Abstract
Cell surface receptor membrane localization is strongly dependent on protein-protein interactions often involving regulation by phosphorylation/dephosphorylation of the intracellular domains of membrane proteins. The present study was carried out to identify metabotropic glutamate receptor (mGluR) 3 regulatory binding proteins. Using the yeast two-hybrid technique, we found that the 50-aa C-terminal cytoplasmic tail of mGluR3 interacts specifically with protein phosphatase 2Cα (PP2Cα). This interaction was confirmed by GST pull-down and coimmunoprecipitation assays. mGluR3 interacts with PP2Cα, β, γ, and δ isoforms; however, among the mGluR family only mGluR3 interacted with PP2C. The minimal interacting domain of mGluR3 comprised residues 836-855. Alignment between mGluR3 and mGluR2, a closely related group II receptor, indicated that this domain is not conserved between the two receptors. The mGluR3 cytoplasmic C-terminal tail contains one phosphorylation site for protein kinase A (Ser-845), but the phosphatase that dephosphorylates this site has not been previously identified. We find that PP2C, but not PP1, PP2A, or PP2B, dephosphorylates the mGluR3 cytoplasmic tail in vitro. The dephosphorylated form of the mGluR3 cytoplasmic tail, but not the equivalent region of mGluR2, inhibited PP2C assayed by using [32P]casein as a substrate. However, phosphorylation of the mGluR3 cytoplasmic tail at Ser-845 inhibits the interaction with PP2C. These results indicate distinct functions for mGluR2 and mGluR3 and suggest a dynamic regulation of mGluR3 by PP2C.
The vast majority of excitatory synapses in the mammalian central nervous system use the neurotransmitter glutamate to activate two different receptor groups, namely the “ionotropic” and the “metabotropic” glutamate receptor families. Although ionotropic stimulation is commonly associated with fast neurotransmission, metabotropic glutamate receptor (mGluR) activation drives slower and longer-lasting events. The metabotropic receptor family is divided into three groups based on pharmacological properties, sequence homology, and coupling to intracellular messengers. Group I receptors (mGluR1 and -5) are positively coupled to phospholipase C, whereas group II (mGluR2 and -3) and group III (mGluR4, -6, -7, and -8) receptors are negatively coupled to adenylyl cyclase.
mGluRs are expressed in most areas of the brain, and extensive data show their importance in major CNS functions and especially neuroplasticity (1, 2). In addition, mGluRs are emerging as new therapeutic targets for neurodegenerative diseases and addiction (3). A variety of drugs acting on group II mGluRs, for instance, have therapeutic potential in the treatment of anxiety, Parkinson disease, schizophrenia, and drug addiction (for reviews, see refs. 4-6).
The important role of group II mGluRs in synaptic plasticity, especially in long-term depression, has been extensively characterized (7-9). However, basic and specific properties of group II mGluRs, such as the modulation of glutamate release, origin (vesicular or nonvesicular) of the released glutamate, and respective roles of mGluR2 versus mGluR3, remain unclear. To date, no pharmacological way exists to distinguish between mGluR2 and mGluR3 activation, and little is known about the molecular events that take place downstream of receptor activation. Especially at the surface expression level, nothing is known about desensitization, endocytosis, and recycling of the group II mGluRs. In an attempt to better understand specific group II mGluR functions, we sought to identify proteins that interact with the C-terminal intracellular tail of the mGluR3 receptor.
We report here that a 20-aa region at the cytoplasmic C terminus of mGluR3 is a binding domain for protein phosphatase 2C (PP2C), and show that this interaction is specific to mGluR3. We also show that phosphorylation at Ser-845 by cAMP-dependent protein kinase (PKA), a site contained within the 20-residue binding domain of mGluR3, can be dephosphorylated by PP2C but not by protein phosphatase 1 (PP1), PP2A, or PP2B. In addition, PKA phosphorylation at Ser-845, and a Ser-845 to Asp mutation, dramatically reduce PP2C binding. Our data thus present the possibility of distinguishing between mGluR2 and mGluR3 functions.
Materials and Methods
Plasmid Constructs. DNA constructs were generated by standard PCR amplification methods, and the subcloned DNA fragments were systematically checked by sequencing. For the mGluR3-myc expression construct, full-length mGluR3 was PCR-amplified from a QUICK-clone rat brain cDNA library (Clontech) and subcloned into pcDNA3.1 (Invitrogen).
Yeast Two-Hybrid Screen. The C-terminal domain of mGluR3 (amino acids 830-879) was PCR amplified from the full-length construct and subcloned into the NcoI/BamHI sites of a bait pAS2-derived vector, for expression as a GAL4 DNA-binding domain fusion protein (for details, see ref. 10).
The mGluR3-bait plasmid was transformed, by using the lithium acetate method, into yeast strain CG1945 (Clontech). The size and expression level of the fusion protein were checked by immunoblot by using an anti-GAL4 DNA binding domain antibody (Clontech). No activation of his3 and LacZ reporter genes was detected for the mGluR3-bait construct in the presence of the control prey plasmid. The pACT2 rat brain cDNA library was transformed into yeast strain Y187 (Clontech). Bait and prey transformants were mated on YPD medium and plated on medium selective for the expression of the histidine reporter gene. After growth on this medium, a 5-bromo-4-chloro-3-indolyl β-d-galactoside overlay assay was performed. Yeast extracts were prepared from double-positive clones. Prey inserts were amplified by PCR and sequenced by using prey vector oligonucleotides. After characterization of the clones by BLAST, certain prey plasmid clones were selectively rescued from the yeast, transformed into Escherichia coli for DNA amplification and retransformed into the yeast strain Y187 with (i) the original mGluR3-bait vector, (ii) the bait-control vector to test for transactivation, or (iii) three other irrelevant bait constructs to test the specificity of the interaction.
The C-terminal domains of other mGluRs were PCR-amplified from the rat brain cDNA library and subcloned into the pAS2-derived vector. mGluR1a was a generous gift from J. Kitano (Kyoto University, Kyoto), and mGluR6 and mGluR8 were a generous gift from R. Enz (Institut für Biochemie, Erlagen, Germany).
Production of Recombinant Proteins. Full-length cDNAs corresponding to the four rat PP2C isoforms (α, β, γ, and δ) were amplified by PCR (data not shown). PCR fragments were purified and digested by the appropriate restriction enzymes and subcloned into the pET21 vector, which expresses the recombinant protein with an N-terminal polyhistidine tag. The resulting expression vectors were transformed into the bacterial host strain BL21 (DE3), and expression of protein was induced at midlog phase (1 mM isopropyl β-d-thiogalactoside, 4 h, 37°C). Recombinant proteins were purified by affinity chromatography by using standard methods.
PP2B was purified from rat brain (11). Recombinant rat PP1 and PP2A (AC and Bδ″ subunits) were expressed and purified from Sf9 cells by using a baculovirus system (12).
GST Pull-Down Assays and Affinity Chromatography. The C termini of mGluR2 (amino acids 821-872) and mGluR3 (amino acids 830-879) were ligated in-frame to the coding sequence of GST by using the EcoRI and BamHI restriction sites of pGEX-4T1 (Amersham Pharmacia Biotech). Plasmids were transformed into E. coli BL21 (DE3) and expression of protein was induced at midlog phase, by adding 1 mM isopropyl β-d-thiogalactoside. GST-fusion proteins were purified by affinity chromatography by using glutathione-Sepharose beads and were used for pull-down assays as described (13), except that buffers were supplemented with 2 mM MgCl2. Bound proteins were eluted by boiling in SDS sample buffer, separated by SDS/PAGE, and detected by immunoblotting with a monoclonal antihistidine antibody.
Brain homogenates were prepared from female Sprague-Dawley rats after they were euthanized and decapitated. One brain was homogenized in 5 ml of homogenization buffer (20 mM Tris, pH 7.4/150 mM NaCl/2 mM MgCl2/0.5 mM DTT) to which a protease inhibitor mixture (Roche Applied Science) was added. Total homogenate was spun at 14,000 rpm for 30 min at 4°C. The supernatant was saved as brain extract. All preparations were carried out on ice or at 4°C. Recombinant GST or GST-fusion proteins were incubated with glutathione-Sepharose beads overnight at 4°C and washed three times for 15 min at 4°C with PBS. The beads were equilibrated with homogenization buffer. Brain extract was precleared with GST protein for 1 h at 4°C, then incubated with GST or GST-fusion proteins (15 μg) bound to glutathione beads. After 2 h of incubation at 4°C, the beads were washed four times for 20 min in homogenization buffer. Proteins bound to the beads were eluted into sample buffer, separated by SDS/PAGE, and processed for immunoblotting.
In Vitro Phosphatase Assays. Hydrolyzed and partially dephosphorylated casein (Sigma) was phosphorylated by using [γ-32P]ATP (6,000 Ci/mmol) and the catalytic subunit of PKA in a buffer containing 50 mM Hepes (pH 7.2), 10 mM magnesium acetate, and 1 mM EGTA. After 30 min at 30°C, PKA was inactivated by heating at 60°C for 10 min. Phosphorylated casein was separated from ATP by using a Sephadex G-50 NICK column (Amersham Pharmacia Biotech).
The phosphatase activities of PP1, PP2A, PP2B, and PP2Cβ were assayed with a phospho-mGluR3 synthetic peptide (residues 830-859) by using the malachite green detection method (14). Phosphatases were assayed in 50 mM Tris·HCl (pH 7.5)/5 mM DTT/1 mg/ml BSA at 30°C for 10 min as described (15). The PP2B assays were supplemented with 1 μM calmodulin and 1 mM CaCl2 (added 5 min before the assay). The activities of PP2C isoforms were assayed by one of two methods: (i) a malachite green assay by using a 100 μM concentration of either a phospho-GluR3 peptide or a phospho-moesin synthetic peptide (Cys-Lys-Tyr-Lys-Thr(P)-Leu-Arg, residues 554-560); or (ii) 32P-labeled phospho-casein.
In Vitro Phosphorylation of mGluR3 Polypeptide. Purified recombinant C-terminal mGluR3 polypeptide (residues 830-879) (0.5 μg) was incubated with 0.05 mg/ml PKA, 500 μM ATP plus 0.05 mCi/ml [γ-32P]ATP (Perkin-Elmer, specific activity, 3,000 Ci/mmol) in buffer A (50 mM Hepes, pH 7.2/10 mM magnesium acetate/1 mM EGTA) (total reaction volume, 50 μl). Reactions were stopped by adding acetic acid to a final concentration of 10%. An aliquot of the reaction mixture (50 μl) was spotted on P81 Whatman filter paper discs and the discs washed with a continuous flow of water for 20 min to remove unincorporated ATP. Radioactivity recovered in the dried filters was measured by Cerenkov counting.
Coimmunoprecipitation and Antibodies. Transfection of Cos7 cells was performed by standard methods. Cells were grown to 60% confluence and then transfected with pcDNA3.1-mGluR3-myc or pcDNA3.1-myc constructs with FuGENE according to the manufacturer's protocol (Roche Applied Science). After transfection, cell extracts were prepared (receptor membrane solubilization for 6 h at 4°C; 50 mM Tris, pH 7.4/150 mM NaCl/2 mM EDTA/2 mM EGTA/0.1% Triton and protease inhibitors). Equal amounts of cell extracts were immunoprecipitated with myc monoclonal antibody 9E10 coupled to agarose beads (Covance, Princeton). 6His monoclonal antibody was purchased from Clontech. Anti-PP2Cα antibodies were purchased from Upstate Biotechnology (Lake Placid, NY).
Results
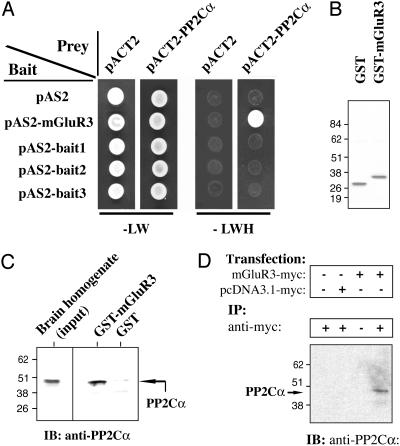
Identification of PP2C as an mGluR3-Interacting Protein by Using the Yeast Two-Hybrid System. To identify specific molecular mechanisms that might distinguish mGluR3 and mGluR2, we used the yeast two-hybrid method to identify proteins that could selectively bind the intracellular C terminus of mGluR3 and not of mGluR2. The last 50 aa of the C-terminal sequence of mGluR3 (residues 830-879) were subcloned into a pAS2-GAL4-derived plasmid and used as bait (mGluR3-830-879). We screened 1 × 106 clones from a pACT2 cDNA library. Of 94 histidine-positive clones, 12 clones were also positive for the β-galactosidase reporter gene. On sequencing, three clones of 12 double-positive clones shared an identical sequence that encoded almost the full-length (minus 21 residues) of protein phosphatase 2Cα (PP2Cα). To determine the specificity of interaction, the PP2Cα prey plasmid was rescued from the yeast, amplified in E. coli, and transformed into yeast that harbored (i) the original mGluR3-830-879 bait plasmid, (ii) the empty bait plasmid, or (iii) one of three irrelevant baits (one viral proteins and two yeast proteins). The interaction was specific, as only the PP2Cα and mGluR3-830-879 interaction was positive (Fig. 1A).
Fig. 1.
Interaction of PP2Cα with mGluR3 in a yeast two-hybrid assay (A), in a GST pull-down assay (B and C), and by coimmunoprecipitation (D). (A)Onthe left, bait constructs are indicated (pAS2 corresponds to the empty-bait plasmid). On the top, prey constructs are indicated (pACT2 corresponds to the empty-prey plasmid). Double transformants were spotted and grown for 3 days at 30°C on -LW medium (Left)oron -LWH medium (Right) to check for histidine auxotrophy. All the clones grew on -LW medium (growth control), but only coexpression of pAS2-mGluR3 and pACT2-PP2Cα allowed growth on histidine-dropout plates. (B) GST or GST-mGluR3-830-879 (last 50 C-terminal amino acids of mGluR3 fused to the GST domain) were expressed in E. coli, purified, and analyzed by SDS/PAGE and staining with Coomassie blue. (C) Total brain homogenate (2 mg) was incubated with immobilized GST (negative control) or GST-mGluR3-830-879 (GST-mGluR3). Retained proteins were analyzed by SDS/PAGE and immunoblotting (IB) by using an anti-PP2Cα antibody. Brain homogenate input (50 μg) was also analyzed (Left). (D) mGluR3-myc or the empty plasmid (pcDNA3.1-myc) was expressed in Cos7 cells. Cells were lysed and immunoprecipitation (IP) was carried out by using a monoclonal anti-myc antibody. Proteins were analyzed by SDS/PAGE and immunoblotting with anti-PP2Cα antibody.
GST-mGluR3-830-879 Pulls Down PP2Cα from Brain Extracts. The specificity of the interaction between mGluR3-830-879 and PP2Cα was biochemically investigated by a GST pull-down assay with immobilized GST-mGluR3-830-879. PP2Cα-6His-tagged protein was incubated with GST-Sepharose beads bound to either GST or GST-mGluR3-830-879 (Fig. 1B) in the presence of 2 mM MgCl2. Purified PP2Cα interacted with GST-mGluR3 but not GST, as demonstrated by immunoblotting by using an anti-PP2Cα antibody and an anti-6His antibody (data not shown). Next the capability of native brain PP2Cα to interact with GST-mGluR3-830-879 was analyzed. GST or GST-mGluR3-830-879 immobilized on Sepharose beads were incubated with rat brain extract in the presence of 2 mM MgCl2, and bound PP2C was analyzed by immunoblotting (Fig. 1C). PP2Cα from rat brain effectively and selectively bound to GST-mGluR3-830-879 but not to GST.
Coimmunoprecipitation of mGluR3 and PP2Cα. To investigate the interaction between full-length mGluR3 and native PP2Cα in mammalian cells, full-length myc-tagged mGluR3 receptor was expressed in Cos-7 cells. mGluR3-myc was immunoprecipitated by using an anti-myc antibody, and bound PP2C was detected by immunoblotting with an anti-PP2Cα antibody (Fig. 1D). mGluR3-myc was coimmunoprecipitated with endogenous PP2Cα (Fig. 1D, lane 4), whereas no PP2C was detected in immunoprecipitates from nontransfected cells (Fig. 1D, lane 1), from cells transfected with the empty plasmid (Fig. 1D, lane 2), or in the absence of the anti-myc antibody (Fig. 1D, lane3).
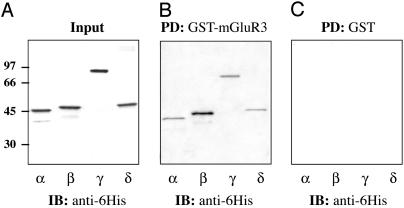
Various PP2C Isoforms Bind to mGluR3-830-879. Numerous PP2C isoforms have been identified, and several of these, including the α, β, γ, and δ isoforms, are expressed in brain (16, 17). As with PP2Cα, the β, γ, and δ isoforms were expressed as 6His-tagged proteins and used in pull-down assays with immobilized mGluR3-830-879. Bound PP2C isoforms were analyzed by immunoblotting with an anti-6His antibody (Fig. 2). Identical amounts of each recombinant PP2C isoform were used (Fig. 2 A). Equal amounts of GST-mGluR3 were used in each pull-down assay as determined by immunoblotting with an anti-GST antibody (not shown). GST-mGluR3-830-879 (Fig. 2B) but not GST (Fig. 2C) bound to all four PP2C isoforms. Approximately equal amounts of PP2Cα, γ, and δ isoforms were recovered, but about 2-fold more PP2Cβ bound to GST-mGluR3, suggesting that this isoform may bind with higher affinity.
Fig. 2.
The four PP2C isoforms (α, β, γ, and δ) interact with GST-mGluR3-830-879. (A) His-tagged PP2C α, β, γ,or δ isoforms were expressed individually in E. coli, purified by affinity chromatography, and analyzed by SDS/PAGE and immunoblotting with an anti-6His antibody (Input, 20%). (B and C) Each PP2C isoform (1 μg) was incubated with 10 μg of either immobilized GST-mGluR3-830-879 (B) or GST (C) (PD, pull down). Retained proteins were analyzed by SDS/PAGE and immunoblotting with an anti-6His antibody.
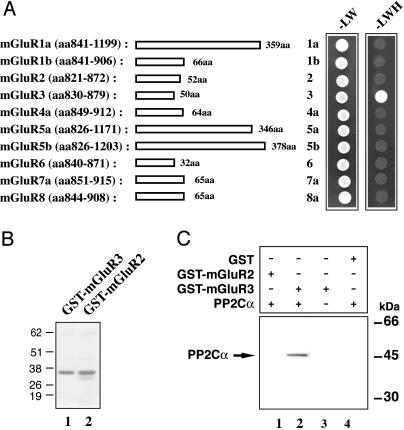
Among mGluRs, PP2Cα Binds Only to mGluR3. We next addressed the question of the specificity of the interaction of PP2C with various members of the mGluR family. The C-terminal regions of all of the mGluR receptors (mGluR1 to mGluR8) were subcloned into the bait plasmid. Each mGluR isoform bait was cotransformed into yeast with the PP2Cα prey construct (originally isolated in the mGluR3-830-879 screen), and the interaction analyzed by using -LW or -LWH medium (Fig. 3A). A positive interaction with PP2Cα was detected only with mGluR3. We further analyzed the interaction of PP2Cα with the group II isoforms, mGluR3 and mGluR2, which have very similar cytoplasmic domains. GST-mGluR2 and -mGluR3 fusion proteins (Fig. 3B) were incubated with purified PP2Cα, and the interaction was analyzed by a GST pull-down assay (Fig. 3C). Only the GST-mGluR3 fusion protein was able to pull down PP2Cα (Fig. 3C, lane 2). All together these results indicate that the interaction of PP2C with mGluR3 is highly specific.
Fig. 3.
PP2C binds specifically to mGluR3 and not to other mGluRs. (A) The domains of mGluR1-8 used as bait in a yeast two-hybrid analysis are indicated on the left (amino acid residue numbers and the total length of each mGluR fragment are indicated). On the right, double transformants were spotted and grown on -LW or -LWH medium (see Fig. 1). (B) GST-mGluR3-830-879 or GST-mGluR2-821-872 were expressed in E. coli and purified by affinity chromatography; proteins were analyzed by SDS/PAGE and staining with Coomassie blue. (C) 6His-PP2Cα (1 μg) was incubated with 10 μg of immobilized GST-mGluR2-821-872 (GST-mGluR2) or GST-mGluR3-830-879 (GST-mGluR3). Retained proteins were analyzed by SDS/PAGE and immunoblotting with an anti-6His antibody.
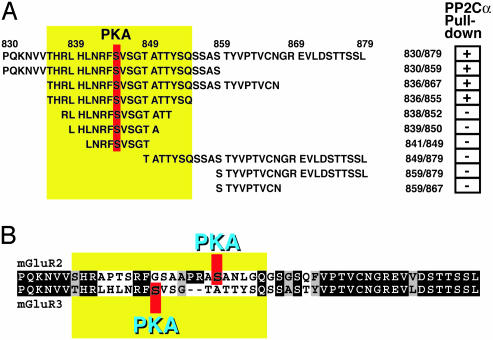
Characterization of the Interaction Between mGluR3 and PP2Cα. We next analyzed the details of the interaction between mGluR3 and PP2Cα by expressing a variety of pieces of the cytoplasmic domain of mGluR3 as GST fusion proteins (Fig. 4A). The interaction of each purified GST-fusion protein with PP2Cα-6His was analyzed by using the GST pull-down assay (results are summarized as + or - in Fig. 4A). The minimal subdomain that bound to PP2Cα comprised residues 836-855 of mGluR3. The amino acid sequences of the group II mGluRs, mGluR2 and mGluR3, are very similar (Fig. 4B), both containing a C-terminal TSSL sequence, a class I PDZ-binding motif (18, 19). Residues 836-855 represent exactly the region of mGluR3 that is not conserved in mGluR2 (Fig. 4 A and B, note position of yellow box).
Fig. 4.
Characterization of the domain of mGluR3 that interacts with PP2C. (A) Various truncated GST-mGluR3 fusion proteins were expressed in E. coli and used in GST pull-down assays. The full-length cytoplasmic region is shown at the top. The results obtained from the GST pull-down assays are summarized on the right side of the panel (+, effective PP2Cα pull down; -, no pull down). The minimal region (residues 836-855) found to interact with PP2Cα is highlighted by the yellow box. Ser-845, a PKA phosphorylation site, is highlighted in red. (B) Alignment of group 2 mGluR cytoplasmic regions. The complete cytoplasmic regions of mGluR2 and mGluR3 are shown. Identical residues are highlighted in black, and homologous residues are highlighted in gray. -, a gap.
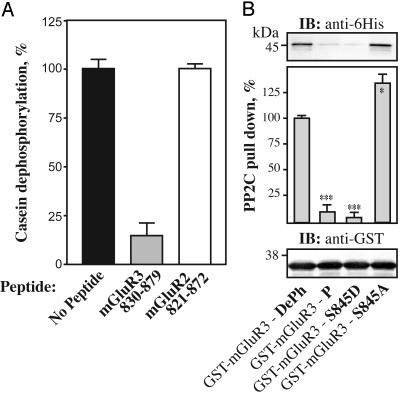
Because the cytoplasmic domain of mGluR3 was able to interact with PP2C isoforms, we examined whether this interaction had any effect on PP2C phosphatase activity. A mGluR3-830-879 polypeptide was an effective inhibitor of PP2C when assayed by using [32P]casein as substrate (Fig. 5A). In the absence of peptide, PP2C effectively dephosphorylated [32P]casein (3 μM). Addition of 3 μM polypeptide mGluR3-830-879 inhibited by >80% the activity of PP2C toward [32P]casein (3 μM). In contrast, a control polypeptide corresponding to mGluR2-821-872 (3 μM) had no effect on PP2C activity.
Fig. 5.
Characterization of the interaction of the dephospho and phospho forms of the mGluR3 cytoplasmic tail with PP2Cα. (A) Positive control (black bar), dephosphorylation of [32P]casein by PP2Cα in the absence of competing polypeptide; mGluR3-830-879 (gray bar), dephosphorylation of [32P]casein (3 μM) by PP2Cα in the presence of 3 μM of mGluR3-830-879; negative control (white bar), dephosphorylation of [32P]casein (3 μM) by PP2Cα in the presence of 3 μM mGluR2-821-872 prepared in the same way as the mGluR3 polypeptide. Data for three experiments (mean ± SEM). (B) 6His-PP2Cα (1 μg) was incubated with 10 μg of immobilized GST-mGluR3-830-879 fusion proteins (GST-mGluR3-wt, no modification; GST-mGluR3-P, Ser-845 phosphorylated in vitro by PKA; GST-mGluR3-S845D and GST-mGluR3-S845A, mutation of Ser-845 to Asp and Ala, respectively). Retained proteins were analyzed by SDS/PAGE and immunoblotting by using an anti-6His antibody. Results were normalized after immunoblotting with an anti-GST antibody. Data for three experiments (mean ± SEM) were compared with the values obtained for the wild-type DePh mGluR3 (*, P < 0.5; ***, P < 0.001, Student's t test).
PP2Cα, but Not PP1, PP2A, and PP2B, Dephosphorylates Phospho-Ser-845 of mGluR3. The region of mGluR3 that interacts with PP2C contains a putative PKA phosphorylation site (Ser-845) (Fig. 4B). We therefore examined if phosphorylation of Ser-845 by PKA might occur. After expression in E. coli and purification, the GST-mGluR3-830-879 fusion protein was cleaved by thrombin to release the mGluR3-830-879 polypeptide. The cleaved polypeptide was efficiently phosphorylated in vitro by PKA (see also ref. 20), and mutation of Ser-845 to Ala or Asp abolished phosphorylation (data not shown).
Given that PP2C can bind to mGluR3 close to the site phosphorylated by PKA, it seemed possible that phospho-Ser-845 might serve as a substrate for PP2C and/or that phosphorylation of Ser-845 might influence the interaction between PP2C and mGluR3. We examined the ability of PP2C to dephosphorylate phospho-Ser-845 of mGluR3 and compared the activity of PP2C with that of the other serine/threonine phosphatases, PP1, PP2A, and PP2B (Table 1). A synthetic phosphopeptide (phospho-mGluR3-830-879) was used as substrate and the relative rate of dephosphorylation by PP2Cβ, the catalytic subunit of PP1, native PP2B, and PP2A (A, C, and Bδ″) was compared. Synthetic peptides derived from moesin, phosphorylase a, and phospho-Thr-34 DARPP-32, were used as control substrates (Table 1). The phospho-mGluR3 substrate was an effective substrate for PP2Cβ (which was the most active of the various PP2C isoforms; data not shown). In contrast, PP1, PP2B, and PP2A did not effectively dephosphorylate phospho-Ser-845, despite being able to efficiently dephosphorylate their standard substrates.
Table 1. PP2C, but not PP1, PP2A, or PP2B, dephosphorylates phospho-Ser-845 of mGluR3.
| Dephosphorylation, % phosphate removed
|
||||
|---|---|---|---|---|
| PP2Cβ | PP1 | PP2B | PP2A | |
| P-mGluR3 peptide | 16 | 1 | 0 | 0 |
| P-moesin peptide | 20 | |||
| P-T34 DARPP 32 | 25 | 18 | ||
| Phosphorylase a | 26 | |||
The activities of purified PP1, PP2A, PP2B, and PP2C were measured under initial-rate conditions by using a mGluR3 peptide (residues 830–859) phosphorylated at Ser-845 by PKA (100 μM), a phosphomoesin peptide (residues 554–560) (100 μM), 32P-labeled Thr-34 DARPP-32 (1 μM), and 32P-labeled phosphorylase a (10 μM) as described (see Materials and Methods). Relative rates of dephosphorylation are expressed as percentage of total phosphate released in the presence of phosphatase in 10 min at 30°C. Data represent means of two independent experiments, performed in duplicate.
We next addressed the question of whether phosphorylation of Ser-845 by PKA might influence the interaction between mGluR3 and PP2C. Phosphorylation of GST-mGluR3 by PKA almost abolished the interaction with PP2Cα when measured by using the GST pull-down assay (Fig. 5B). Mutation of Ser-845 to Asp also prevented the interaction with PP2Cα, likely by acting as a mimic of phospho-Ser-845. In contrast, mutation of Ser-845 to Ala slightly increased the binding to PP2Cα.
Discussion
In the present study, the 50-aa mGluR3 cytoplasmic tail has been found to interact with PP2C. Results were confirmed by several methods, including GST pull-down assays with recombinant and native PP2Cα, and coimmunoprecipitation of the two proteins from cells transfected with PP2Cα and mGluR3-HA. Four well characterized PP2C isoforms (α, β, γ, and δ) were able to bind the mGluR3 cytoplasmic tail, with a slightly greater preference being exhibited by PP2Cβ. By using several different techniques, PP2C was found to bind specifically to mGluR3 and not to the closely related mGluR2 cytoplasmic tail. In addition, none of the other group I or III mGluR receptors were able to bind PP2C. Only a short stretch of the cytoplasmic domain of mGluR3 was needed to interact with PP2C. This 20-aa region between residues 836 and 855 corresponds to the part of the cytoplasmic domain of mGluR3 that is not conserved between mGluR3 and mGluR2. The C-terminal cytoplasmic tails of several mGluRs are phosphorylated, with both mGluR2 and mGluR3 being phosphorylated by PKA at distinct sites within the nonconserved region (20, 21). We confirmed that Ser-845, situated within residues 836-855 of mGluR3, was an effective substrate for PKA in vitro. Furthermore, a synthetic peptide (residues 830-879) phosphorylated at Ser-845 was an effective substrate for PP2C.
Although the phosphorylated form of the C-terminal cytoplasmic domain of mGluR3 was an effective substrate for PP2C, the dephosphorylated domain was a very effective inhibitor of PP2C when assayed by using phospho-casein as substrate. Although this action may occur in part by way of the dephospho-mGluR3 peptide acting as a competitive pseudosubstrate, the inhibitory properties may more likely result from the interaction that occurs between the dephospho-mGluR3 cytoplasmic tail and PP2C. To date, there are no inhibitors of PP2C available, and as a result it has been difficult to examine the role(s) of PP2C in intact cells. Detailed analysis of the interaction of PP2C with the C-terminal region of mGluR3 may help in the future preparation of high-affinity PP2C inhibitors.
Phosphorylation of Ser-845 by PKA or mutation of Ser-845 to Asp greatly inhibited the binding of PP2C to the C-terminal tail of mGluR3. Based on those observations, it seems likely that PP2C interacts with the dephospho form of mGluR3 in intact cells and that it is a complex of dephospho-mGluR3 and PP2C that is immunoprecipitated from cell lysates. In turn, PP2C bound to mGluR3 in intact cells would be inhibited and unable to dephosphorylate cellular substrates. In intact cells, activation of adenylyl cyclase by a variety of neurotransmitters or other agents would result in activation of PKA and presumably lead to an increase in the steady-state phosphorylation level of Ser-845. This would then lead to the release of active PP2C from the C-terminal tail of mGluR3, which would be able to act on phospho-Ser-845 of mGluR3 itself, with the result that the phosphorylation of this site would be transient. The dephosphorylation of Ser-845 would then allow a reassociation of inhibited PP2C with the C-terminal tail of mGluR3. However, it is also possible that PP2C bound to mGluR3 would act to prevent the phosphorylation of Ser-845, both by physically blocking the interaction of PKA with mGluR3 and by rapidly reversing any phosphorylation at Ser-845. Moreover, released PP2C could act locally on other substrates that are closely associated with mGluR3 receptors, or on more distant cellular substrates.
A variety of recent studies have indicated that protein-protein interactions are critical for regulating the cellular localization and substrate specificity of protein phosphatases, including PP1 and PP2A. In the PP1, binding of certain regulatory subunits can inhibit the general activity of the PP1 catalytic subunit and/or switch specificity to a different phospho substrate. Our results indicate that PP2C can both bind to and dephosphorylate the same region of mGluR3, but we do not know if both events can take place at the same time. Because mGluR receptors may dimerize (22), it is possible that PP2C could interact with one subunit of a dimerized receptor and dephosphorylate the other subunit. Moreover, like PP1 bound to certain regulatory subunits, we cannot exclude the possibility that, although inactive toward phospho-casein in vitro, PP2C bound to mGluR3 could still dephosphorylate another region of mGluR3 or a closely associated substrate. The PP1γ isoform has been shown to interact specifically with mGluR7b (23). Thus, an important component of mGluR-dependent signal transduction may involve if and how various mGluRs interact with distinct protein phosphatases.
For PP1, a consensus docking sequence, the so-called “RVxF motif,” is found in many PP1-binding proteins. This motif interacts with an acidic/hydrophobic groove on the surface of PP1, and, in several cases, phosphorylation of serine or threonine residues close to the motif can regulate the interaction of the binding proteins with the PP1 catalytic subunit. Ser-845 is situated within the sequence RFS845V, which resembles the RVxF motif found in PP1-binding proteins. Moreover, phosphorylation of Ser-845 inhibits the interaction of mGluR3 with PP2C. Conceivably, although the structures of the catalytic subunits of PP2C and PP1 are distinct, binding of mGluR3 (and perhaps other proteins) might use a charged/hydrophobic motif for interaction with PP2C.
Except for the G proteins themselves, the regulatory proteins that are known to interact directly with G-coupled protein receptors do so mainly by interacting with C-terminal intracellular regions. For example, the PSD-95/SAP90/GRIP/PICK family of proteins, which has been implicated in the organization of synaptic structure, interact by way of their PDZ domains with the extreme C terminus of receptors, including mGluR3 (24, 25). Other examples of proteins that bind the C-terminal tails of G protein-coupled receptors include the Homer family (26) and the calcyon protein (27). In some cases, such as the interaction of mGluR7b, multiple proteins may be able to bind to overlapping regions of C-terminal tails of mGluRs (23).
The precise function of these protein-receptor interactions has not been fully clarified in most cases, but it seems likely that these protein-protein interactions will inf luence signal-transduction mechanisms, and/or trafficking and localization of the receptor proteins (28). Phosphorylation of receptors or of their binding proteins is likely to influence the actions of the hormones and neurotransmitters that function by way of G protein-coupled receptors. For example, phosphorylation has been found to affect desensitization, internalization, and degradation/recycling of various receptors (for a review, see ref. 29). PP2C may play a role in the recycling of internalized mGluR3 receptors through removal of the phosphate from its cytoplasmic tail (for a general review, see ref. 30). In contrast, mGluR2 would not be subject to the same type of regulation. Phosphorylation of mGluR2 by PKA has been shown to inhibit mGluR2/G protein coupling and attenuate its function as a presynaptic receptor (20, 31). Given the close relationship between mGluR2 and mGluR3, it is possible that phosphorylation by PKA also limits signaling by mGluR3. In this case, the interaction of PP2C with the mGluR3 cytoplasmic tail may function to limit the extent of desensitization. In the prefrontal cortex, group II mGluR agonists can induce postsynaptic Ca2+-dependent long-term depression at glutamatergic synapses, and this can be blocked by PKA inhibitors (9). It will be interesting to examine if mGluR3-Ser-845 phosphorylation by PKA or dephosphorylation by PP2C might play a role in this form of synaptic plasticity.
PP2C in the brain is believed to play an important role in the regulation of synaptic activity, although few neuronal substrates of PP2C have been well characterized. DARPP-32, a key component for dopaminergic and glutamatergic signaling in the neostriatum and other brain regions, is a substrate for PP2C (when phosphorylated at a specific site by CK1) (15). mGluR3 has been shown to be highly expressed in the striatum and in other brain structures, including cerebral cortex, dentate gyrus of the hippocampus, olfactory tubercule, lateral septal nucleus, lateral and basolateral amygdaloid nuclei, and nucleus of the olfactory tract (32). Various PP2C isoforms are expressed in many regions of the brain, including striatum (data not shown), suggesting that the interaction of mGluR3 and PP2C isoforms will have widespread functional relevance to glutamatergic signaling in the brain. PP2C has been found to interact with the cystic fibrosis transmembrane conductance regulator chloride channel (33, 34) and, in Arabidopsis thaliana, PP2C has been shown to interact with the AKT3 potassium channel (35). Together these and the present results suggest an important role for PP2C isoforms in the regulation of receptors and channels.
Acknowledgments
This work was supported by National Institutes of Health Grants DA10044 and MH40899, the F. M. Kirby Foundation, and the Michael Stern Parkinson's Research Foundation. M.F. received an award from the Philippe Foundation.
Abbreviations: mGluR, metabotropic glutamate receptor; PKA, cAMP-dependent protein kinase; PP1, protein phosphatase 1; PP2C, protein phosphatase 2C.
References
- 1.Anwyl, R. (1999) Brain Res. Brain Res. Rev. 29, 83-120. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho, V. & Knopfel, T. (2002) Neuroscientist 8, 551-561. [DOI] [PubMed] [Google Scholar]
- 3.Cornish, J. L. & Kalivas, P. W. (2000) J. Neurosci. 20, RC89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conn, P. J. & Pin, J. P. (1997) Annu. Rev. Pharmacol. Toxicol. 37, 205-237. [DOI] [PubMed] [Google Scholar]
- 5.Aghajanian, G. K. & Marek, G. J. (2000) Brain Res. Brain Res. Rev. 31, 302-312. [DOI] [PubMed] [Google Scholar]
- 6.Rouse, S. T., Marino, M. J., Bradley, S. R., Awad, H., Wittmann, M. & Conn, P. J. (2000) Pharmacol. Ther. 88, 427-435. [DOI] [PubMed] [Google Scholar]
- 7.Bortolotto, Z. A., Fitzjohn, S. M. & Collingridge, G. L. (1999) Curr. Opin. Neurobiol. 9, 299-304. [DOI] [PubMed] [Google Scholar]
- 8.Kahn, L., Alonso, G., Robbe, D., Bockaert, J. & Manzoni, O. J. (2001) Neurosci. Lett. 316, 178-182. [DOI] [PubMed] [Google Scholar]
- 9.Otani, S., Daniel, H., Takita, M. & Crepel, F. (2002) J. Neurosci. 22, 3434-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromont-Racine, M., Rain, J. C. & Legrain, P. (1997) Nat. Genet. 16, 277-282. [DOI] [PubMed] [Google Scholar]
- 11.Sihra, T. S., Nairn, A. C., Kloppenburg, P., Lin, Z. & Pouzat, C. (1995) Biochem. Biophys. Res. Commun. 212, 609-616. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe, T., Huang, H. B., Horiuchi, A., da Cruze Silva, E. F., Hsieh-Wilson, L., Allen, P. B., Shenolikar, S., Greengard, P. & Nairn, A. C. (2001) Proc. Natl. Acad. Sci. USA 98, 3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flajolet, M., Rotondo, G., Daviet, L., Bergametti, F., Inchauspe, G., Tiollais, P., Transy, C. & Legrain, P. (2000) Gene 242, 369-379. [DOI] [PubMed] [Google Scholar]
- 14.Baykov, A. A., Evtushenko, O. A. & Avaeva, S. M. (1988) Anal. Biochem. 171, 266-270. [DOI] [PubMed] [Google Scholar]
- 15.Desdouits, F., Siciliano, J. C., Nairn, A. C., Greengard, P. & Girault, J. A. (1998) Biochem. J. 330, 211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura, S., Yasui, A. & Tsuiki, S. (1989) Biochem. Biophys. Res. Commun. 163, 131-136. [DOI] [PubMed] [Google Scholar]
- 17.Tong, Y., Quirion, R. & Shen, S. H. (1998) J. Biol. Chem. 273, 35282-35290. [DOI] [PubMed] [Google Scholar]
- 18.Songyang, Z., Fanning, A. S., Fu, C., Xu, J., Marfatia, S. M., Chishti, A. H., Crompton, A., Chan, A. C., Anderson, J. M. & Cantley, L. C. (1997) Science 275, 73-77. [DOI] [PubMed] [Google Scholar]
- 19.Sheng, M. & Sala, C. (2001) Annu. Rev. Neurosci. 24, 1-29. [DOI] [PubMed] [Google Scholar]
- 20.Cai, Z., Saugstad, J. A., Sorensen, S. D., Ciombor, K. J., Zhang, C., Schaffhauser, H., Hubalek, F., Pohl, J., Duvoisin, R. M. & Conn, P. J. (2001) J. Neurochem. 78, 756-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alagarsamy, S., Sorensen, S. D. & Conn, P. J. (2001) Curr. Opin. Neurobiol. 11, 357-362. [DOI] [PubMed] [Google Scholar]
- 22.Kunishima, N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H. & Morikawa, K. (2000) Nature 407, 971-977. [DOI] [PubMed] [Google Scholar]
- 23.Enz, R. (2002) J. Neurochem. 81, 1130-1140. [DOI] [PubMed] [Google Scholar]
- 24.Xia, J., Chung, H. J., Wihler, C., Huganir, R. L. & Linden, D. J. (2000) Neuron 28, 499-510. [DOI] [PubMed] [Google Scholar]
- 25.Hirbec, H., Perestenko, O., Nishimune, A., Meyer, G., Nakanishi, S., Henley, J. M. & Dev, K. K. (2002) J. Biol. Chem. 277, 15221-15224. [DOI] [PubMed] [Google Scholar]
- 26.Kammermeier, P. J., Xiao, B., Tu, J. C., Worley, P. F. & Ikeda, S. R. (2000) J. Neurosci. 20, 7238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lezcano, N., Mrzljak, L., Eubanks, S., Levenson, R., Goldman-Rakic, P. & Bergson, C. (2000) Science 287, 1660-1664. [DOI] [PubMed] [Google Scholar]
- 28.Xiao, B., Tu, J. C. & Worley, P. F. (2000) Curr. Opin. Neurobiol. 10, 370-374. [DOI] [PubMed] [Google Scholar]
- 29.Kohout, T. A. & Lefkowitz, R. J. (2003) Mol. Pharmacol. 63, 9-18. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson, S. S. (2001) Pharmacol. Rev. 53, 1-24. [PubMed] [Google Scholar]
- 31.Schaffhauser, H., Cai, Z., Hubalek, F., Macek, T. A., Pohl, J., Murphy, T. J. & Conn, P. J. (2000) J. Neurosci. 20, 5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamaru, Y., Nomura, S., Mizuno, N. & Shigemoto, R. (2001) Neuroscience 106, 481-503. [DOI] [PubMed] [Google Scholar]
- 33.Travis, S. M., Berger, H. A. & Welsh, M. J. (1997) Proc. Natl. Acad. Sci. USA 94, 11055-11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, T., Dahan, D., Evagelidis, A., Zheng, S., Luo, J. & Hanrahan, J. W. (1999) J. Biol. Chem. 274, 29102-29107. [DOI] [PubMed] [Google Scholar]
- 35.Vranova, E., Tahtiharju, S., Sriprang, R., Willekens, H., Heino, P., Palva, E. T., Inze, D. & Van Camp, W. (2001) J. Exp. Bot. 52, 181-182. [PubMed] [Google Scholar]