) chow diet before and 4 wk after gavage. 16S rRNA analysis of fecal samples collected from these mice at the end of the 4-wk period indicated that the complete and the cultured communities were influenced similarly by host selection: 91 ± 3% of the 16S rRNA reads identified from mice colonized with a human donor's complete fecal microbiota were derived from genus-level taxa that also were identified in the mice colonized with the cultured microbial community from the same donor (Fig. 1A Lower). Importantly, control experiments demonstrated that the harvested, actively growing colonies gavaged into each germfree mouse are able to prevent nongrowing species that might be present on GMM plates from establishing themselves in recipient animals (details are given in SI Materials and Methods).
Luminal material was collected from the proximal, central, and distal portions of the small intestine, cecum, and colon of mice colonized with either the complete or cultured communities from each of the two human donors. V2-directed bacterial 16S rRNA sequencing revealed similar geographic variations in community structures (Fig. 1C and Fig. S3 A–C).
To determine whether the similarities in community composition in vivo extend to similarities in community gene content, the same fecal DNA samples that had been prepared from these mice after 4 wk on the LF/PP diet for 16S rRNA analyses were subjected to shotgun pyrosequencing (n = 4 samples; 87,357 ± 30,710 reads per sample). As with the 16S rRNA analysis, comparisons of the representation of KOs in the various microbiome samples revealed an even greater correlation between complete and cultured communities after they had been subjected to in vivo selection than before their introduction into mice (Fig. 1B, Fig. S2 A–F, and Tables S3–S5).
Previous comparisons of adult germfree mice with those that harbor gut microbial communities (either conventionally raised animals or formerly germfree animals colonized from mouse or human donors) have shown that the presence of a complete gut microbiota is associated with increased adiposity (6, 7). In comparison, colonization of germfree mice with a single, readily cultured, prominent human gut symbiont (Bacteroides thetaiotaomicron) or with a defined community of 12 bacterial species prominently represented in the distal human gut (8) is insufficient to restore epididymal fat pad stores to levels observed in conventionally raised animals (data not shown). To assess whether a complex community of cultured microbes could restore epididymal fat pad weights to the levels associated with complete microbial communities, we evaluated mice colonized with the complete or the cultured fecal communities from the two human donors. All animals displayed significantly greater fat pad to body weight ratios than germfree controls, and no significant difference was observed in adiposity between mice colonized with the donors’ complete or cultured microbiota (Fig. S3D).
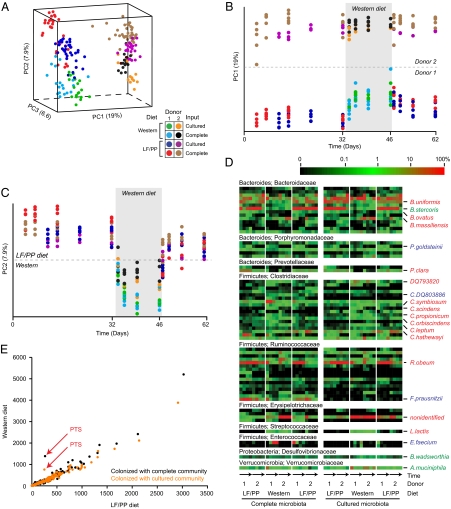
We have reported that mice colonized with a complete human microbiota undergo marked changes in microbial community structure (even after a single day) when shifted from LF/PP chow to a high-fat, high-sugar Western diet (7). To test whether a microbial community consisting only of cultured members recapitulates this behavior in vivo, the four groups of gnotobiotic mice colonized with the complete or cultured microbes from two unrelated human donors were monitored by fecal sampling before, during, and after a 2-wk period when they were placed on the Western diet (samples were collected at days 4, 7, and 14 of the first LF/PP phase, then 1 d before and 3, 7, and 14 d after initiation of the Western diet phase, and finally 1, 3, 8, and 15 d after the return to the LF/PP diet). 16S rRNA-based comparisons of fecal communities were performed using both phylogenetic and nonphylogenetic distance metrics. With either metric, principal coordinates analysis (PCoA) revealed that mice colonized with the complete or cultured samples maintain communities that cluster first by donor (principal coordinate 1; PC1) and that the complete and cultured communities from both donors respond to the diet shift in a similar manner [principal coordinate 2 (PC2); Fig. 2 A–C and Fig. S4 A and B]. Like the transplanted complete microbiota examined here and in previous reports (7), the cultured microbiota responded to this Western diet by increasing the relative proportion of representatives of one class of Firmicutes (the Erysipilotrichi) and decreasing the relative proportion of the Bacteroidia class (Fig. S4C). Notably, of the 18 species-level phylotypes significantly affected by diet shift in the mice containing the complete microbiota of both human donors, 14 were detected and demonstrated the same statistically significant response in mice colonized with readily cultured taxa (Fig. 2D and Fig. S5).
Fig. 2.
Human gut microbial communities composed only of cultured members exhibit in vivo dynamics similar to those observed in their complete counterparts. (A) PCoA of UniFrac distances between 16S rRNA datasets generated from fecal samples from gnotobiotic mice, colonized with complete or cultured human fecal microbial communities from two unrelated donors and sampled over time. From day 33–46, mice were switched from their standard LF/PP chow to a high-fat, high-sugar Western diet. Time series analysis of community structure as viewed along the first two principal coordinates from A shows that interpersonal (donor) differences separated communities on PC1 (B), and host diet separated communities on PC2 (C). Principal coordinate 3 (PC3) separated samples from mice colonized with complete communities from those colonized with cultured populations (Fig. S4A). Nonphylogenetic distance metrics produced similar results (Fig. S4 D–H). (D) Evidence that the community response to diet is driven by readily cultured bacteria and that members of the same taxonomic group manifest distinct responses to diet perturbations. Species-level taxa significantly influenced by diet (Student's t test P ≤ 0.01 after Bonferroni correction; n = 97 taxa tested) in either the complete communities (blue names), the cultured communities (green names), or both (red names) are plotted over time (arrows). Each column represents the average relative abundance in fecal samples harvested from three to five individually caged mice that were sampled at various times: (i) during the initial LF/PP diet phase; (ii) during the subsequent shift to the Western diet; and (iii) upon return to LF/PP chow. Members of family-level groups with at least one diet-responsive species are shown (excluding rare species with average abundance <0.1% across each time point). The names of all taxa are shown in Fig. S5. (E) The functional gene repertoire in the fecal microbiomes of humanized gnotobiotic mice. Each point represents a KEGG level 2 pathway; the number of hits to each pathway per 100,000 shotgun pyrosequencing reads is plotted for mice consuming LF/PP (x axis) or Western (y axis) diets. Data represent the averages of mice colonized with microbial communities from two unrelated donors. The results show that the fecal microbiome associated with the Western diet is enriched for genes in pathways associated with carbohydrate phosphotransferase systems (PTS; red arrows) both in mice colonized with complete (uncultured) human gut communities (black points) and mice colonized with communities of readily cultured members (orange points). Donor-specific data and results from alternate annotation schemes are shown in Fig. S6 and Tables S3–S5.
The fecal microbiomes of LF/PP-fed mice harboring complete or cultured communities from each of the two unrelated donors were compared with microbiomes sampled after these mice consumed the Western diet for 14 d (101,222 ± 24,271 reads per sample). The representation of level 2 KEGG pathway functions was highly concordant on both diets, with one exception: Genes encoding phosphotransferase system (PTS) pathways for carbohydrate transport were significantly overrepresented in Western diet-fed mice harboring complete or cultured communities from either donor (Fig. 2E and Fig. S6 A–D). Higher-resolution KO-level annotations confirmed that the diet-based PTS pathway enrichment reflected increased representation of multiple carbohydrate transporters (Fig. S6 E and F). These results emphasize that the similar taxonomic restructuring of complete and cultured communities in response to diet is accompanied by similar changes in community gene content.
Because human gut communities composed of readily cultured members exhibit responses to host diet that mirror those characteristic of a complete microbiota, we explored the possibility that gnotobiotic mice can be used as biological filters to recover collections of readily cultured microbes, obtained from selected human hosts, that are enriched for certain properties [e.g., the ability to prosper (bloom) when exposed to specific foods or food ingredients]. To this end, fecal samples from mice colonized with the complete or corresponding cultured human gut microbial communities from the two unrelated donors and fed the LF/PP or Western diets were collected directly into anaerobic medium and then plated on prereduced GMM plates (Fig. 3A). After 7-d incubation, V2-directed 16S rRNA profiling of these plated microbial collections confirmed that these populations of cultured microbes can be reshaped deliberately in vivo and then recovered in vitro (Fig. 3B and Fig. S7). On either diet, cultured populations showed significantly greater resemblance to the in vivo communities from mice consuming the same diet than to the in vivo communities from the same mice consuming the alternate diet (P < 10−11, unpaired two-tailed Student's t test of within-donor distances shown in Fig. 3B, assuming equal variances).
Fig. 3.
The community composition of microbes cultured from humanized gnotobiotic mice can be reshaped by altering host diet. (A) Culture collections were generated from fecal samples obtained from gnotobiotic mice colonized with complete or cultured gut microbial communities from either of two unrelated human donors and maintained on LF/PP or Western diets. (B) PCoA of nonphylogenetic (binary Jaccard) distances between cultured samples indicates that manipulation of host diet can be used to shape the composition of communities recovered in culture from these animals. Analysis of phylogenetic (UniFrac) distances between samples produced similar clustering by donor and host diet (Fig. S7).
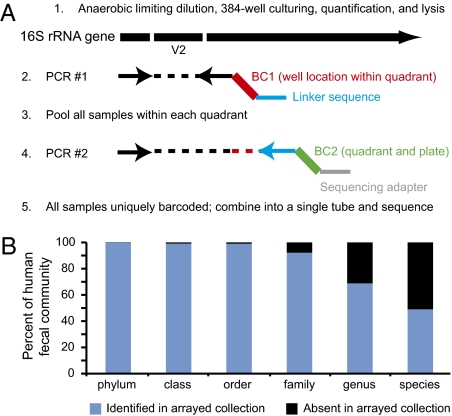
We next turned our attention to developing ways to dissect these recovered populations that display properties of interest in vivo. The strict anaerobic techniques used here, compounded with highly diverse colony morphologies across taxa, complicate efforts to pick and isolate individual colonies at a scale sufficient to capture the bacterial diversity represented on the culture plates. Therefore, we used a most probable number (MPN) technique for creating arrayed species collections in a multiwell format without colony picking. We first empirically determined the dilution point for a fecal sample that yields 70% empty wells (no detectable growth) after inoculation into 384-well trays and 7-d anaerobic incubation. Assuming that the distribution of cells into the wells follows a Poisson distribution, a dilution that leaves 70% of wells empty should yield nonclonal wells (that is, wells that received more than one cell in the inoculum) only 5% of the time; the remainder should be clonal (Fig. S8 F and G). At this dilution, ten 384-well trays should yield ∼1,000 clonal wells. We developed the two-step barcoded pyrosequencing scheme outlined in Fig. 4A to assign a 16S rRNA sequence to the isolate(s) present in each turbid well.
Fig. 4.
Personal culture collections archived in a clonally arrayed, taxonomically defined format. (A) After limiting dilution of the sample into 384-well trays to the point at which most turbid wells are clonal, a two-step, barcoded pyrosequencing scheme allows each culture well to be associated with its corresponding bacterial 16S rRNA sequence. In the first round of PCR, one of the V2-directed 16S rRNA primers incorporates 1 of 96 error-correcting barcodes (BC1, highlighted in red) that designates the location (row and column) within a quadrant of the 384-well tray where the sample resides. The primer also contains a 12-bp linker (blue). All amplicons generated from all wells in a given quadrant from a single plate then are pooled and subjected to a second round of PCR in which one of primers, which targets the linker sequence, incorporates another error-correcting barcode (BC2; green) that designates the quadrant and plate from which the samples were derived, plus an oligonucleotide (gray) used for 454 pyrosequencing. Amplicons generated from the second round of PCR then are pooled from multiple trays and subjected to multiplex pyrosequencing. This approach allows unambiguous assignment of 16S rRNA reads to well and plate locations using a minimum number of barcodes and primers; e.g., 96 BC1 primers and 96 BC2 primers allow 962 (9,216) wells to be analyzed. (B) Representation of the original (complete) microbial community in the arrayed strain collection.
We used this approach to create an archived, personalized culture collection of ten 384-well trays from one of the human donors. 16S rRNA sequences could be assigned to more than 99% of growth-positive wells (Table S8). One advantage of clonally arrayed collections is that the effects of 16S rRNA primer bias encountered using DNA templates prepared from complex microbial populations are minimized when wells contain a single taxon. This point is illustrated by the known bias of most commonly used primers against Bifidobacteria spp. (9). Members of this genus were better represented among the set of 16S rRNA genes produced from individual wells than among those observed in complex communities harvested from GMM plates.
After the archived trays had been frozen under anaerobic conditions and stored at −80 °C for 7 mo, recovery of organisms from wells exceeded 60%. Full-length 16S rRNA sequences generated from these recovered strains matched the assignments from the barcoded pyrosequencing data in every case, suggesting that the dilutions did follow a Poisson distribution as predicted. Like 16S rRNA-based community profiling, such collections may miss rare, but important, members of the microbiota; seeding additional 384-well trays with the diluted sample will capture additional phylotypes (Fig. S8H). In total, this individual's culture collection contained 1,172 taxonomically defined isolates from four different phyla, seven classes, eight orders, 15 families, 23 genera, and 48 named bacterial species. Novel isolates were encountered at the family-, genus-, and species- levels (Table S8), and 69% of the complete community had a genus-level representative in the arrayed collection (Fig. 4B). As a frame of reference, we identified a total of 159 human fecal or gut bacterial species from humans worldwide (including pathogens) in the German Resource Centre for Biological Material (DSMZ) culture collection (SI Materials and Methods). As such, personalized microbiota collections can complement those of international repositories by capturing strains that coexist in a shared habitat where community structure and host parameters can be measured.
Our ability to capture this level of diversity after MPN dilution in these arrayed collections indicates that it is unlikely that interspecies syntrophic relationships by themselves are sufficient to explain the diversity observed on the GMM agar plates. On the other hand, these personalized arrayed culture collections should help identify obligate syntrophic relationships (e.g., by analyzing the patterns of co-occurrence of taxa in wells harboring more than one phylotype or by comparing arrayed collections in which one set of trays contains a candidate syntroph deliberately added to all wells).
Discussion
We find that it is possible to capture a remarkable proportion of a person's fecal microbiota using straightforward anaerobic culturing conditions and easily obtained reagents. Variations in culturing conditions, including components that are not commercially available (e.g., sterile rumen or human fecal extracts) and other approaches for more closely approximating a native gut habitat, undoubtedly will allow additional members of the human gut microbiota to be cultured in vitro (10, 11). These personal culture collections can be generated from humans representing diverse cultural traditions and various physiologic or pathophysiologic states. A key opportunity is provided when anaerobic culture initiatives are combined with gnotobiotic mouse models, thereby allowing culture collections to be characterized and manipulated in mice with defined (including engineered) genotypes who are fed diets comparable to those of the human donor, or diets with systematically manipulated ingredients. Temporal and spatial studies of these communities can be used to identify readily cultured microbes that thrive in certain physiological and nutritional contexts, creating a discovery pipeline for new probiotics and for preclinical evaluation of the nutritional value of food ingredients. Based on their in vivo responses, clonally archived cultured representatives of a person's microbiota can be selected for complete genome sequencing (including multiple strains of a given species-level phylotype) to identify potential functional variations that exist or evolve within a species occupying a given host's body habitat. Coinciding with the introduction of yet another generation of massively parallel DNA sequencers, this approach should also allow vast scaling of current sequencing efforts directed at characterizing human (gut) microbial genome diversity, evolution, and function. In addition, recovered organisms could also be used as source material for functional metagenomic screens (bio-prospecting). Guided by the results of metagenomic studies of human microbiota donors, components of a personalized collection that have coevolved in a single host can be reunited in varying combinations in gnotobiotic mice, potentially after genome-wide transposon mutagenesis of selected taxa of interest (8), for further mechanistic studies of their interactions and impact on host phenotypes.
Materials and Methods
Culturing of Fecal Microbiota.
The Washington University Human Studies Committee reviewed the study design. Freshly discarded fecal samples from two anonymous unrelated human donors were transferred into an anaerobic chamber (Coy Laboratory Products) within 5 min of their collection as described in SI Materials and Methods and Tables S2 and S9.
Gnotobiotic Mouse Husbandry.
All experiments involving mice were performed with protocols approved by the Washington University Animal Studies Committee. Germfree adult male C57BL/6J mice were maintained in plastic gnotobiotic isolators. Colonization, housing, diet manipulations, and control experiments to evaluate the contribution of uncultured cells to microbial communities from gnotobiotic mice are described in SI Materials and Methods.
16S rRNA Sequencing and Analysis.
The V2 region of bacterial 16S rRNA genes was subjected to PCR amplification (DNA extraction and PCR protocols are described in SI Materials and Methods). Metadata for all 500 samples, including barcodes, are provided in Table S10. All 16S rRNA pyrosequencing datasets have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (accession no. SRA026269).
16S rRNA sequences were filtered to remove low-quality or chimeric sequences, de-noised, and analyzed using QIIME v1.1 (3) with parameters described in SI Materials and Methods. To quantify the representation of cultured and uncultured lineages in microbial communities, the presence or absence of each phylum-, class-, order-, family-, genus-, or species-level phylotype assigned to sequences in the complete sample(s) was determined in the cultured sample(s). Taxonomy was assigned with both SILVA-VOTE (SI Materials and Methods and Table S11) and RDP Bayesian classifiers. Data were normalized by the abundance of each taxonomic group in the original (uncultured) sample. For analysis of microbial communities from mice, taxonomic groups observed in fewer than two replicate animals were omitted. Protocols for creation and 16S rRNA sequencing of arrayed culture collections are described in SI Materials and Methods and Tables S12 and S13.
Shotgun Pyrosequencing.
Aliquots (500 ng) of DNA prepared from selected complete and cultured microbiota were sheared and ligated to the default 454 Titanium multiplex identifiers (MIDs; Roche Rapid Library Preparation Method Manual, GS FLX Titanium Series, October 2009) and sequenced using 454 Titanium pyrosequencing chemistry. After filtering of low-quality or host DNA sequences, reads were queried against the KEGG KO database (v52) using parameters described in SI Materials and Methods; metadata for each sample are provided in Table S14, and all shotgun pyrosequencing datasets are available in the NCBI SRA under accession no. SRA026270. Procedures for bio-prospecting for antibiotic resistance genes in complete and cultured microbial communities are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank B. Muegge, S. Wagoner, D. O'Donnell, M. Karlsson, M. Gonzalez, A. Keel, T. Ellison, B. Wang, J. Symington, V. Wagner, M. Dunne, and R. Knight for assistance and comments. This work was supported by National Institutes of Health Grants DK30292, DK70977, and DK78669 (to J.I.G), F32AI078628 and K01DK089121 (to A.L.G.), and T32-HD043010 (to A.M.) and by the Crohn's and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession nos. SRA026269, 026270, and 026271).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102938108/-/DCSupplemental.
References
- 1.Razumov AS. Mikrobiologija. 1932;1:131–146. [Google Scholar]
- 2.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäckhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6–14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill JE, et al. Improvement of the representation of bifidobacteria in fecal microbiota metagenomic libraries by application of the cpn60 universal primer cocktail. Appl Environ Microbiol. 2010;76:4550–4552. doi: 10.1128/AEM.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols D, et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollmann A, Lewis K, Epstein SS. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 2007;73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.