Abstract
Elucidating the molecular mechanisms that regulate human stromal (mesenchymal) stem cell (hMSC) differentiation into osteogenic lineage is important for the development of anabolic therapies for treatment of osteoporosis. MicroRNAs (miRNAs) are short, noncoding RNAs that act as key regulators of diverse biological processes by mediating translational repression or mRNA degradation of their target genes. Here, we show that miRNA-138 (miR-138) modulates osteogenic differentiation of hMSCs. miRNA array profiling and further validation by quantitative RT-PCR (qRT-PCR) revealed that miR-138 was down-regulated during osteoblast differentiation of hMSCs. Overexpression of miR-138 inhibited osteoblast differentiation of hMSCs in vitro, whereas inhibition of miR-138 function by antimiR-138 promoted expression of osteoblast-specific genes, alkaline phosphatase (ALP) activity, and matrix mineralization. Furthermore, overexpression of miR-138 reduced ectopic bone formation in vivo by 85%, and conversely, in vivo bone formation was enhanced by 60% when miR-138 was antagonized. Target prediction analysis and experimental validation by luciferase 3′ UTR reporter assay confirmed focal adhesion kinase, a kinase playing a central role in promoting osteoblast differentiation, as a bona fide target of miR-138. We show that miR-138 attenuates bone formation in vivo, at least in part by inhibiting the focal adhesion kinase signaling pathway. Our findings suggest that pharmacological inhibition of miR-138 by antimiR-138 could represent a therapeutic strategy for enhancing bone formation in vivo.
Keywords: regulatory RNA, bone biology, osteoblastic differentiation
Bone marrow-derived human stromal (mesenchymal and skeletal) stem cells (hMSCs) are a population of self-renewing multipotent cells that have significant clinical potential in cellular therapies for tissue regeneration. hMSCs can differentiate along several lineages, including the osteogenic lineage, in response to stimulation by multiple environmental factors (1–4), and they involve complex pathways regulated at both transcriptional and posttranscriptional levels. However, the regulation of these cellular pathways is not fully understood. The ERK-dependent pathway plays a key role in the transcriptional control of bone formation, including phosphorylation of Runt-related transcription factor 2 (Runx2)/Cbfa-1 (5), induction of Osterix gene expression, and alkaline phosphatase activity (6, 7). Furthermore, contact with ECM proteins can induce osteogenic differentiation of hMSCs through the ERK-dependent pathway. The focal adhesion kinase (FAK) signaling pathway is suggested to provide a link between activation of ERK1/2 by ECM proteins (7), thereby stimulating subsequent phosphorylation of Runx2/Cbfa-1, a key transcription factor controlling osteogenic gene expression (7). Furthermore, disruption of FAK signaling in osteoblasts indicates that FAK is a critical factor in the mechanotransduction pathway for osteoblast differentiation (8).
MicroRNAs (miRNAs) function at the posttranscriptional level by negatively regulating translation of their target mRNAs by imperfect binding to their 3′ UTRs. miRNAs have emerged as key regulators of diverse physiological and pathological processes, including cell proliferation, apoptosis, and cancer (9, 10). Although 1,048 miRNAs have been identified in the human genome, the biological functions of relatively few miRNAs have been characterized in detail (11). The impact of miRNAs on osteoblastic differentiation of various cell types has been investigated by modulation of miRNA function (12). These approaches successfully showed that miRNA-204/211 (miR-204/211) targets Runx2 in bone marrow-derived MSC, stimulates adipocyte differentiation, and inhibits osteoblastic differentiation (13). However, Runx2 has been shown to negatively regulate expression of the miR cluster 23a∼27a∼24-2, thereby increasing expression of bone markers and enhancing osteoblast differentiation (14). In human adipose tissue-derived MSCs (hADSCs), miR-26a has been reported to repress the translation of the osteogenic marker, SMAD1 (15), whereas overexpression of miR-196a regulates HOXC8, enhances osteogenic differentiation, and decreases hADSC proliferation (16). miR-125b has been shown to inhibit osteoblast differentiation in the mouse MSC line ST2 (17), whereas miR-133 and -135 directly target Runx2 and Smad5, respectively, and inhibit differentiation of osteoprogenitors of C2C12 mesenchymal cells (18). Moreover, miR-141 and -200a were recently found to be involved in preosteoblast differentiation through regulation of their common target Dlx5 (19), whereas miR-206 inhibits osteogenesis in vitro and in vivo by targeting Cx43 (20). Additionally, miR-29b and -2861 were characterized as positive regulators by targeting inhibitors of osteoblast differentiation (21, 22).
In this study, we screened for differentially expressed miRNAs during osteoblast differentiation of hMSCs and identified miR-138 as a potential inhibitor of osteogenesis. By modulating miR-138 activity, we show that inhibition of miR-138 by an antimiR oligonucleotide markedly increased osteogenic differentiation in vitro and enhanced ectopic bone formation in vivo, whereas miR-138 overexpression reversed these effects. FAK was identified as a direct target of miR-138. Our findings that inhibition of miR-138 by antimiR-138 leads to derepression of FAK and stimulation of FAK signaling that coincides with increased bone formation strongly suggest that miR-138 suppresses osteogenic differentiation by repression of FAK and its downstream signaling.
Results
Identification of Differentially Expressed miRNAs During Osteoblast Differentiation.
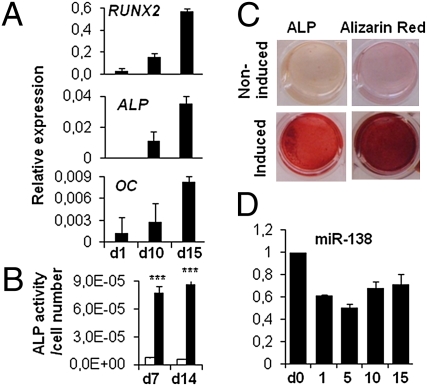
Osteoblast differentiation of hMSCs was induced by using standard osteoblast-induction medium and evidenced by increased expression of genes associated with osteoblast differentiation, RUNX2, alkaline phosphatase (ALP), and osteocalcin (OC), at days 1, 10, and 15 after induction (Fig. 1A). The osteoblast phenotype was confirmed by demonstration of increased alkaline phosphatase activity (Fig. 1 B and C) and Alizarin Red staining for matrix mineralization (Fig. 1C). Increased expression of osteoblast-associated genes and the observed osteoblast phenotype were in accordance with previous reports describing hMSC differentiation into osteoblasts (1–3).
Fig. 1.
Osteoblast differentiation of hMSC. hMSCs were induced to osteoblast differentiation. (A) Osteoblast differentiation was confirmed by qRT-PCR analysis of osteoblast marker genes (RUNX2, ALP, and OC normalized to β-actin). (B) ALP activity was measured during the course of differentiation. miR-138 expression was measured during osteoblast differentiation. White bars represent noninduced samples, and black bars represent induced samples. ***P < 0.001 between noninduced and induced samples. (C) ALP and Alizarin Red staining were performed at day 15. (D) miR-138 expression (n = 3 for all experiments).
To identify differentially expressed miRNAs during osteoblast differentiation, we carried out miRNA array profiling of hMSCs 10 d after induction to osteoblasts using locked nucleic acid (LNA) microarrays (23, 24). Expression levels of 33 miRNAs were significantly altered between differentiated and nondifferentiated cells (Table S1). Of these, seven miRNAs were selected for validation by quantitative RT-PCR (qRT-PCR) based on their relative difference score (Table S1). Microarray analysis and qRT-PCR data showed that the expression of miR-26a, -26b, -30c, -101, and -143 were up-regulated and the expression of miR-138 and -222 were down-regulated during osteoblast differentiation of hMSCs.
Effect of miR-138 on the Osteoblast Differentiation of hMSC.
To evaluate the biological effect of the differentially expressed miRNAs, antimiRs and miRNA overexpression for selected miRNAs were applied in a functional screening for their impact on osteoblast differentiation in vitro and ectopic bone formation in vivo. The screening identified miR-138 as a potential negative regulator of osteoblastic differentiation. Expression level of miR-138 was measured at different time points during osteoblast differentiation. miR-138 was down-regulated at day 1 and remained down-regulated up to day 15 of osteoblast differentiation in hMSC-tert (Fig. 1D), primary hMSC, and mouse calvarial MC3T3-E1 cells (Fig. S1A). Furthermore, in a detailed analysis of miR-138 expression in various mouse tissues, we detected high expression of miR-138 in muscle. Additionally, miR-138 was expressed in calvaria, liver, heart, and brain and at a low level in long bones and bone marrow. miR-138 was absent in cartilage (Fig. S1B).
For functional evaluation of miR-138, we overexpressed or inhibited miR-138 levels using synthetic premiR-138 and LNA-modified antimiR-oligonucleotides complementary to mature miR-138. The degree of miRNA inhibition and overexpression was monitored by qRT-PCR after transfection of antimiR-138 or premiR-138, respectively, to hMSCs at 25 nM. Mature miR-138 levels were elevated ∼900-fold relative to control-treated cells 24 h posttransfection, with the levels remaining 16- and 8-fold higher than in the control 10 and 15 d after premiR-138 transfection, respectively (Fig. S2A). By comparison, treatment of hMSCs with antimiR-138 led to inhibition of endogenous miR-138 by 1.1-, 1.5-, and 2-fold by 1, 10, and 15 d after transfection (Fig. S2A).
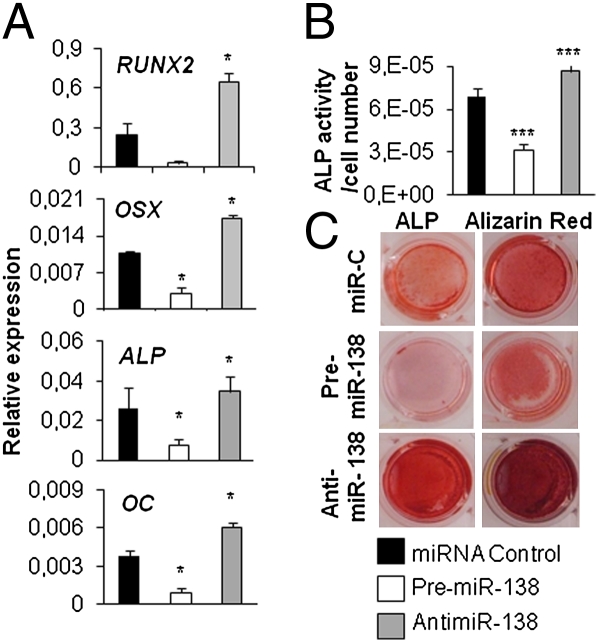
To study the impact of miR-138 on hMSC osteoblast differentiation, hMSCs were induced to differentiate to osteoblasts after transfection with either premiR-138 or antimiR-138. Inhibition of miR-138 significantly enhanced osteoblastic differentiation, which was indicated by higher expression of the osteoblast-specific genes RUNX2, Osterix (OSX), ALP, and OC (Fig. 2A), increased ALP activity, and enhanced in vitro matrix mineralization visualized by Alizarin red staining (Fig. 2 B and C) in antimiR-138–transfected hMSCs compared with cells transfected with control miR. In contrast, ALP activity, matrix mineralization, and osteoblast marker gene expression were reduced in premiR-138–treated hMSCs (Fig. 2 B and C). Similar results were obtained when miR-138 levels were manipulated in primary hMSCs (Fig. S2B). Modulation of miR-138 levels did not alter the proliferation or morphology of hMSCs, indicating that the decreased differentiation potential is not a consequence of decreased proliferation capacity of the cells (Fig. S2 C and D). Taken together, our results suggest that miR-138 is a negative regulator of osteoblast differentiation of hMSCs.
Fig. 2.
miR-138 inhibits osteoblast differentiation. To study the effect of miR-138 on osteoblast differentiation, hMSCs transfected with 25 nM miR-C, premiR-138, or antimiR-138 were induced to osteoblast differentiation for 15 d. (A) Osteoblast differentiation was evaluated with qRT-PCR analysis of osteoblast marker genes (RUNX2, OSX, ALP, and OC normalized to β-actin) at day 15. (B) Alkaline phosphatase activity was measured at day 10 of osteoblast differentiation. (C) ALP and Alizarin Red staining were performed at day 15. *P < 0.05; ***P < 0.001 (n = 3 for all experiments).
Effect of miR-138 on in Vivo Ectopic Bone Formation.
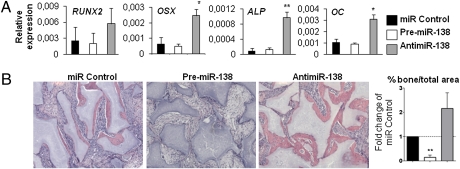
To study whether silencing of miR-138 enhances in vivo ectopic bone formation, untransfected control hMSCs and hMSCs transfected with miR-138, antimiR-138, or miR control were loaded on hydroxyapatite-tricalcium phosphate scaffold and implanted s.c. in NOD/SCID mice for 1 or 8 wk. No major changes were observed by histology of the implants in untransfected hMSCs compared with miR control-transfected hMSCs after 8 wk. Gene expression of osteoblast marker genes was analyzed after 1 wk implantation. qRT-PCR analysis revealed up-regulation of RUNX2, OSX, ALP, and OC in the antimiR-138–treated implants compared with implants transfected with miR control (Fig. 3A), corroborating the results obtained from in vitro cell culture assays. Additionally, we determined the ability of miR-138 inhibition or overexpression to regulate ectopic in vivo bone formation by quantifying the area of bone per total area after 8 wk. Bone formation was increased 2.2-fold in implants treated with the antimiR-138 compared with miR control (Fig. 3B), indicating that inhibition of miR-138 enhances bone formation of hMSCs. Furthermore, overexpression of miR-138 decreased bone formation by 6.7-fold compared with miR control implants (Fig. 3B), supporting the notion that miR-138 negatively regulates osteoblast differentiation and bone formation in vivo.
Fig. 3.
miR-138 inhibits ectopic bone formation in vivo. hMSCs were transfected with 25 nM miR-C, premiR-138, or antimiR-138 and implanted into NOD/SCID mice. (A) qRT-PCR analysis of osteoblast marker genes (RUNX2, OSX, ALP, and OC normalized to β-actin) was performed after 1 wk of implantation. (B) H&E staining was performed after 8 wk of implantation. Bone formation was quantified as total bone volume per total volume from H&E staining, and it was expressed as fold change of miR-C. Four implants per treatment were engrafted into mice, and three sections of each implant were quantified to minimize variations within the implants. *P < 0.05; **P < 0.01.
miR-138 Targets FAK and Regulates FAK Downstream Signaling.
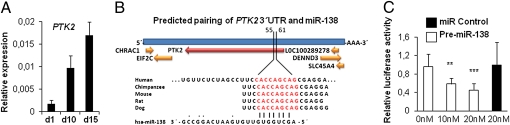
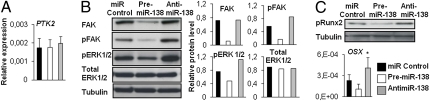
To understand the molecular mechanisms that underlie miR-138–mediated regulation, we searched for potential targets of miR-138 implicated in osteoblast differentiation using the miRNA target prediction algorithms TargetScan and PicTar (25). Among the predicted targets, we identified FAK, a kinase with an established role in ostegenesis (Fig. S3). To confirm the involvement of FAK in osteoblast differentiation of hMSCs, we studied the expression pattern of FAK during differentiation. qRT-PCR analysis revealed that expression of protein tyrosine kinase 2 (PTK2), the gene encoding FAK, was increased during osteoblast differentiation of hMSC-tert (Fig. 4A) and in primary hMSCs (Fig. S1C), similar to osteoblast marker genes and coinciding with down-regulation of miR-138 (Fig. 1).
Fig. 4.
FAK is a potential target of miR-138. (A) PTK2 gene expression profile during osteoblast differentiation of hMSCs was quantified with qRT-PCR. (B) Computational analysis was performed for the complementarities of miR-138 seed sequence to the 3′ UTR of PTK2 and conservation of the putative binding site in vertebrates. (C) Huh7 cells were transfected with 20 ng empty PsiCHECK-2 or PsiCHECK-2-PTK2 vector. Cells were cotransfected with 0, 10, or 20 nM of the premiR-138 or 2 0nM of a negative control. Firefly and Renilla luciferases were measured in cell lysates, and values are normalized to the PsiCHECK vector and presented as fold change of miR-C. **P < 0.01; ***P < 0.001.
According to in silico analysis, PTK2 has a 7-nt seed match site for miR-138 within its 3′ UTR, and this putative target site is highly conserved among the vertebrates (Fig. 4B). To determine whether miR-138 inhibits PTK2 gene expression by binding to the predicted target site in the 3′ UTR, we used a dual luciferase reporter gene system (26). Cotransfection of the PTK2 3′ UTR luciferase reporter with premiR-138 resulted in concentration-dependent down-regulation of luciferase activity compared with the mock or scrambled oligonucleotide controls (Fig. 4C). In comparison, premiR-138 had no effect on the luciferase control reporter without the PTK2 3′ UTR, implying that PTK2 is a direct target of miR-138.
Gene expression analysis revealed no significant changes in PTK2 mRNA levels when miR-138 was either overexpressed or antagonized (Fig. 5A). However, Western blot analysis showed reduced FAK protein levels in the premiR-138–treated cells at day 2 compared with miR control samples (Fig. 5B). Because the FAK signaling pathway is suggested to provide a link between activation of Runx2 and ERK1/2 in osteogenesis (7), we assessed phosphorylation of FAK, Runx2, and ERK1/2. Western blot analysis showed markedly decreased phosphorylation of both FAK and ERK1/2 in miR-138 overexpressing hMSCs, whereas levels of pFAK and pERK1/2 were increased in the antimiR-138–transfected hMSCs (Fig. 5B). Accordingly, phosphorylation of Runx2 and expression of OSX, a downstream target gene of the ERK1/2 pathway (7), were decreased when miR-138 was overexpressed and increased in the absence of miR-138 (Fig. 5C), supporting that miR-138 suppresses FAK downstream signaling by negatively regulating FAK at the posttranscriptional level.
Fig. 5.
miR-138 targets FAK and inhibits ERK pathway. hMSCs were transfected with 25 nM miR-C, premiR-138, or antimiR-138 and induced to osteoblast differentiation. (A) PTK2 gene expression at day 2. (B) Western blot analysis for FAK protein and phosphorylation of FAK and ERK1/2 were performed on day 2. Graphs represent quantifications of Western blot results relative to tubulin. (C) Phosphorylation of Runx2 was analyzed by Western blot at day 10. qRT-PCR analysis of OSX gene expression at day 10.
Discussion
Bone marrow contains a population of stromal (skeletal and mesenchymal) stem cells (hMSCs) that, under appropriate conditions, differentiate into osteoblastic cells (27). In the present study, we identified miR-138 as a negative regulator of hMSC osteoblast differentiation. Data obtained from in vitro experiments revealed that inhibition of miR-138 function enhanced osteoblast differentiation of hMSCs, whereas miR-138 overexpression inhibited their osteogenic potential. Moreover, silencing of miR-138 by antimiR-138 led to increased ectopic bone formation, whereas overexpression of miR-138 significantly diminished bone formation in vivo. These findings suggest that miR-138 plays a pivotal role in bone formation in vivo by negatively regulating osteogenic differentiation in hMSCs leading to reduced ectopic bone formation.
Recently, miR-138 was reported as a negative regulator of adipocyte differentiation of human adipose tissue-derived MSCs (28). These data, together with our findings, suggest a general role of miR-138 in the inhibition of hMSC differentiation and maintenance of the undifferentiated state of MSC. Supporting this statement, we find that miR-138 is also down-regulated during differentiation of hMSCs into other mesoderm-type lineages (e.g., adipocytes and chondrocytes) (Fig. S4).
To study the molecular mechanism by which miR-138 regulates osteoblast differentiation of hMSC, we searched for potential target genes that have an established function in promoting osteogenesis. Interestingly, the 3′ UTR of PTK2 possesses a 7-nt perfect match site to the miR-138 seed region. The protein encoded by PTK2, FAK, has been reported to function as an activator of extracellular signal-related kinase (ERK1 and ERK2) through the Grb2-Sos-Ras pathway during osteoblast differentiation of hMSCs (29, 30). However, the exact organization of these pathways in developing bone is not well-understood, although osteoblasts obtained from patients with osteoporosis and osteoarthritis exhibit reduced FAK activity (31).
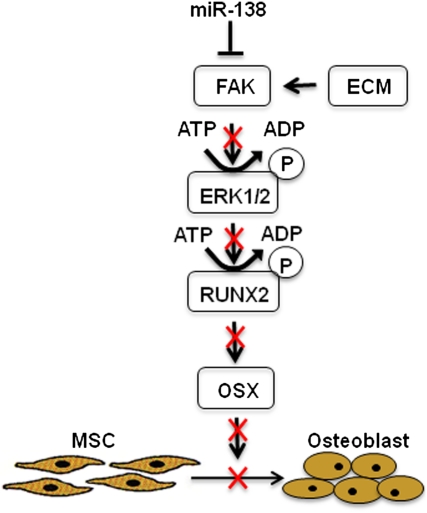
We showed that miR-138 overexpression results in down-regulation of FAK at the protein level, whereas functional inhibition of miR-138 by antimiR-138 leads to derepression of FAK, strongly suggesting that FAK is regulated by miR-138 during osteoblast differentiation. Indeed, PTK2 3′ UTR luciferase reporter assays confirmed that FAK is a direct target of miR-138. Furthermore, overexpression of miR-138 decreased phosphorylation of FAK and subsequently attenuated activation of FAK downstream signaling, as shown by decreased phosphorylation of ERK1/2 in hMSCs. Activation of the ERK1/2 pathway has emerged as an important regulator of osteoblast differentiation, because it regulates Runx2 phosphorylation and subsequently, expression of OSX (7). Increased phosphorylation of Runx2 and significant up-regulation of OSX in the absence of miR-138 support the notion that inhibition of osteoblast differentiation by miR-138 is caused by suppression of the FAK downstream pathway (Fig. 6).
Fig. 6.
A proposed model for miR-138–mediated suppression of osteoblast differentiation. miR-138 is expressed in undifferentiated MSC and suppresses FAK translation, thereby decreases phosphorylation of FAK and its downstream target ERK1/2. Inhibition of the cascade results in decreased phosphorylation of Runx2 and expression of OSX; subsequently, this results in suppression of osteoblast differentiation of MSC.
The impact of miRNAs on osteoblastic differentiation of a number of cell types has been investigated by modulation of miRNA function using antimiRs and miRNA overexpression. These approaches successfully showed that miRNAs (i.e., miR-204/211, the miR-23a∼27a∼24–2 cluster, miR-125b, miR-2861, miR-26a, and miR-196a) are involved in the differentiation of various cell types into mesoderm-type lineages through targeting transcription factors and signaling molecules in the FAK-ERK1/2 pathway.
Obviously, other undisclosed miR-138 targets may contribute to the phenotypic effects observed on inhibition or overexpression of miR-138. There are more than 300 predicted targets for miR-138 in PicTar, some of which are very relevant in the process of osteoblast differentiation (e.g., ROCK2, Ankyrin, MUCDHL, MAP2K7, EFNB3, and PPARGC1A). The involvement of other potential osteogenic targets should be elucidated in future studies.
In conclusion, we find that miR-138 functions as a negative regulator of osteogenic differentiation of hMSCs by repressing FAK expression, which in turn, results in suppression of the FAK-ERK1/2 signaling pathway. Importantly, our results show that functional inhibition of miR-138 can accelerate osteogenic differentiation of hMSCs and lead to increased bone formation in vivo, suggesting that therapeutic approaches targeting miR-138 could be useful in the enhancing bone formation and treatment of pathological conditions of bone loss.
Materials and Methods
Cell Culture and Osteogenic Differentiation.
Telomerase immortalized bone marrow-derived hMSCs (3, 32) were cultured in regular growth medium [Minimum Essential Medium (MEM), 10% FBS, antibiotics, and glutamax I; GIBCO Invitrogen Corporation] as previously described (3, 32). Primary hMSCs were cultured as described (33). Osteogenesis was induced using differentiation medium (growth medium supplemented with 10−8 M dexamethasone, 0.2 mM l-ascorbic acid, 10 mM β-glycerophosphate, and 10 mM 1.25-vitamin-D3). Mouse preosteoblastic MC3T3-E1 calvarial cells were maintained and differentiated as described (14). The osteoblast phenotype was evaluated by determining ALP activity as previously described (34). Alizarin Red staining was performed to detect matrix mineralization with 40 mM alizarin red S (AR-S; Sigma-Aldrich), pH 4.2, for 10 min at room temperature. Cell viability and proliferation was measured with Alamar Blue assay (AbD Serotec).
Microarray and Data Processing.
Total RNA was extracted using the TRIzol method (Invitrogen) according to the manufacturer's protocol. miRNA microarray was performed using Exiqon’s microarray platform (version 8.0; containing 330 human miRNAs) in quadruplicates with LNA-modified oligonucleotide (mercury; Exiqon) capture probes. Slides were scanned using a Genepix 4000B laser scanner (Axon Instruments). Artifact-associated spots were eliminated by software (TIGR spotfinder 3.1.1). Image intensities were measured as a function of the median of foreground minus background. Negative values and values below 50 were normalized to one. Additional data analysis was performed using Microsoft Excel with Significant Analysis of Microarrays (SAM) excel software with multiclass response dataset analysis. The data were normalized using the Limma package for statistical programming language R (version 2.5.1). Medians of four background-corrected replicas for each miRNA capture probe were uploaded into the microarray analysis software for more advanced analysis.
qRT-PCR Analysis.
For qRT-PCR analysis, total RNA was extracted with TRIzol, and cDNA was prepared using revertAid H minus first strand cDNA synthesis kit (Fermentas). SYBR (N',N'-dimethyl-N-[4-[(E)-(3-methyl-1,3-benzothiazol-2-ylidene)methyl]-1-phenylquinolin-1-ium-2-yl]-N-propylpropane-1,3-diamine) green mRNA qRT-PCR was performed using the primers listed in Table S2. Expression levels were analyzed by qRT-PCR (SYBR Green supermix and iCycler IQ detection system; Bio-Rad) using conventional protocols.
For miRNA qRT-PCR primers specific for human miR-26a, -26b, -30c, -101, -138, -143, and -222 and internal control snoRNU44 were purchased from Applied Biosystems. Amplification and detection were performed using 7500HT Fast Real-Time PCR System (Applied Biosystems).
Transfection of Oligonucleotides.
LNA oligonucleotides were synthesized as unconjugated LNA/DNA mixmers with a complete phosphorothioate backbone (IDT). The antimiR control (miR control) was purchased from IDT. LNA-modified antimiRs sequences were AntimiR-138 3′-AtTcaCAacAcCaGC-5′ and AntimiR control 3′-TgtAacAcGTcTAtA-5′, where uppercase letters refer to LNA and lowercase letters refer to DNA. Synthetic premiR-138 sequence 3′-AGCUGGUGUUGUGAAUCAGGCCG-5′ was RNA oligonucleotides. Transfections of 25 nM antimiR oligonucleotide (IDT) or premiR (Ambion) with lipofectamine 2000 (Invitrogen) were performed according to the manufacturer's instructions in OPTI-MEM I Reduced Serum (Invitrogen) The transfection efficiency was evaluated by using 5′ Carboxyfluorescein (FAM)-labeled LNA oligonucleotides.
Immunoprecipitation of Runx2 and Western Blot Analysis.
hMSCs were lysed with Radioimmunoprecipitation (RIPA) buffer (Sigma-Aldrich), and total protein concentration was determined with Pierce Coomassie Plus Bradford assay kit (Thermo Fisher Scientific Inc). For immunoprecipitation of Runx2, 500 μg cell lysate were incubated with 2 μg mouse monoclonal Runx2 antibody (MBL International) and thereafter, were incubated with Protein A/G-Sepharose beads (Santa Cruz Biotechnology). Proteins were separated by 10% SDS/PAGE and electrotransferred into nitrocellulose filters; membranes were incubated with primary antibodies against anti-rabbit FAK (Santa Cruz Biotechnology), pFAK (Santa Cruz Biotechnology), anti-mouse ERK (Cell Signaling), pERK (Cell Signaling), anti-rabbit α-tubulin (Cell Signaling), and anti-rabbit phosphoserine (Millipore) overnight at 4 °C. Membranes were incubated with HRP conjugated anti-mouse or anti-rabbit secondary antibody for 45 min at room temperature, and protein bands were visualized with Amersham ECL chemiluminescence detection system (GE Healthcare Bio-Sciences Corp).
Dual Luciferase Reporter Gene Construct.
A 655-bp fragment of the FAK (PTK2) 3′ UTR containing the predicted binding site for hsa-miR-138 was amplified from human genomic DNA using primers with a short extension containing cleavage sites for XhoI (5′ end) and NotI (3′ end): PTK2 forward (5′-ATACTCGAGAAACTGGCCCAGCAGTATG-3′) and PTK2 reverse (5′-ATAGCGGCCGCTTGCAACTGAAGGGTGTTC-3′). Amplicons were cleaved with XhoI and NotI and cloned in between the XhoI and NotI cleavage sites of the PsiCHECK-2 vector (Promega) downstream of the Renilla luciferase reporter gene.
Luciferase Assay.
Huh7 cells, chosen based on their low endogenous expression of miRNAs (26), were grown to 85–90% confluence in white 96-well plates in DMEM (Invitrogen) supplemented with 10% FBS, 1% nonessential amino acids, l-glutamine, and penicillin/straptamicin at 37 °C under 5% CO2. Cells were transfected with 20 ng empty PsiCHECK-2 vector or PsiCHECK-2-PTK2 3′ UTR reporter for 4 h in reduced serum and antibiotics-free Opti-MEM with Lipofectamine 2000. Cells were cotransfected with the premiR-138 or a negative control (miR control) (Applied Biosystems) at concentrations of 0, 10, or 20 nM. Firefly and Renilla luciferase were measured in cell lysates using a Dual-Luciferase Reporter Assay System (Promega) on a Fusion plate reader (PerkinElmer). Firefly luciferase activity was used for normalization and as an internal control for transfection efficiency.
Ectopic in Vivo Bone Formation Assay of Transfected hMSC Cells.
hMSCs were transfected as described above, loaded on hydroxyapatite/tricalcium phosphate (HA/TCP) ceramic powder (Zimmer Scandinavia), and implanted s.c. into 8-wk-old NOD/MrkBom Tac-Prkdcscid mice (Taconic) as previously described (35, 36). Briefly, cells (5 × 105) were resuspended in 500 μL medium, transferred to 40 mg wet HA/TCP, incubated at 37 °C overnight, and implanted in NOD/SCID mice. Each mouse received four identical implants (two on each side). Mice were anesthetized by i.p. injection of Ketaminal (100 mg/kg ketamine; Intervet) and Rampun (10 mg/kg xylazine; Bayer). After the surgery, mice received an s.c. injection of Temgesic (0.3 mg/mL buprenorphin; Schering-Plough) for pain relief.
Implants were removed after 1 or 8 wk; 1-wk implants were subjected to RNA extraction, whereas 8-wk implants were fixed in 4% paraformaldehyde (Bie & Berntsen), decalcified in formic acid (Local Pharmacy, Odense University Hospital), and embedded in paraffin. Samples were cut into 4-μm sections and stained with H&E Y (Bie & Berntsens). Total bone volume per total volume was quantified as previously described (24).
miRNA Target Site Prediction.
A search for predicted target mRNAs was performed using the databases TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/) (27). Sequence conservation was examined using University of California Santa Cruz genome browser (http://genome.ucsc.edu/).
Statistical Analysis.
Data are presented as mean ± SD. Comparisons were made by using a two-tailed t test or one-way ANOVA for experiments with more than two subgroups. Probability values were considered statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
We are grateful to Lone Christiansen, Bianca Jørgensen, Tina Kamilla Nielsen, and Jane Hinrichsen for excellent technical assistance. Dr. Anna-Marja Säämänen is thanked for helpful discussions. Salla Suomi (MSc) is acknowledged for help with qRT-PCR analysis. H.T. is a graduate student in the Turku Graduate School of Biomedical Sciences, Turku, Finland. The Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation. These studies were supported by the Danish National Advanced Technology Foundation (S.K.), the Danish Medical Research Council (S.K.), the Novo Nordisk Foundation (M.K.), the Lundbeck Foundation (M.K.), and the Region of Southern Denmark (M.K.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016758108/-/DCSupplemental.
References
- 1.Jaiswal RK, et al. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J Biol Chem. 2000;275:9645–9652. doi: 10.1074/jbc.275.13.9645. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Abdallah BM, et al. Maintenance of differentiation potential of human bone marrow mesenchymal stem cells immortalized by human telomerase reverse transcriptase gene despite [corrected] extensive proliferation. Biochem Biophys Res Commun. 2005;326:527–538. doi: 10.1016/j.bbrc.2004.11.059. [DOI] [PubMed] [Google Scholar]
- 4.Abdallah BM, Kassem M. Human mesenchymal stem cells: From basic biology to clinical applications. Gene Ther. 2008;15:109–116. doi: 10.1038/sj.gt.3303067. [DOI] [PubMed] [Google Scholar]
- 5.Xiao G, et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275:4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi Y, et al. Differentiation and transforming growth factor-beta receptor down-regulation by collagen-alpha2beta1 integrin interaction is mediated by focal adhesion kinase and its downstream signals in murine osteoblastic cells. J Biol Chem. 1997;272:29309–29316. doi: 10.1074/jbc.272.46.29309. [DOI] [PubMed] [Google Scholar]
- 7.Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Exp Cell Res. 2007;313:22–37. doi: 10.1016/j.yexcr.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young SR, Gerard-O'Riley R, Kim JB, Pavalko FM. Focal adhesion kinase is important for fluid shear stress-induced mechanotransduction in osteoblasts. J Bone Miner Res. 2009;24:411–424. doi: 10.1359/JBMR.081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 10.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 11.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 12.Hu R, et al. Targeting miRNAs in osteoblast differentiation and bone formation. Expert Opin Ther Targets. 2010;14:1109–1120. doi: 10.1517/14728222.2010.512916. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan MQ, et al. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luzi E, et al. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Bae SW, Yu SS, Bae YC, Jung JS. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno Y, et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem. 2009;284:19272–19279. doi: 10.1074/jbc.M109.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inose H, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–3677. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castoldi M, et al. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bork S, et al. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol. 2010 doi: 10.1002/jcp.22557. [DOI] [PubMed] [Google Scholar]
- 25.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn DE, et al. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3:393–403. doi: 10.1111/j.1365-2184.1970.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, et al. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011;20:259–267. doi: 10.1089/scd.2010.0072. [DOI] [PubMed] [Google Scholar]
- 29.Schaller MD. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim Biophys Acta. 2001;1540:1–21. doi: 10.1016/s0167-4889(01)00123-9. [DOI] [PubMed] [Google Scholar]
- 30.Schlaepfer DD, Hanks SK, Hunter T, van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372:786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 31.Perinpanayagam H, et al. Early cell adhesion events differ between osteoporotic and non-osteoporotic osteoblasts. J Orthop Res. 2001;19:993–1000. doi: 10.1016/S0736-0266(01)00045-6. [DOI] [PubMed] [Google Scholar]
- 32.Simonsen JL, et al. Telomerase expression extends the proliferative life-span and maintains the osteogenic potential of human bone marrow stromal cells. Nat Biotechnol. 2002;20:592–596. doi: 10.1038/nbt0602-592. [DOI] [PubMed] [Google Scholar]
- 33.Alm JJ, et al. Circulating plastic adherent mesenchymal stem cells in aged hip fracture patients. J Orthop Res. 2010;28:1634–1642. doi: 10.1002/jor.21167. [DOI] [PubMed] [Google Scholar]
- 34.Qiu W, et al. Tumor necrosis factor receptor superfamily member 19 (TNFRSF19) regulates differentiation fate of human mesenchymal (stromal) stem cells through canonical Wnt signaling and C/EBP. J Biol Chem. 2010;285:14438–14449. doi: 10.1074/jbc.M109.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenderup K, et al. Aged human bone marrow stromal cells maintaining bone forming capacity in vivo evaluated using an improved method of visualization. Biogerontology. 2004;5:107–118. doi: 10.1023/B:BGEN.0000025074.88476.e2. [DOI] [PubMed] [Google Scholar]
- 36.Abdallah BM, Ditzel N, Kassem M. Assessment of bone formation capacity using in vivo transplantation assays: Procedure and tissue analysis. Methods Mol Biol. 2008;455:89–100. doi: 10.1007/978-1-59745-104-8_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.