Abstract
Although β2-adrenergic receptors (β2AR) are expressed on most cell types, mechanisms that establish expression levels and regulate expression by chronic agonist remain unclear. The 3′ UTR of ADRB2 has a conserved 8-nucleotide seed region that we hypothesized is targeted by the let-7 family of miRNAs leading to translational repression. In luciferase assays with transfected cells, luc-β2WT3′UTR had decreased expression when cotransfected with let-7f, but a mutated luc-β23′UTR lacking the seed was unaffected by let-7f; a mutated let-7f also had no effect on luc-β2WT3′UTR expression. ADRB2 mRNA was in greater abundance in immunoprecipitates of Ago2, a core component of the miRNA-induced silencing complex, when cells were transfected with let-7f, but not with a mutated let-7f, indicating a direct interaction with the silencing mechanism. H292 cells transfected with let-7f caused ∼60% decrease in native β2AR expression, but transfection with let-7f–specific locked nucleic acid anti-miRNA increased β2AR expression by ∼twofold. We considered that an increase in let-7f leading to greater repression of translation contributes to agonist-promoted down-regulation. Paradoxically, in cells and in lungs from mice treated in vivo, an ∼50% decrease in let-7f occurs during long-term agonist exposure, indicating a counterregulatory event. Consistent with this notion, let-7f locked nucleic acid transfection caused depressed agonist-promoted down-regulation. Thus, let-7f miRNA regulates baseline β2AR expression and decreases in let-7f evoked by agonist attenuate down-regulation. This positive feedback loop has not previously been described for a G protein-coupled receptor and its miRNA. Methods to decrease let-7f expression in targeted cells may increase therapeutic responses to β-agonist by increasing β2AR expression or minimizing tachyphylaxis.
Keywords: desensitization, airways, noncoding RNA, gene regulation
G protein-coupled receptors (GPCRs) regulate a large repertoire of physiological functions and their pathways are the most commonly targeted by current therapeutic agents (1). Most GPCRs undergo agonist-promoted desensitization, defined as a loss of signaling during continuous activation by agonist (2). Such regulation limits potential deleterious effects from overstimulation and is considered critical to the integration of the multiple signals received by the cell. Early events in agonist-promoted desensitization are phosphorylation of the receptor by G protein-coupled receptor kinases, which promote the binding of β-arrestins that interdict between receptor and G protein, acting to partially uncouple receptor signaling to effectors (2). For some GPCRs, these events also lead to internalization of the receptor to the intracellular space, where routing back to the cell surface or to degradation pathways take place. With more prolonged exposure to an agonist, a loss of the net complement of cellular receptors is frequently observed, which is termed down-regulation. In physiologic settings where stimulation is persistent, down-regulation is readily quantified, such as with elevated catecholamines in heart failure and the down-regulation of cardiac β-adrenergic receptors (βAR) (3, 4). The down-regulation process can also limit the therapeutic effectiveness of chronic agonist administration, termed tachyphylaxis, as has been demonstrated at the cellular and physiologic levels for β-agonists in the treatment of asthma (5, 6). The molecular basis for the various components of agonist-promoted GPCR down-regulation, and in particular for that of the prototypic β2AR, is not well-defined. Indeed, the mechanisms in play that establish baseline expression of β2AR in a given cell, an obvious determinant of the response to endogenous or exogenous agonist, are also not known. Differences (or changes) in transcription (7), mRNA stability (8, 9), and receptor processing (10, 11) have been described as potential mechanisms that establish baseline β2AR expression or agonist-promoted down-regulation. At the level of transcriptional control, multiple transcription factors appear to interact with the promoter and 5′-upstream regions of the intronless ADRB2 gene (12).
Recently, a different level of regulation of many genes has been delineated, which involves RNA–RNA interactions rather than the canonical protein-dependent mechanisms. Indeed, nearly half of all mammalian genes are now predicted to be regulated by small noncoding RNAs (13, 14). Within the small noncoding RNAs, the miRNAs represent one such group with regulatory potential. MicroRNAs are small, single-stranded molecules approximately 21 nucleotides in length. They have the capacity to modulate protein expression levels in response to changing environmental stimuli by controlling protein synthesis in a highly specific spatiotemporal pattern (15, 16). This type of miRNA:mRNA interaction, usually occurring at the 3′ UTR, leads to translational repression, likely through steric hindrance of RNA polymerase actions (17, 18). It has been proposed that this mode of action provides cells the capacity to regulate protein expression levels in a controlled manner yet maintain a full complement of mRNA for a given gene (19). Here we define a unique mechanism by a member of the let-7 family of miRNAs that sets the baseline level of expression of β2AR, and is regulated by agonist leading to a counteraction to down-regulation.
Results
ADRB2 Is a Target for let-7f.
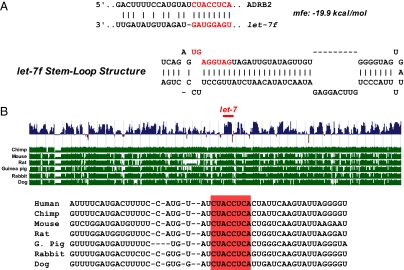
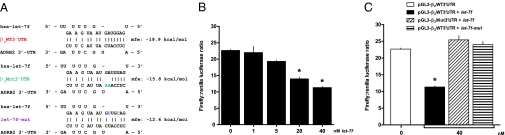
To determine if β2AR mRNA is potentially targeted by miRNAs, we used bioinformatic analyses (TargetScan, PicTar, MicroCosm, and the microRNA.org resource) deriving structural and minimum free energy (mfe) binding predictions. Fig. 1A shows the predicted let-7f stem loop structure and a mfe of −19.9 kcal/mol associated with the interaction between ADRB2 mRNA and let-7f. This β2AR sequence was found to be fully conserved across multiple species, with the let-7f binding site located at virtually the same location within the 3′ UTR (Fig. 1B). To test these prediction models that the let-7 family of miRNAs can functionally interact with the 3′ UTR of ADRB2, HEK293 cells were used for transfecting β2AR-3′UTR–based constructs. HEK293 were transfected with a construct consisting of firefly luciferase followed by the 3′ UTR of ADRB2 (pGL3-β2WT3′UTR) and a mutated 3′ UTR (mfe = −15.8 kcal/mol) lacking the predicted let-7f binding domain (pGL3-β2Mut3′UTR), as well as WT or a mutated let-7f (mfe = −12.6 kcal/mol). The characteristics of these mutated constructs are shown in Fig. 2A. As shown in Fig. 2B, luciferase activity decreased with cotransfection of WT let-7f in a dose-dependent fashion, consistent with let-7f–mediated translational repression. In contrast, let-7f had no effect on firefly luciferase expression when the mutated 3′ UTR construct lacking the predicted let-7f binding site was transfected (Fig. 2C). In addition, the mutated let-7f that lacked the seed region for the ADRB2 3′UTR interaction (let-7f-mut) had no effect when coexpressed with the pGL3-β2WT3′UTR construct (Fig. 2C). These studies with WT and mutated ADRB2 3′ UTR, and WT and mutated let-7f, indicate that the observed effects of let-7f on ADRB2 3′ UTR-based repression are not a result of off-target or nonspecific interactions that might occur because of miRNA transfection. Taken together, these studies point toward an interaction between let-7f and a specific region of the ADRB2 3′ UTR that acts to repress expression.
Fig. 1.
Sequence analysis of ADRB2 3′ UTR. (A) Comparison of the predicted let-7 stem-loop structure with a predicted 8-nucleotide binding site within the 3′ UTR of ADRB2. The mfe for the binding was calculated to be −19.9 kcal/mol. (B) Conservation plot of the 8-nulceotide predicted let-7 binding region within ADRB2 3′ UTR among various species generated from the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu).
Fig. 2.
let-7f alters expression of ADRB2 3′ UTR reporter genes. (A) Characteristics of the mutated ADRB2 3′UTR and let-7f constructs with mfe predictions. (B) Cotransfection of let-7f decreased pGL3-β2WT3′UTR in a dose-dependent fashion in HEK293 cells. *P = <0.05 vs. mock transfection, n = 4 experiments. (C) Mutation of the seed region of ADRB2 3′UTR, or of let-7, ablates effects on reporter expression. *P < 0.05 vs. mock transfection, n = 4 experiments.
Let-7f Promoted Interaction of ADRB2 mRNA and the RNA Silencing Complex Component Ago2.
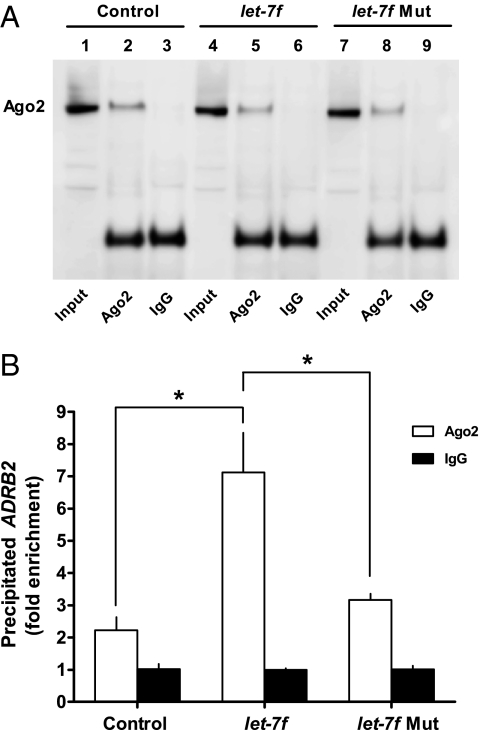
Most miRNAs are transcribed as pri-miRNA. These molecules have a characteristic stem-loop structure and are the target of cleavage by the microprocessor complex consisting of Drosha and DGCR8 (20). The resulting 60 to 70 nucleotide pre-miRNA is then exported to the cytoplasm by the Ran-GTP–dependent nuclear receptor Exportin-5, where it is further processed by the Rnase III enzyme Dicer into ∼21 nucleotide miRNA:miRNA* duplexes. Finally, one strand of this duplex (termed guide miRNA) is assembled into a ribonucleoprotein known as miRNA-induced silencing complex (miRISC); the other strand (termed “passenger strand”) is degraded. The core component of miRISC is Ago2, a member of the Argonaute endonuclease family, which binds the remaining guide strand to silence target mRNAs (21). If β2AR is regulated by the proposed miRNA pathway, then overexpression of let-7f should lead to an increase in Ago2-bound ADRB2 mRNA. To test this, H292 (a human airway epithelial cell line that natively express β2AR) extracts were immunoprecipitated with an Ago2 antibody, and the immunoprecipitates probed quantitatively for ADRB2 mRNA by real-time RT-PCR. The specificity of the antibody and the immunoprecipitation method is shown in Fig. 3A, using Western blotting of the immunoprecipitates where the precipitating antibody was either the Ago2 antibody or IgG. Control (mock-transfected), let-7f-, and let-7f-mut–transfected cells were studied. Immunoprecipitated Ago2 protein was observed only with Ago2 antibody and not with IgG, and Ago2 was expressed at comparable levels under the conditions of all three transfections. When these complexes were then subjected to ADRB2-specific real-time RT-PCR, marked differences in ADRB2 mRNA content were observed (Fig. 3B). In these cells, a modest enrichment of ADRB2 mRNA was observed in Ago2 vs. IgG immunoprecipitates in the nontransfected cells. With cotransfection of let-7f, a significant increase in ADRB2 mRNA was detected in Ago2 immunoprecipitates. In contrast, extracts from cells transfected with the let-7f-mut construct showed no enrichment of ADRB2 mRNA from Ago2 immunoprecipitates over that of the nontransfected controls.
Fig. 3.
let-7 increases Ago2-associated ADRB2 mRNA. (A) Western blots from lysates of H292 cells transfected with either let-7f of let-7f-mut. Extracts were immunoprecipitated with either Ago2 or IgG and blotted with Ago2 antibody. The upper band represents the 97-kDa endogenous input (lanes 1, 4, 7) and immunoprecipitated (lanes 2, 5, 8) Ago2 protein. Lower bands are 55-kDa IgG heavy chains. (B) RNA from immunoprecipitated materials was converted to cDNA by reverse transcriptase and quantified by real-time PCR using ADRB2 specific primers. *P < 0.05, n = 3 experiments.
Let-7f Regulates Endogenous β2AR Expression and Limits Agonist-Promoted Down-Regulation.
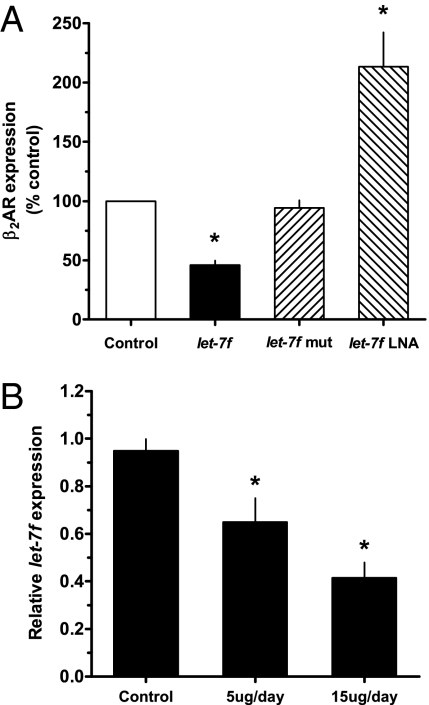
The above studies indicate a specific interaction between let-7f miRNA and the 3′ UTR of ADRB2, which results in repression of a reporter gene. In addition, the studies indicate that there is a direct correlation between the amount of transfected let-7f and the extent of expression. The enrichment of ADRB2 mRNA from Ago2 immunoprecipitates from cells transfected with let-7f further validates a miRNA-based effect of let-7 for β2AR expression. We next assessed whether let-7 regulates baseline expression of β2AR protein natively expressed on H292 cells by increasing let-7f expression and functional let-7f knock-downs. β2AR protein expression on cell membranes was determined by quantitative 125I-cyanopindolol (125I-CYP) radioligand binding. Transfection of let-7f resulted in an ∼60% decrease in β2AR expression (Fig. 4A). No change in expression was found when let-7f-mut was transfected. Endogenous let-7f activity was inhibited by transfection of a let-7f–specific locked nucleic acid (LNA) anti-miRNA. As shown in Fig. 4A, let-7f LNA increased native β2AR expression by ∼twofold over control cells. These studies confirm that native β2AR protein expression is specifically modulated by let-7, with a range of influence amounting to fourfold (∼50 to ∼200 fmol/mg).
Fig. 4.
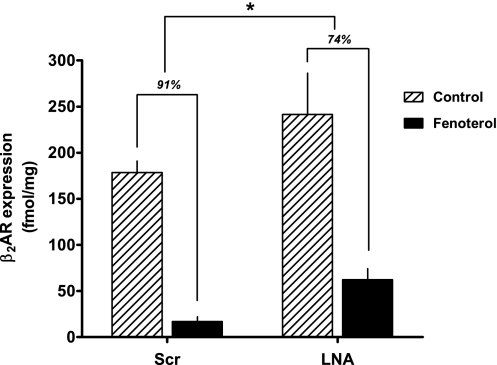
Native β2AR expression is modulated by increasing and decreasing let-7. (A) H292 cells were transfected with let-7f, let-7f-mut, let-7f LNA, or scrambled (scr) LNA. β2AR protein expression was quantitated by 125I-CYP radioligand binding. Control cells are either mock-transfected or, for the LNA experiments, cells transfected with scr LNA. *P < 0.01 vs. control, n = 4. (B) Long-term in vivo agonist treatment decreases let-7f miRNA expression in mouse lung. Mice were treated by intraperitoneal doses of 5 or 15 μg fenoterol daily for 7 d and let-7 miRNA determined by real-time PCR. *P < 0.05 vs. control, n = 3 for each condition.
We hypothesized that let-7f might be up-regulated with agonist exposure, and thus the increased repression of translation would be a component of the agonist-promoted down-regulation process. However, we found a 33 ± 7% decrease in let-7f expression in H292 cells after 12 h of exposure to 10 μM of the β2AR agonist fenoterol (P < 0.05, n = 7). Using pharmacologically relevant doses of fenoterol delivered for 7 d to mice, we noted a dose-dependent decrease of up to 50% in lung let-7f expression compared with lungs from sham-treated mice (Fig. 4B). Given that these conditions readily evoke a loss of β2AR expression (Fig. 5) (22, 23), we then considered that agonist-promoted decreases in let-7f are a mechanism to limit the extent of agonist-promoted down-regulation. Thus, the net effect on β2AR expression from prolonged agonist exposure includes a counterregulatory loss of miRNA-mediated translational repression. To further examine this effect, H292 cells were transfected with let-7f LNA or a scrambled LNA control in the absence or presence of 12-h agonist exposure. We expected that the effect of lowering let-7f by agonist exposure and also decreasing the effect of the remaining let-7f by LNA would lead to decreased down-regulation, as translational repression would be minimized by the two mechanisms. As shown in Fig. 5, this was indeed the case, with agonist-promoted down-regulation amounting to ∼91% in control cells and agonist-promoted down-regulation in the face of let-7 LNA transfection being ∼74%.
Fig. 5.
Effects of let-7f miRNA on agonist-promoted β2AR down-regulation. H292 cells were transfected with scr let-7f LNA (representing the control condition) or let-7f LNA to depress let-7f effects. Cells were treated with carrier (0.1 mM ascorbic acid, control) or carrier with 10 μM fenoterol for 12 h, washed, and 125I-CYP binding performed to quantitate β2AR expression. *Down-regulation less than scr control, P < 0.05, n = 3.
Discussion
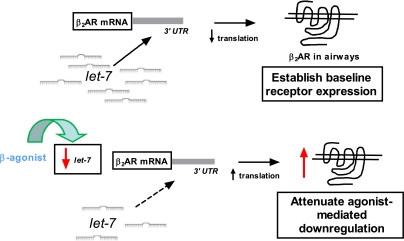
GPCRs serve as critical elements in the maintenance of homeostasis at the cell, organ, and organism levels under normal physiologic states and during pathophysiologic conditions. These receptors are targets for pharmacologic therapy with receptor agonists and antagonists for a wide range of diseases, and have also been implicated in aberrant responses contributing to disease. A key aspect of signaling by most GPCRs is the capacity to adapt to a changing environment. This plasticity is a necessary attribute because the influences that impact a given state are highly dynamic, thus requiring receptors to adjust signaling along timeframes ranging from seconds to days. Of particular interest have been the mechanisms by which a cell regulates the level of receptor expression and how expression is affected by chronic agonist exposure. In regards to the former, “basal” receptor expression is considered a major determinant of the response to endogenously generated or exogenously administered agonist (24). With prolonged agonist exposure, many GPCRs display a decrease in expression, which is termed down-regulation, and is one component of tachyphylaxis to administered agonists; down-regulation from elevated levels of endogenous agonist is also apparent (3, 4). For the prototypic β2AR, these adaptive responses, such as rapid phosphorylation by G protein-coupled receptor kinases, and many of the mechanisms that are apparent over longer time courses have been documented (2, 25–27). The mechanisms responsible for β2AR down-regulation have been shown to occur at the level of transcription (7), mRNA stability (8, 9), and protein degradation (10, 11). Here we show an additional component, translational repression by let-7f, which has a two-pronged effect on receptor expression (Fig. 6). Basal levels of β2AR expression appear to be effected by this mechanism, and indeed let-7–dependent regulation resulted in protein expression levels that span at least a fourfold range. Of note, measured β2AR expression varies between individuals by about this same extent in many tissues, such as heart (28), skeletal muscle (29), peripheral lung (30), lymphocytes (30), and airway epithelial cells (5), supporting the notion that this variation in expression is within the physiologic range. We do not claim, however, that this let-7 mechanism is the only means by which basal levels of β2AR are set, but rather it is one mechanism that results in changes within the known range in relevant tissues. Interestingly, let-7f binding sites are also predicted in the 3′ UTRs of the β1AR and β3AR subtypes (704–710 and 521–527 nucleotides from the stop codons, respectively) but are not found in genes for the three α1AR subtypes or the three α2AR subtypes. This finding suggests that the let-7 mechanism defined in the present report may be a βAR-specific mechanism within the adrenergic receptor family.
Fig. 6.
Dual role of let-7f in regulation of β2AR protein expression. Under static conditions, let-7f actively represses translation and establishes basal levels of β2AR expression. During agonist activation, let-7f levels decrease, resulting in depressed ADRB2 gene silencing and thus an attenuation of agonist-promoted down-regulation.
An unexpected finding was the effect of prolonged agonist exposure on let-7f expression. Given the efficient control of β2AR expression exhibited by let-7f, we assumed that it participated in agonist-mediated down-regulation by enhanced expression and thus increased gene silencing. However, we found that let-7f expression was decreased by prolonged agonist exposure. Given that the net effect of such exposure is decreased β2AR expression, the decrease in let-7f, which causes an increase in β2AR expression, appears to be a counterregulatory event that attenuates down-regulation (Fig. 6). Several reports indicate that GPCR activation can alter miRNA expression, which has been linked to altered expression of other proteins (31). Furthermore, forskolin treatment of cardiomyocytes has been shown to increase miR-1 miRNA expression (32). However, we are unaware of a feedback loop as reported here for the β2AR being previously shown between any GPCR and miRNAs that regulate its expression. In the present report, activated β2AR leading to decreased let-7 would be considered a positive feedback loop, in that decreased let-7 diminishes ADRB2 translational repression and thus shifts the equilibrium toward an increase in β2AR expression. To our knowledge, no positive feedback loops regulating human β2AR expression by any mechanism have been reported. The previously characterized regulatory events that accompany agonist activation result in a loss of expression or function. It is not entirely unexpected, however, that “buffering” mechanisms would also be present, essentially providing additional control nodes to moderate negative feedback. Finally, we note that only two other groups of miRNAs, represented by miR-15 and miR-30, have predicted binding sites within the 3′ UTR of the β2AR (Fig. S1). The functional effects of these miRNAs will require additional study.
In summary, we have shown here that translational repression imposed by the binding of let-7f to the 3′ UTR of the β2AR regulates baseline expression. Such regulation can be demonstrated in transfected cells and natively expressing cells, and is highly specific because mutations of the 3′ UTR or let-7f abolish the effect. The range of control over β2AR expression is within the physiological range. In addition, agonist activation of the β2AR decreases let-7f expression, leading to an attenuation of agonist-promoted down-regulation. Taken together, these studies suggest that genetic or pharmacologic means of modulating let-7 expression in target tissues, such as heart or lung, may improve therapeutic outcome by increasing baseline β2AR expression or attenuating tachyphylaxis.
Methods
Constructs and Luciferase.
To generate the ∼600-bp β2 3′ UTR, the following PCR primers were used to amplify this fragment from human genomic DNA: forward primer: 5′-AGCAGTTTTTCTACTTTTTA AAGAC-3′ and reverse primer: 5′-AGGCAACAGCACTCCAGTCAAG-3′. This PCR product was subsequently cloned into the firefly luciferase pGL3-Control vector downstream of the luciferase ORF at the XbaI site to generate pGL3-β2WT3′UTR. Orientation and sequence accuracy were confirmed by sequencing. To generate the β2mut3′UTR, site-directed mutagenesis (Strategene) was used to mutate the first two nucleotides (CU to AA) within the let-7 seed region of β2WT3′UTR. The let-7f-mut miRNA containing mutations within its seed region was obtained from Invitrogen. For luciferase assays, HEK293 cells were seeded in 96-well plates. Transfections used lipofectamine 2000 (Invitrogen) with pGL3-β2WT3′UTR, pGL3-β2mut3′UTR, let-7f, or let-7f-mut, and renilla luciferase (pRL-SV40), which was used as a transfection control. Luciferase activity was measured using the dual-luciferase reporter assay (Promega). Forty-eight hours after transfection, cells were lysed and luciferase activities were measured on a Victor3 Multilabel Counter (Perkin-Elmer). Data are reported as the firefly:renilla luciferase activity ratios.
Immunoprecipitation.
Immunoprecipitation studies were performed on H292 cells (ATCC) transfected with let-7f or a scrambled control, using a modification of previously described methods (33). Briefly, 1 mg of whole-cell lysate was precleared with protein A/G agarose and then incubated with 1:50 Ago2 antibody (Cell Signaling) or control IgG (Santa Cruz) for 1 h at 4 °C. After addition of 50 μL of protein G agarose, the lysates were incubated for 12 h at 4 °C. Agarose beads were washed by serial centrifugation with lysis buffer, high salt, and then low-salt buffer. RNA was extracted with TRIzol (Invitrogen), precipitated with isopropanol in the presence of glycogen to aid in pellet recovery, and treated with DNase I (Applied Biosystems). Complementary DNA was generated using SuperScript III reverse transcriptase and quantitated by real-time PCR as described elsewhere (34). The 18s rRNA was used for normalization. Western blots were performed on the immunoprecipitated material, as previously described (35, 36), with the following modifications: 25 μL of gel-loading buffer was directly loaded on the agarose pellet and the protein was denatured for 3 min at 100 °C. Membranes were blotted with antihuman Ago2 (Wako) at a titer of 1:200. The ECL Advanced Western Blotting kit (GE Healthcare) was used for detection and images were acquired using the Fuji LAS-3000 camera (Fujifilm Medical Systems) and quantitated using the associated software.
Radioligand Binding and Down-Regulation.
β2AR expression levels were determined by quantitative 125I-CYP radioligand binding, as previously described (37). Briefly, H292 cells were transfected with either let-7f (40 nM final concentration) (Invitrogen) or let-7f LNA (25 nM final concentration) (Exiqon) and detached after 48 h by scraping in 5 mM Tris, 2 mM EDTA, pH 7.40 and centrifuged at 30,000 × g for 10 min at 4 °C. Lysates were then homogenized in 500 μL of buffer containing 75 mM Tris, 12 mM MgCl2, and 2 mM EDTA, pH 7.40, which was also used as the incubation buffer for radioligand binding. 125I-CYP binding was carried out for 2 h at 25 °C, and bound radioligand separated from free by vacuum filtration over glass-fiber filters (Whatman), which were counted in a γ counter. Nonspecific binding was determined by coincubations with 10 μM propranolol, and all tubes contained 100 μM GTP. For receptor down-regulation studies, H292 cells were transfected with either let-7f LNA or an LNA scrambled control; after 48 h, cells were treated with the β2AR-specific full-agonist fenoterol for an additional 12 h followed by washing three times with cold PBS and 125I-CYP binding, as described above. Results are expressed as femtomole per milligram or as a percentage of the expression of cells under a control condition.
Mouse Studies.
The mouse studies were approved by the Institutional Animal Care and Use Committee at the University of Maryland School of Medicine. Twelve- to 18-wk-old FVB mice (Taconics) were administered intraperitoneal injections of fenoterol at 5 μg/d or 15 μg/d for 7 consecutive days; controls were administered saline. Following drug treatment, lungs were harvested and RNA extracted using TRIzol reagent. For detecting let-7f levels, real-time PCR as described above was used to determine expression levels with U6 snRNA used as an internal control.
MicroRNA Target Prediction and Sequence Alignment.
Multiple species alignment of the β2AR 3′ UTR was performed using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu) (38). To identify potential miRNA binding sites within the 3′ UTR of β2AR, the following bioinformatic databases were used: TargetScan (http://www.targetscan.org) (39), the microRNA.org resource (http://www.microrna.org) (40), MicroCosm (http://www.ebi.ac.uk/enright-srv/microcosm) (41), and PicTar (http://pictar.mdc-berlin.de) (42). MicroRNA-mRNA free energies were determined using RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) (43).
Data Analysis and Statistics.
All statistical calculations were performed using Prism (GraphPad). Comparisons were by two-way paired or unpaired t tests, with P < 0.05 considered significant. Comparisons among multiple conditions were performed by ANOVA followed by post hoc t tests. Data are expressed as mean ± SEM of n experiments.
Supplementary Material
Acknowledgments
We thank Rachel Schillinger and Molly Malone for technical assistance, and Esther Moses for manuscript preparation. This work was funded by National Institutes of Health Grants HL104119 (to W.C.H.W.) and HL045967, HL071609, and HL065899 (to S.B.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101439108/-/DCSupplemental.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Reiter E, Lefkowitz RJ. GRKs and beta-arrestins: Roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17(4):159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Bristow MR, et al. Decreased catecholamine sensitivity and β-adrenergic-receptor density in failing human hearts. N Engl J Med. 1982;307:205–211. doi: 10.1056/NEJM198207223070401. [DOI] [PubMed] [Google Scholar]
- 4.Bristow MR, Hershberger RE, Port JD, Minobe W, Rasmussen R. β1- and β2-adrenergic receptor-mediated adenylate cyclase stimulation in nonfailing and failing human ventricular myocardium. Mol Pharmacol. 1989;35:295–303. [PubMed] [Google Scholar]
- 5.Turki J, Green SA, Newman KB, Meyers MA, Liggett SB. Human lung cell β2-adrenergic receptors desensitize in response to in vivo administered β-agonist. Am J Physiol. 1995;269:L709–L714. doi: 10.1152/ajplung.1995.269.5.L709. [DOI] [PubMed] [Google Scholar]
- 6.van der Woude HJ, Winter TH, Aalbers R. Decreased bronchodilating effect of salbutamol in relieving methacholine induced moderate to severe bronchoconstriction during high dose treatment with long acting beta2 agonists. Thorax. 2001;56:529–535. doi: 10.1136/thorax.56.7.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosoda K, et al. Regulation of beta 2-adrenergic receptor mRNA and gene transcription in rat C6 glioma cells: Effects of agonist, forskolin, and protein synthesis inhibition. Mol Pharmacol. 1995;48:206–211. [PubMed] [Google Scholar]
- 8.Hadcock JR, Wang H, Malbon CC. Agonist-induced destabilization of β-adrenergic receptor mRNA. Attenuation of glucocorticoid induced upregulation of β-adrenergic receptors. J Biol Chem. 1992;267:4740–4746. [PubMed] [Google Scholar]
- 9.Tholanikunnel BG, Granneman JG, Malbon CC. The M(r) 35,000 β-adrenergic receptor mRNA-binding protein binds transcripts of G-protein-linked receptors which undergo agonist-induced destabilization. J Biol Chem. 1995;270:12787–12793. doi: 10.1074/jbc.270.21.12787. [DOI] [PubMed] [Google Scholar]
- 10.Bouvier M, et al. Two distinct pathways for cAMP-mediated down-regulation of the β2-adrenergic receptor. Phosphorylation of the receptor and regulation of its mRNA level. J Biol Chem. 1989;264:16786–16792. [PubMed] [Google Scholar]
- 11.Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 12.Panebra A, Schwarb MR, Glinka CB, Liggett SB. Allele-specific binding of airway nuclear extracts to polymorphic β2-adrenergic receptor 5′ sequence. Am J Respir Cell Mol Biol. 2007;36:654–660. doi: 10.1165/rcmb.2006-0394OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condorelli G, Latronico MV, Dorn GW., 2nd microRNAs in heart disease: Putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 20.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swift SM, Schwarb MR, Mihlbachler KA, Liggett SB. Pleiotropic β-agonist-promoted receptor conformations and signals independent of intrinsic activity. Am J Respir Cell Mol Biol. 2007;36:236–243. doi: 10.1165/rcmb.2006-0257OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak JC, Nishikawa M, Shirasaki H, Miyayasu K, Barnes PJ. Protective effects of a glucocorticoid on downregulation of pulmonary beta 2-adrenergic receptors in vivo. J Clin Invest. 1995;96(1):99–106. doi: 10.1172/JCI118084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouvier M, et al. Expression of a human cDNA encoding the beta 2-adrenergic receptor in Chinese hamster fibroblasts (CHW): Functionality and regulation of the expressed receptors. Mol Pharmacol. 1988;33(2):133–139. [PubMed] [Google Scholar]
- 25.Liggett SB. Molecular and genetic basis of β2-adrenergic receptor function. J Allergy Clin Immunol. 1999;104(2 pt 2):S42–S46. doi: 10.1016/s0091-6749(99)70272-1. [DOI] [PubMed] [Google Scholar]
- 26.Collins S, Lohse MJ, O'Dowd B, Caron MG, Lefkowitz RJ. Structure and regulation of G protein-coupled receptors: The beta 2-adrenergic receptor as a model. Vitam Horm. 1991;46:1–39. doi: 10.1016/s0083-6729(08)60681-0. [DOI] [PubMed] [Google Scholar]
- 27.Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 28.Liggett SB, et al. A polymorphism within a conserved β(1)-adrenergic receptor motif alters cardiac function and β-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liggett SB, Shah SD, Cryer PE. Characterization of β-adrenergic receptors of human skeletal muscle obtained by needle biopsy. Am J Physiol. 1988;254:E795–E798. doi: 10.1152/ajpendo.1988.254.6.E795. [DOI] [PubMed] [Google Scholar]
- 30.Liggett SB, Marker JC, Shah SD, Roper CL, Cryer PE. Direct relationship between mononuclear leukocyte and lung β-adrenergic receptors and apparent reciprocal regulation of extravascular, but not intravascular, α- and β-adrenergic receptors by the sympathochromaffin system in humans. J Clin Invest. 1988;82(1):48–56. doi: 10.1172/JCI113600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra PK, Metreveli N, Tyagi SC. MMP-9 gene ablation and TIMP-4 mitigate PAR-1-mediated cardiomyocyte dysfunction: A plausible role of dicer and miRNA. Cell Biochem Biophys. 2010;57(2-3):67–76. doi: 10.1007/s12013-010-9084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, et al. MicroRNA-1 downregulation by propranolol in a rat model of myocardial infarction: A new mechanism for ischaemic cardioprotection. Cardiovasc Res. 2009;84:434–441. doi: 10.1093/cvr/cvp232. [DOI] [PubMed] [Google Scholar]
- 33.McGraw DW, et al. Airway smooth muscle prostaglandin-EP1 receptors directly modulate beta2-adrenergic receptors within a unique heterodimeric complex. J Clin Invest. 2006;116:1400–1409. doi: 10.1172/JCI25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deshpande DA, et al. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med. 2010;16:1299–1304. doi: 10.1038/nm.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Small KM, et al. Alpha2A- and alpha2C-adrenergic receptors form homo- and heterodimers: The heterodimeric state impairs agonist-promoted GRK phosphorylation and beta-arrestin recruitment. Biochemistry. 2006;45:4760–4767. doi: 10.1021/bi052074z. [DOI] [PubMed] [Google Scholar]
- 36.Wang WCH, Schillinger RM, Malone MM, Liggett SB. Paradoxical attenuation of β2-AR function in airway smooth muscle by Gi-mediated counterregulation in transgenic mice overexpressing type 5 adenylyl cyclase. Am J Physiol Lung Cell Mol Physiol. 2011;300:L472–L478. doi: 10.1152/ajplung.00273.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panebra A, et al. Common ADRB2 haplotypes derived from 26 polymorphic sites direct β2-adrenergic receptor expression and regulation phenotypes. PLoS ONE. 2010;5:e11819. doi: 10.1371/journal.pone.0011819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. UCSC genome browser tutorial. Genomics. 2008;92(2):75–84. doi: 10.1016/j.ygeno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 43.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.