Abstract
Caspase-1 plays a key role in inflammatory pathways by processing pro-IL-1β into the active cytokine mature IL-1β. Given its sequence similarity with the Caenorhabditis elegans cell death gene ced-3,it has long been speculated that caspase-1 may also play a role in cell death. However, an unequivocal role for caspase-1 in cell death has been questioned, and not definitively demonstrated. Furthermore, if caspase-1 does play a role in cell death, its position in the apoptotic hierarchy has not been clearly defined. Previous studies have shown that caspase-1 knockout (KO) mice and transgenic mice expressing a dominant-negative caspase-1 construct are resistant to ischemic brain injury. We provide direct evidence that caspase-1 plays a key role in neuronal cell death and that caspase-1 is an apical activator of the cell death pathway in the premitochondrial collapse stage. Furthermore, we demonstrate that Rip2/Cardiak/Rick is a stress-inducible upstream modulator of pro-caspase-1 apoptotic activation. We provide evidence that Bid cleavage appears to be an important downstream effector of caspase-1-mediated cell death. Our data demonstrate that caspase-1 is an apical mediator of neuronal cell death during in vitro hypoxia, and confirmed in vivo in ischemia, and provide insights into the sequence of events involved in this pathological cell death process.
Traditionally, the caspase family of proteases has been divided into two main subfamilies, one involved in inflammation and one involved in apoptosis. Members of the inflammatory caspase subfamily include caspase-1, -4, -5, -11, -12, and -13 (1, 2). The apoptotic caspase subfamily is divided between initiators (caspase-2, -8, -9, and -10) and effectors of cell death (caspase-3, -6, -7, and -14) (1, 2). Several caspases play a role in both inflammatory and apoptotic cascades, including caspase-11 and -12 (3, 4). Evidence for caspase-1 activation has been demonstrated in a variety of in vitro and in vivo cell death paradigms (5-10). Previous work using caspase-1-deficient or caspase-1 dominant-negative transgenic mice has provided evidence for a role of caspase-1 in ischemic brain injury in vivo (9, 11). However, the possibility that inflammatory pathways (in the caspase-1-deficient mouse) or caspases in addition to caspase-1 (in the caspase-1 dominant-negative mouse) may play a role in the observed neuroprotection has precluded the ability to directly implicate caspase-1 in neuronal cell death. Therefore, direct evidence for a functional role of caspase-1 in cell death has not been demonstrated. In particular, caspase-1-deficient mice have no apparent defects in developmental programmed cell death (12), suggesting that this protease is not required for normal physiological cell death. It remains possible, however, that caspase-1 could be important for pathological cell death in some conditions where cell stress occurs.
Given the evidence that caspase-1 is activated in a variety of cell death paradigms, we evaluated whether caspase-1 plays a functional role in neuronal cell death (13). In this study, we demonstrate that caspase-1 is an important contributor to ischemia/hypoxia-mediated neuronal cell death. We also delineated some of the upstream and downstream mediators of the cell death pathway involving caspase-1. We provide evidence that caspase-1 is an apical caspase in neuronal cell death pathways, mediating Bid cleavage, release of mitochondrial apoptogenic factors [cytochrome c, Smac/Diablo, and apoptosis-inducing factor (AIF)], and activation of caspase-9 and -3. Furthermore, we demonstrate that Rip2/Cardiak/Rick plays a key role in regulating caspase-1 apoptotic activity in neurons.
Materials and Methods
Animals. Caspase-1-deficient mice were created and characterized by BASF Bioresearch (Worcester, MA) and were generously provided by W. Wong (12). These mice contain a null mutation in the caspase-1 gene that was generated by homologous recombination in embryonic stem cells.
Plasmid Construction, Transfection, and Cell Death Count. Plasmids encoding wild-type caspase-1, Rip2, and GFP have been described (14). The transfection efficiency of primary cortical neurons was analyzed under the microscope of 12 fields (×200 magnification) at 48 h after transfection with GFP plasmid, using Lipofectamine 2000 (Life Technologies). Primary cortical neurons cultured in six-well plates were cotransfected with plasmids encoding GFP in combination with plasmids encoding mouse caspase-1, Rip2, or both by using Lipofectamine 2000. At 48 h after transfection, supernatants were collected for lactate dehydrogenase (LDH) determination. Cell death were quantified as described by Khursigara et al. (15). Briefly, cells were stained with Hoechst for 15 min at 48 h after transfection, and GFP expressing apoptotic morphology cells were identified and quantitated for the percentage of cell death. All wells were counted double blindly in each group.
Primary Cortical Neuron Culture and Oxygen/Glucose Deprivation (OGD) Treatment. Cerebral cortex of mouse embryos at day 15 (E15) were freed from meninges and separated from olfactory bulb and hippocampus. Trypsinized cells were suspended in medium [neurobasal medium (NBM) with 2% (vol/vol) B27 supplement, 2 mM glutamine, 100 units/ml penicillin, and streptomycin (GIBCO)] and seeded at a density of 2 × 104 per cm2 on polylysin-coated dishes. Cells were used for experiments on day seven of culture. OGD on primary cortical cell cultures was performed as described (8), with minor modifications. Briefly, the culture medium was replaced by glucose-free Earle's balanced salt solution, and the cells were placed in an anaerobic chamber with BBL GasPak Plus (BD), which reduces the oxygen concentration to <100 ppm within 90 min. Control cells were incubated in Earle's balanced salt solution with glucose in a normoxic incubator for the same period. After 3 h, OGD was terminated by returning to normal culture conditions.
Immunocytochemistry. Primary mouse cortical neurons isolated from E15 embryos were plated on chamber slide covers (Nalge Nunc) precoated with polylysin and laminin. After OGD treatment, immunocytochemistry of caspase-1 and -3 activation was performed as described (5), with minor modification. Digital images were scanned with a Nikon ECLIPSE TE 200 fluorescence microscope. Caspase-1 antibody was generously provided by Junying Yuan (Harvard Medical School). Caspase-3 (CM1) antibody was purchased from BD Pharmingen, and Rip2 antibody was purchased from Alexis (positive and negative controls for Rip2 antibody were provided in Fig. 8, which is published as supporting information on the PNAS web site).
Mature IL-1β Determination. Mature IL-1β quantification was performed as previously described by using an ELISA kit specific for the mature form of the cytokine (R & D Systems).
Measurement of LDH Activity. Released LDH activity was measured according to the protocol provided by the manufacturer (Roche). Briefly, reaction mixture (100 μl) was added to conditioned media (100 μl) removed from 24-well plates after centrifugation at 250 × g for 10 min. Absorbance of samples at 490 nm was measured in an ELISA reader after 15 min of incubation at room temperature. The same volume of medium was used as the background control.
Middle Cerebral Artery Occlusion. Focal cerebral ischemia was induced by introduction of an intraluminal nylon thread into the cervical internal carotid artery. Mice were initially anesthetized with 1.5% (vol/vol) isoflurane (70% N2O/30% O2). Isoflurane concentration was thereafter titrated to 0.5%. Rectal temperature was maintained between 37.0 and 37.5°C with a heating pad (Harvard Apparatus). The right common carotid artery was exposed, and the external carotid artery was ligated. A 7-0 monofilament nylon thread with the tip blunted with silicone was inserted 9 ± 1.0 mm into the internal carotid artery up to the middle cerebral artery. After 120 min of ischemia, the middle cerebral artery blood flow was restored by removing the thread. Blood flow was documented with an infrared Doppler (16). Blood pressure was not different in wild-type or caspase-1-deficient mice. After 5 h reperfusion, the brains were removed and the ischemic territory was dissected for caspase activation and cytochrome c, Smac/Diablo, and AIF release assay.
Immunoblotting. Cells were collected in lysis buffer (20 mM Tris, pH 8.0/137 mM NaCl/10% glycerol/1% Nonidet P-40/2 mM EDTA with 5 mM Na2VO4/protease inhibitor mixture (Roche)/0.2 mM phenylmethylsulfonyl fluoride) on ice, centrifuged at 19,720 × g for 15 min at 4°C and directly analyzed by immunoblotting. Mouse brain samples were lysed in RIPA buffer with protease inhibitors. Caspase-1 antibody was generously provided by Junying Yuan. Caspase-9 and -3 antibodies were purchased from Cell Signaling Technology, caspase-8 antibody was purchased from BD Pharmingen, Bid antibody was purchased from R & D systems, Rip2 antibody and blocking peptide for Rip2 were purchased from Alexis, and β-actin antibody was purchased from Sigma. For analysis of cytosolic components, cytochrome c antibody was purchased from BD Pharmingen, Smac/Diablo antibody was purchased from Novus Biologicals (Littleton, CO), and AIF antibody was purchased from Sigma.
Cell and Tissue Cytosolic Fraction. Cell and tissue cytosolic fraction were performed as described (17). Released cytochrome c, Smac/Diablo, and AIF were analyzed by immunoblotting.
Data Analysis. Densitometric quantification was performed by using the QUANTITY ONE program (Bio-Rad). Data are presented as mean ± SD. Statistical comparisons were performed by using Student's t test. A value of P < 0.05 was considered significant.
Results
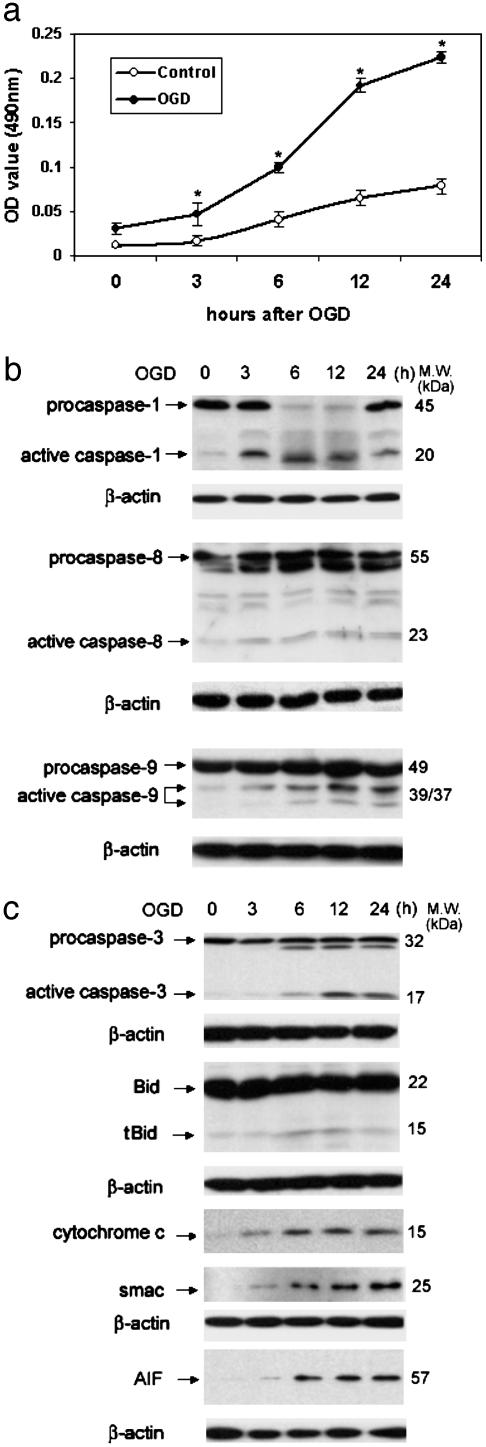
Hypoxia Recruitment of Neuronal Caspase and Mitochondrial Apoptotic Pathways. To evaluate whether caspase-1 plays a direct role in cell death, we isolated primary cerebrocortical neurons (90-95% neurons; see Fig. 9, which is published as supporting information on the PNAS web site) and evaluated activation of this protease after OGD. We characterized the time course of activation of different members of the caspase family, and also monitored proteolytic processing of Bid, and release of mitochondrial apoptogenic factors. After hypoxia (Fig. 1a), we detected caspase-1, -3, -8, and -9 activation (Fig. 1b). We also detected cleavage of Bid and generation of truncated Bid (tBid), a proapoptotic fragment of this Bcl-2 family member (Fig. 1b). Furthermore, recruitment of mitochondrial cell death pathways was demonstrated by detection of cytochrome c, Smac/Diablo, and AIF in cytosolic extracts (Fig. 1b). These data suggest a role for the involvement of the mitochondrial (“intrinsic”) cell death pathway in hypoxia-induced cell death of cultured neurons.
Fig. 1.
Activation of caspase-1, -3, -8, and -9, cleavage of Bid, and release of cytochrome c, Smac/Diablo, and AIF during OGD-induced neuronal cell death. (a) Primary cortical neurons from wild-type mice cultured for a week were treated with 3 h of OGD. At the indicated time points after completion of OGD, culture media was collected and analyzed for LDH release as an indicator of cell death. Results are the average of three independent experiments. *, P < 0.05 versus control. (b) Cell extracts were subjected to immunoblotting with antibodies against caspase-1, -3, -8, and -9 and Bid. Cytosolic extracts were evaluated for release of cytochrome c, Smac/Diablo, AIF, and β-actin. The blots are representative of three independent experiments.
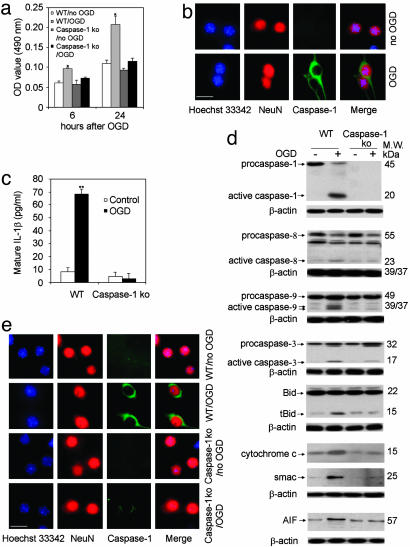
Critical Role of Caspase-1 in OGD-Induced Neuronal Death. To evaluate whether caspase-1 plays a critical role in the OGD-induced neuronal cell death, we isolated cerebrocortical neurons from wild-type and caspase-1-deficient mice and compared their responses to OGD. In previous work, caspase-1-deficient mice have been demonstrated to be resistant to cerebral ischemic damage (11), although this effect was proposed to be due to its role in inflammation rather than in cell death (12, 18). Cultured neurons from caspase-1-deficient mice were highly resistant to OGD (Fig. 2a), with only half as much LDH release occurring in cultures of caspase-1-deficient cells compared to wild-type neurons. The caspase-1-deficient mice used in this study are the same as the ones previously used in the above-referenced report (11). To confirm that caspase-1 activation occurs in neurons (rather than contaminating glial cells), we localized caspase-1 to neurons by immunofluorescence (Figs. 2b and 9a). Consistent with our previous findings in amyotrophic lateral sclerosis and spinal cord injury, the caspase-1 antibody used appears to have higher affinity for the active form of caspase-1 as compared to the proform (5, 6). Because caspase-1 is the key mediator of IL-1β processing, we evaluated whether IL-1β processing was inhibited in caspase-1-deficient neurons. As shown on Fig. 2c, caspase-1-deficient neurons demonstrate inhibition of hypoxia-mediated generation of mature IL-1β. As expected, we did not detect caspase-1 activation in neurons from caspase-1-deficient mice, in contrast to wild-type neurons (Fig. 2d). To evaluate the mechanisms of neuroprotection associated with caspase-1 deficiency, we compared neurons from caspase-1-deficient and wild-type mice with respect to proteolytic processing of various members of the caspase pathway, cleavage of Bid, and release of mitochondrial apoptogenic factors. Interestingly, neurons from caspase-1-deficient mice demonstrated inhibition of OGD-mediated caspase-9 and -3 processing, Bid cleavage, and cytochrome c, Smac/Diablo, and AIF release (Fig. 2 d and e). Hypoxia-associated loss of mitochondrial transmembrane potential was also inhibited in caspase-1-deficient neurons (Fig. 3). Note that in these cultured neurons, we only detected a small amount of caspase-8 processing, which was not significantly reduced in caspase-1-deficient neurons. These data are consistent with a model in which caspase-1 is an apical mediator of hypoxia-induced cell death.
Fig. 2.
Role of caspase-1 in OGD-induced neuronal death. (a) Primary cortical neurons from wild-type mice and caspase-1 KO mice cultured for a week were treated with 3 h of OGD. Supernatants were collected and assayed for LDH release 6 and 24 h after the completion of OGD. Results are the average of three independent experiments. *, P < 0.05 versus control. (b) Six hours after completion of OGD, wild-type cells were processed for immunofluorescence stained for active caspase-1 (green). Blue, nuclei stained by Hoechst; red, neurons stained with NeuN. (Scale bar, 2.5 μm.) (c-e) Wild-type and caspase-1 KO neurons were treated with OGD. (c) Supernatants were collected and assayed for mature IL-1β release 6 h after the completion of OGD. Results are the average of three independent experiments. **, P < 0.01 versus control. (d) Cell extracts were subjected to immunoblotting with antibodies against caspase-1, -3, -8, and -9, bid, cytochrome c, Smac/Diablo, AIF, and β-actin. The blots are representative of three independent experiments. (e) Cells were also processed for immunofluorescence staining with antibodies against active caspase-3 (green). Blue, nuclei stained by Hoechst; red, neurons stained with NeuN. (Scale bar, 2.5 μm.)
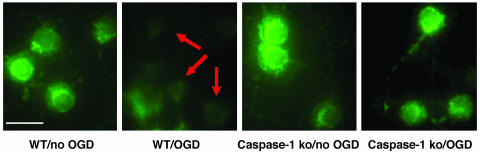
Fig. 3.
Caspase-1-deficient neurons demonstrate resistance to hypoxia-mediated loss of mitochondrial membrane potential. Live primary cortical neurons were stained directly with 2 μM Rhodamine-123 over 6 h after 3 h of OGD. Arrows show loss of mitochondrial transmembrane potential after OGD treatment of wild-type cortical neurons. Mitochondrial transmembrane potential is retained after OGD in caspase-1-deficient neurons. (Scale bar, 2.5 μm.)
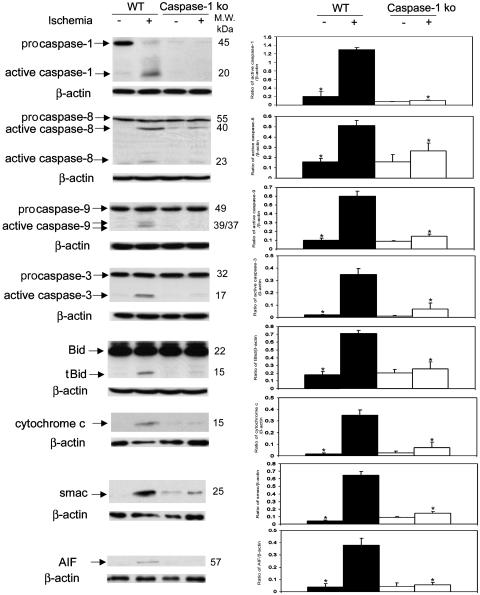
Role of Caspase-1 in Ischemia-Induced Neuronal Cell Death. To explore the in vivo relevance of the above-described observations, we evaluated the role of caspase-1 in cell death pathways activated in a cerebral ischemia model. Caspase-1-deficient mice have been previously demonstrated to be protected from ischemic injury (11). In wild-type mice, we detected cerebral ischemia-associated processing of caspase-1, -3, -8, and -9, as well as Bid cleavage and release of mitochondrial apoptogenic factors. Similar to the results in the primary cerebrocortical neuron culture, caspase-1-deficient mice demonstrated reduced caspase-9 and -3 processing, Bid cleavage, and release of mitochondrial apoptogenic factors. We detected low levels of caspase-8 processing in ischemic brain, which was not significantly altered in caspase-1-deficient mice (Fig. 4). Given that, both caspase-8 and -1 can cleave Bid (19), our data provide indirect evidence that caspase-1, rather than caspase-8, is the mediator of Bid cleavage in neuronal ischemia/hypoxia. Furthermore, our findings support a model in which caspase-1 is a key apical activator of downstream ischemia-triggered cell death pathways in vivo.
Fig. 4.
Role of caspase-1 in ischemia-induced neuronal cell death. Caspase-1 KO mice and wild-type mice were subjected to 2 h of focal ischemia. After 5 h of reperfusion, the ischemic territory was isolated. Whole tissue lysates were subjected to immunoblotting with antibodies against caspase-1, -3, -8, and -9 and Bid. Cytosolic extracts of ischemic tissue were evaluated for release of cytochrome c, Smac/Diablo, and AIF. (Right) Densitometry with three samples for each condition. *, P < 0.05 versus wild-type ischemia group.
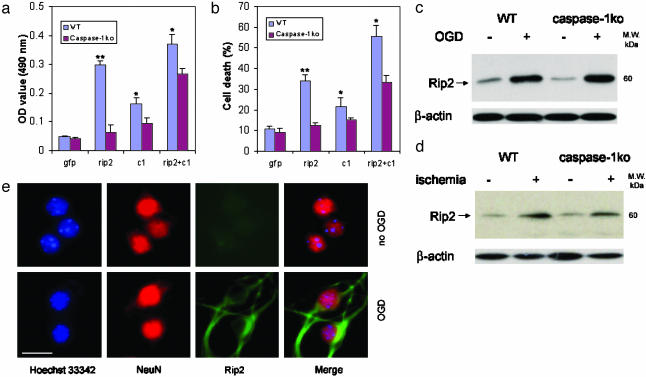
Role of Rip2 in Caspase-1-Mediated Neuronal Cell Death. Activity of long prodomain caspases (i.e., caspase-1, -2, -4, -5, -9, -11, -12, and -13) can be regulated by direct protein-protein interactions. The interaction occurs at the N-terminal prodomain called the caspase-recruitment domain (CARD) (13). In vitro studies have shown that the CARD of caspase-1 can associate with a variety of other CARD-containing molecules through CARD-CARD interactions (20-24). Rip2 is a CARD-containing protein with demonstrated ability to bind and activate caspase-1 in inflammatory conditions (21, 22). We therefore transfected primary cerebrocortical neurons with Rip2 to determine whether it can induce cell death (transfection efficiency 17 ± 2%; Fig. 10a, which is published as supporting information on the PNAS web site). As shown in Figs. 5 a and b and 10b, Rip2 transfection resulted in significant induction of cell death. To determine whether endogenous caspase-1 is responsible for Rip2-mediated cell death, we transfected Rip2 into wild-type and caspase-1-deficient primary neurons. Caspase-1-deficient neurons were highly resistant to Rip2-mediated cell death (Fig. 5 a and b). Transfecting caspase-1 into caspase-1-deficient neurons restored sensitivity to Rip2-induced apoptosis (Fig. 5 a and b), providing further support that Rip2-mediated cell death is dependent on caspase-1.
Fig. 5.
Role of Rip2 in caspase-1-mediated neuronal cell death. Primary cortical neurons were transfected with caspase-1 (c1), Rip2, or both. Forty-eight hours after transfection, media were collected and assayed for LDH release (a) and evaluation of cell death (b). Results are the average of three independent experiments. *, P < 0.05 versus caspase-1 KO. (c and d) The same samples as in Figs. 2d and 4 were subjected to immunoblotting with antibodies against Rip2 and β-actin. (e) The same cells as Fig. 2b used were also processed for immunofluorescence staining with antibodies against Rip2 (green). Blue, nuclei stained by Hoechst; red, neurons stained with NeuN. (Scale bar, 2.5 μm.)
Rip2 expression is induced in macrophages on stimulation with lipopolysaccharide (25). Because caspase-1 is activated during hypoxia and ischemia in neurons, we evaluated whether Rip2 is expressed endogenously in neurons subjected to hypoxia/ischemia. Immunoblotting showed that levels of Rip2 protein are up-regulated in vitro and in vivo after hypoxia/ischemia (Fig. 5 c and d). This induction of Rip2 expression was not impaired in caspase-1-deficient neurons, suggesting that neuroprotection in caspase-1-deficient neurons is not due to defective Rip2 induction. To confirm that Rip2 up-regulation is neuronal in nature, we demonstrate that endogenous Rip2 is specifically up-regulated in NeuN-positive cells (Figs. 5e and 9b).
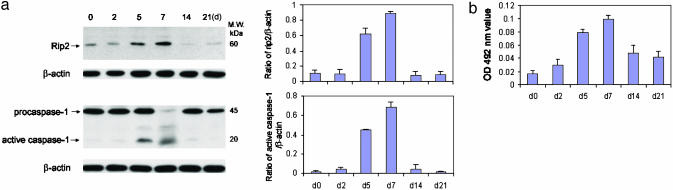
Rip2 Up-Regulation Mediates Caspase-1 Activation. To provide further insight into the role of Rip2 in caspase-1-mediated neuronal cell death, we made use of the phenomenon that Rip2 appears to be differentially regulated with the age of primary cultured cells. Consistent with findings reported by Khursigara et al. (14) in Schwann cells, levels of Rip2 in neurons subjected to hypoxia were highest at 5-7 days after initiation of cultures coinciding with the maximum sensitivity of these cells to hypoxia-mediated processing of caspase-1 and of cell death (Fig. 6 a and b). These data are consistent with a role for Rip2 in regulating caspase-1 activation within neurons subjected to hypoxia/ischemia injury.
Fig. 6.
Rip2 up-regulation results in caspase-1 activation. Primary cortical neurons from wild-type mice cultured for 0, 2, 5, 7, 14, and 21 days were treated with 3 h OGD. (a) Cells were collected 6 h after OGD and lysates were immunoblotted with antibodies against Rip2, caspase-1, and β-actin. (Right) Densitometry using three samples for each condition. (b) Supernatants were assayed for LDH release.
Discussion
A role for caspase-1 in neuronal cell death has often been questioned. Other than the clear role it plays in inflammatory pathways, a function of caspase-1 in neuronal cell death has not been unequivocally demonstrated. Furthermore, if caspase-1 plays a role in cell death, neither its upstream activators nor its downstream mediators have been clearly identified. Our findings provide definitive evidence of the role of caspase-1 in ischemia/hypoxia-mediated neuronal cell death.
We demonstrate that Rip2 is hypoxia-inducible, suggesting an important role for this protein in mediating caspase-1 activation (Fig. 7). We provide evidence that caspase-1 is a mediator of Bid cleavage in hypoxia/ischemia-stimulated neurons. Generation of tBid is known to result in release of mitochondrial caspase-dependent (cytochrome c and Smac/Diablo) and caspase-independent (AIF) cell death pathways. Consistent with a role for Bid in neuronal injury, studies of Bid-deficient mice have shown that Bid-deficiency is associated with reduced infarct size in stroke models (26). Also, we have previously demonstrated inhibition of Bid cleavage in spinal cord of amyotrophic lateral sclerosis mice expressing a dominant-negative version of caspase-1 (27). Because the caspase-1 dominant-negative transgene might not be specific for caspase-1, the current data from caspase-1-deficient mice provide clear evidence that Bid is a substrate within the cell death pathway activated by caspase-1 during ischemic/hypoxic injury in the brain. Similar to other CARD-containing caspases, our data are most consistent with caspase-1 being an apical cell death caspase. Our findings thus provide further validation of Rip2-mediated caspase-1 activation as a target for new agents designed to reduce neuron loss during ischemia.
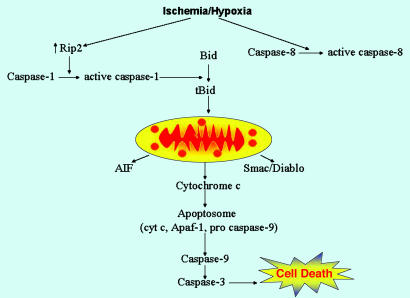
Fig. 7.
The Rip2/Caspase-1 pathway plays a key role in ischemia/hypoxia-associated neuronal death. From the above-described data, a pathway mediating neuronal death involving Rip2 and caspase-1 may be described. After the hypoxia/ischemia insult, increased levels of Rip2 protein are detected within neurons. Rip2 then mediates caspase-1 activation. Caspase-1 thereafter generates the proapoptotic tBid fragment. tBid plays a role in the release of mitochondrial apoptogenic factors, including cytochrome c, AIF, and Smac/Diablo. Released cytochrome c induces apoptosome assembly, resulting in caspase-9 and -3 activation. Activated caspase-3 plays a key role in the final stages of the apoptotic cascade. In our in vivo and in vitro models, we do detect caspase-8 activation. However, it appears that the magnitude of caspase-8 activation is not sufficient to be of functional importance. Furthermore, the mechanism resulting in ischemia/hypoxia-mediated caspase-8 activation, whether it is direct or indirect, is not clearly understood.
Supplementary Material
Acknowledgments
We thank E. Friedlander for editorial assistance and Dr. Dean Hartley for tissue culture technical assistance. Caspase-1 antibody was kindly provided by Dr. Junying Yuan. This work was supported by grants from the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (to R.M.F.), the NIH (to J.C.R.), and the Huntington's Disease Society of America (R.M.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AIF, apoptosis-inducing factor; CARD, caspase-recruitment domain; KO, knockout; LDH, lactate dehydrogenase; OGD, oxygen/glucose deprivation; tBid, truncated Bid.
References
- 1.Thornberry, N. A., Rano, T. A., Peterson, E. P., Rasper, D. M., Timkey, T., Garcia-Calvo, M., Houtzager, V. M., Nordstrom, P. A., Roy, S., Vaillancourt, J. P., et al. (1997) J. Biol. Chem. 272, 17907-17911. [DOI] [PubMed] [Google Scholar]
- 2.Shi, Y. (2002) Mol. Cell 9, 459-470. [DOI] [PubMed] [Google Scholar]
- 3.Wang, S., Miura, M., Jung, Y. K., Zhu, H., Li, E. & Yuan, J. (1998) Cell 92, 501-509. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa, T., Zhu, H., Morishima, N., Li, E., Xu, J., Yankner, B. A. & Yuan, J. (2000) Nature 403, 98-103. [DOI] [PubMed] [Google Scholar]
- 5.Li, M., Ona, V. O., Guegan, C., Chen, M., Jackson-Lewis, V., Andrews, L. J., Olszewski, A. J., Stieg, P. E., Lee, J. P., Przedborski, S. & Friedlander, R. M. (2000) Science 288, 335-339. [DOI] [PubMed] [Google Scholar]
- 6.Li, M., Ona, V. O., Chen, M., Kaul, M., Tenneti, L., Zhang, X., Stieg, P. E., Lipton, S. A. & Friedlander, R. M. (2000) Neuroscience 99, 333-342. [DOI] [PubMed] [Google Scholar]
- 7.Ona, V. O., Li, M., Vonsattel, J. P., Andrews, L. J., Khan, S. Q., Chung, W. M., Frey, A. S., Menon, A. S., Li, X. J., Stieg, P. E., et al. (1999) Nature 399, 263-267. [DOI] [PubMed] [Google Scholar]
- 8.Friedlander, R. M., Gagliardini, V., Rotello, R. J. & Yuan, J. (1996) J. Exp. Med. 184, 717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara, H., Fink, K., Endres, M., Friedlander, R. M., Gagliardini, V., Yuan, J. & Moskowitz, M. A. (1997) J. Cereb. Blood Flow Metab. 17, 370-375. [DOI] [PubMed] [Google Scholar]
- 10.Klevenyi, P., Andreassen, O., Ferrante, R. J., Schleicher, J. R., Jr., Friedlander, R. M. & Beal, M. F. (1999) NeuroReport 10, 635-638. [DOI] [PubMed] [Google Scholar]
- 11.Schielke, G. P., Yang, G. Y., Shivers, B. D. & Betz, A. L. (1998) J. Cereb. Blood Flow Metab. 18, 180-185. [DOI] [PubMed] [Google Scholar]
- 12.Li, P., Allen, H., Banerjee, S., Franklin, S., Herzog, L., Johnston, C., McDowell, J., Paskind, M., Rodman, L., Salfeld, J., et al. (1995) Cell 80, 401-411. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander, R. M. (2003) N. Engl. J. Med. 348, 1365-1375. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. H., Stehlik, C. & Reed, J. C. (2001) J. Biol. Chem. 276, 34495-34500. [DOI] [PubMed] [Google Scholar]
- 15.Khursigara, G., Bertin, J., Yano, H., Moffett, H., DiStefano, P. S. & Chao, M. V. (2001) J. Neurosci. 21, 5854-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedlander, R. M., Gagliardini, V., Hara, H., Fink, K. B., Li, W., MacDonald, G., Fishman, M. C., Greenberg, A. H., Moskowitz, M. A. & Yuan, J. (1997) J. Exp. Med. 185, 933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guegan, C., Vila, M., Rosoklija, G., Hays, A. P. & Przedborski, S. (2001) J. Neurosci. 21, 6569-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuida, K., Lippke, J. A., Ku, G., Harding, M. W., Livingston, D. J., Su, M. S. & Flavell, R. A. (1995) Science 267, 2000-2003. [DOI] [PubMed] [Google Scholar]
- 19.Li, H., Zhu, H., Xu, C. J. & Yuan, J. (1998) Cell 94, 491-501. [DOI] [PubMed] [Google Scholar]
- 20.Poyet, J. L., Srinivasula, S. M., Tnani, M., Razmara, M., Fernandes-Alnemri, T. & Alnemri, E. S. (2001) J. Biol. Chem. 276, 28309-28313. [DOI] [PubMed] [Google Scholar]
- 21.Thome, M., Hofmann, K., Burns, K., Martinon, F., Bodmer, J. L., Mattmann, C. & Tschopp, J. (1998) Curr. Biol. 8, 885-888. [DOI] [PubMed] [Google Scholar]
- 22.Humke, E. W., Shriver, S. K., Starovasnik, M. A., Fairbrother, W. J. & Dixit, V. M. (2000) Cell 103, 99-111. [DOI] [PubMed] [Google Scholar]
- 23.Damiano, J. S., Stehlik, C., Pio, F., Godzik, A. & Reed, J. C. (2001) Genomics 75, 77-83. [DOI] [PubMed] [Google Scholar]
- 24.Druilhe, A., Srinivasula, S. M., Razmara, M., Ahmad, M. & Alnemri, E. S. (2001) Cell Death Differ. 8, 649-657. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K., Inohara, N., Hernandez, L. D., Galan, J. E., Nunez, G., Janeway, C. A., Medzhitov, R. & Flavell, R. A. (2002) Nature 416, 194-199. [DOI] [PubMed] [Google Scholar]
- 26.Plesnila, N., Zinkel, S., Le, D. A., Amin-Hanjani, S., Wu, Y., Qiu, J., Chiarugi, A., Thomas, S. S., Kohane, D. S., Korsmeyer, S. J. & Moskowitz, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 15318-15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guegan, C., Vila, M., Teissman, P., Chen, C., Onteniente, B., Li, M., Friedlander, R. M. & Przedborski, S. (2002) Mol. Cell. Neurosci. 20, 553-562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.