Abstract
Human evolution is characterized by the rapid expansion of brain size and drastic increase in cognitive capabilities. It has long been suggested that these changes were accompanied by modifications of brain metabolism. Indeed, human-specific changes on gene expression or amino acid sequence were reported for a number of metabolic genes, but actual metabolite measurements in humans and apes have remained scarce. Here, we investigate concentrations of more than 100 metabolites in the prefrontal and cerebellar cortex in 49 humans, 11 chimpanzees, and 45 rhesus macaques of different ages using gas chromatography–mass spectrometry (GC-MS). We show that the brain metabolome undergoes substantial changes, both ontogenetically and evolutionarily: 88% of detected metabolites show significant concentration changes with age, whereas 77% of these metabolic changes differ significantly among species. Although overall metabolic divergence reflects phylogenetic relationships among species, we found a fourfold acceleration of metabolic changes in prefrontal cortex compared with cerebellum in the human lineage. These human-specific metabolic changes are paralleled by changes in expression patterns of the corresponding enzymes, and affect pathways involved in synaptic transmission, memory, and learning.
Keywords: cognition, glutamate, development
Human evolution is characterized by rapid expansion of brain size and increase in cognitive capabilities, leading to the emergence of unique and complex cognitive skills. These changes have long been associated with changes in brain metabolism, in particular with respect to increased energy demand (1). Large brains are metabolically costly. Thus, humans allocate approximately 20% of their total energy to the brain, compared with 11–13% for apes and 2–8% for other mammalian species (2). This increased metabolic demand has been associated with elevated expression of genes involved in neuronal functions and energy metabolism (3, 4). These changes may have been evolutionary advantageous, as indicated by signatures of positive selection reported for the amino acid changes, which occur in the mitochondrial electron-transport chain proteins, in anthropoid primates and humans, as well as elevated expression of the energy metabolism pathways in the human brain (5, 6). On the histological level, human brains show the largest density of glia cells relative to neurons in the prefrontal cortex, which provides an indirect indication of increased neuronal metabolic demand (7).
Changes in brain metabolism are also implicated in neuropsychiatric disorders, such as schizophrenia, which affect some of the human-specific cognitive abilities (8, 9). Furthermore, direct measurements of 21 metabolites in the prefrontal cortex of adult human controls compared with human schizophrenia patients, chimpanzees, and rhesus macaques, carried out using proton NMR spectroscopy have shown significant overlap between human-specific evolutionary changes and metabolic differences between controls and schizophrenia patients (10). Notably, this study has identified not only energy metabolites, such as lactate and creatine, but also metabolites related to neurotransmission, such as choline and glycine, as being altered in both human evolution and in disease. Taken together, these observations suggest that diverse metabolic changes might have been important to the establishment and maintenance of the human-specific cognitive abilities.
Here, we used a combination of gas chromatography–mass spectrometry (GC-MS) metabolomics and quantitative label-free proteomics to measure concentrations of more than 100 metabolites and ∼2,000 proteins in brains of humans, chimpanzees, and rhesus macaques. Application of GC-MS methodology to metabolic profiling in the human tissues, including brain, was successfully introduced almost 30 y ago (11). This methodology provides good resolution for relatively small, thermally stable compounds that can be made volatile through derivatization (12). It must be noted, however, that although GC-MS methodology can potentially resolve thousands of metabolites, compound derivatization and fragmentation during the procedure preclude direct metabolite identification. Instead, the methodology relies on using pure compounds as standards, thus leaving a number of metabolites detected in the procedure unidentified.
Using GC-MS methodology, we studied metabolic differences among humans, chimpanzees, and macaques at different ages in two functionally and evolutionary distinct brain regions: superior frontal gyrus of the prefrontal cortex (PFC) and lateral part of the cerebellar cortex (CBC). PFC is a brain region that emerged recently during primate evolution. It is implicated in complex associative functions including reasoning, planning, social behavior, and general intelligence (13–15). Furthermore, the PFC has been shown to undergo greater expansion at the gross anatomy level on the human evolutionary lineage than the CBC (16). Although CBC has also evolved considerably in primates (17, 18), its involvement in processing and execution of human-specific cognitive tasks is not yet well understood.
Results
Estimation of Metabolic Variation in the Primate Brain.
To directly assess the extent of metabolic changes between human brain and brains of other primate species, we measured metabolite levels in postmortem PFC and CBC samples from 49 humans, 11 chimpanzees, and 45 rhesus macaques, using GC-MS (Table S1). Many phenotypic parameters, such as maximal life span, age of sexual maturity, and brain growth curves differ between humans, chimpanzees, and rhesus macaques (19, 20). This makes it difficult to match samples with respect to age among these three species. To overcome this problem and to estimate changes in metabolite levels during brain development, maturation and aging, we sampled individuals of different age, covering most of the species’ lifespan: humans 0–98 y, chimpanzees 0–40 y, and macaques 0–28 y (Fig. S1 and Table S1). To minimize sampling variation, for all individuals we took care to sample gray matter only. To minimize technical variation, we measured each sample twice.
Using GC-MS methodology, we detected a total of 232 metabolites, some present in a single individual. Out of these, we focused on 118 metabolites, 61 annotated and 57 unknown, detected in more than 80% of samples in at least one brain region in one or more of the three species (Table S2). For these metabolites we found good agreement in metabolite measurements between replicates in all three species and all brain regions (median Pearson r > 0.95, P < 0.0001), indicating high reproducibility of the GC-MS measurements (Fig. S2).
Besides technical reproducibility, another concern is the relevance of metabolite measurements made in postmortem brain tissue to in vivo metabolite concentrations. Previous studies that have used proton magnetic resonance spectroscopy in human and rat brain biopsy samples have shown that concentrations of metabolites associated with anaerobic glycolysis, such as glucose, alanine, glutathione and lactate, change after death (21–24). In rat brain, postmortem delay also leads to concentration changes of dopamine, norepinephrine, and serotonin in the occipital cortex. This effect, however, was not shared by all brain regions, as serotonin levels remained stable over an 18-h postmortem interval in the cingulate cortex (25). Furthermore, concentrations of many metabolites, such as myo-inositol, creatine, glutamine, glutamate, N-acetylaspartate, and taurine were shown to remain stable in the postmortem brain tissue over long time intervals (22, 23, 26, 27). Similarly, experiments quantifying brain polyamines, such as spermine, spermidine, and putrescine, in postmortem human brain using GC-MS did not report any significant relationship between these metabolite concentrations and postmortem interval (28). To estimate the effect of postmortem delay on metabolite concentrations in our study, we took advantage of the fact that, with the exception of one sample with approximately a 5-h postmortem delay, all rhesus macaque brain tissues were collected and frozen within 20 min after death (Table S1). By examining the profile of each metabolite among the macaque samples, we identified 26 metabolites with significant differences in concentration between replicate measurements taken from the macaque individual with prolonged postmortem delay and measurements taken from all other samples in at least one brain region (Fig. S3). In agreement with previous studies, many of these metabolites were involved in glucose metabolism (Table S3). Consequently, we excluded these metabolites from all further analyses.
Some of the human and chimpanzee samples used in our study have a postmortem delay exceeding the 5-h interval tested using macaque samples. Still, in macaques, the vast majority of metabolites showed no indication of postmortem-induced concentration change after 5 h, demonstrating their long-term stability (Fig. S4). Furthermore, our assignment of metabolic changes to the evolutionary lineages requires similarity between macaque metabolite measurements over the lifespan and metabolic measurements in either human or chimpanzee brains. As macaque data are not affected by postmortem delay, and as human and chimpanzee samples have no postmortem interval bias, we can largely exclude postmortem delay as a confounding factor in this study.
To estimate the proportions of the total metabolic variance explained by species, age, and brain region identity, we used principal variance component analysis (29). The differences between species explained 49% of the total metabolic variation, differences with age 17%, and differences between two brain regions 9% (Fig. S5). Accordingly, metabolic differences across species, as well as differences with age, can be visualized using principal component analysis in both brain regions (Fig. 1 A and B).
Fig. 1.
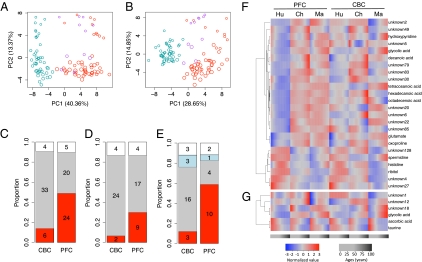
Metabolite variation among species. (A and B) Principal component analysis of CBC and PFC metabolomes of the three species based on 92 detected metabolites. Each circle represents an individual. Size of circles is proportional to the individuals’ ages, with larger circles corresponding to older individuals. Colors represent species: red, human; purple, chimpanzee; blue, rhesus macaque. (C–E) Proportions of human-specific (red), macaque-specific (gray), chimpanzee-specific (blue), and uncategorized metabolites (white) among the following: (C) the 43 and 49 metabolites with significant difference in concentration profiles between humans and rhesus macaques in CBC and PFC, respectively, identified using the full set of individuals; (D) the 30 metabolites with significant difference in concentration profiles between humans and rhesus macaques in both CBC and PFC; (E) the 25 and 17 metabolites with significant difference in concentration profiles among humans, chimpanzees, and rhesus macaques identified in CBC and PFC, respectively, using the subsets of 11 individuals per species matched using stage-of-life approach (SI Appendix). Numbers indicate numbers of metabolites in corresponding category. (F and G) Hierarchical clustering based on concentration profiles of 24 metabolites classified as human specific in PFC and six metabolites classified as human specific in CBC. Column headers indicate species: Ch, chimpanzee; Hu, human; Ma, rhesus macaque. Red and blue color intensities indicate metabolite concentration levels at different age normalized to mean equal a value of 0 and SD equal a value of 1 within each brain region.
Species-Specific Metabolic Changes.
We next identified metabolites showing significant concentration changes with age in each species using polynomial regression (30). At a false discovery rate (FDR) <10% (permutation test; SI Appendix), we identified 48 and 47 metabolites in the CBC and 53 and 26 in the PFC of humans and macaques, respectively (Table S4). In chimpanzees, due to the smaller sample size, we found only two and five age-related metabolites in CBC and PFC, respectively. Among the metabolites that change with age in our study, taurine was previously reported to decrease in concentration during postnatal brain development in the human and macaque occipital brain lobe (31). Similarly, in our data we found an approximately linear decrease with age in taurine concentration in both human and rhesus macaque PFC and CBC (human PFC: r = −0.37, P < 0.01; human CBC: r = −0.53, P < 10−5; macaque PFC: r = −0.28, P < 0.05; macaque CBC: r = −0.51, P < 0.001) (Fig. S6). Other than taurine, we found no reported primate brain time-series data that involved age-related metabolites as identified in our study.
We next tested whether metabolites change differently with age among the three species. Here, we focused on metabolites showing different concentration profiles over the species’ lifespan, rather than on mean concentration differences alone. Given the large and comparable numbers of human and rhesus macaque samples, we first identified metabolites with significant difference in concentration profiles between these two species in each brain region. Using analysis of covariance (ANCOVA) with linear, quadratic, and cubic models, we found 49 metabolites in PFC and 43 in CBC at FDR < 1% (permutation test; SI Appendix). Of these metabolites, 30 were different between humans and macaques in both brain regions (Fig. S7). Thus, both PFC and CBC metabolome show substantial divergence between humans and macaques, with a large proportion of the differences shared between the two brain regions.
We then used chimpanzee metabolite measurements to sort the identified human-macaque metabolic differences into three categories: (i) human-specific changes, (ii) macaque-specific changes, and (iii) uncategorized changes. Human-specific changes were defined based on significant difference between human metabolic profiles and metabolic profiles of (but not between) both chimpanzees and macaques (Wilcoxon test, P < 0.01, FDR < 1%). Macaque-specific changes were defined analogously, and therefore include changes on both the rhesus macaque evolutionary lineage, as well as the lineage between the common ancestor of Old World primates and the common ancestor of humans and chimpanzees. Uncategorized changes included metabolite profiles that are not described by either of the other two groups. We found six human-specific, 33 macaque-specific, and four uncharacterized metabolic changes in CBC. By contrast, in PFC, we found 24 human-specific, 20 macaque-specific, and five uncharacterized metabolic changes. Thus, there is a fourfold excess of human-specific metabolic changes in the human PFC compared with CBC (Figs. 1 C, F, and G and 2 and Table S5). This result was robust with respect to data normalization procedures (Fig. S10).
Fig. 2.
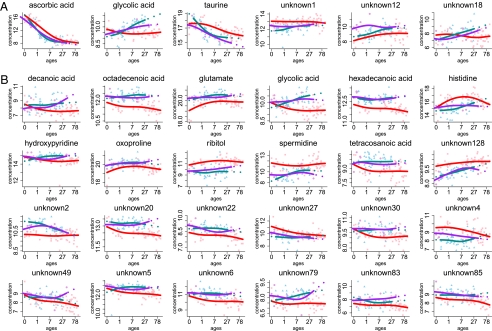
Metabolites with human-specific concentration profiles. Shown are the six metabolites with human-specific concentration profiles in CBC (A) and 24 metabolites with human-specific concentration profiles in PFC (B). Points show metabolite concentrations in each individual. Colors represent species: red, humans; purple, chimpanzees; and blue, rhesus macaques. Lines are spline curves fitted to data points with 3 df. The x axis shows individuals’ ages in years. The y axis shows normalized GC-MS measurements representing metabolite concentrations. Titles show metabolite annotation. Unannotated metabolites are labeled “unknown.”
The additional analyses confirmed the robustness of this result. Excluding unknown metabolites, we found three metabolites with human-specific profiles in CBC and 11 in PFC (Fig. S8). Similarly, limiting our analysis to 30 metabolites with significant concentration difference between humans and macaque in both PFC and CBC, we again found more human-specific changes in PFC (Fig. 1D). Thus, since the separation from the last common ancestor of humans and chimpanzees, many more metabolic changes have taken place on the human evolutionary linage in PFC than in CBC. By contrast, we found no indication for faster PFC metabolome evolution, compared with CBC, on either the macaque evolutionary lineage or the lineage between the common ancestor of Old World primates and the common ancestor of humans and chimpanzees (Fig. 1C).
Furthermore, we repeated the linage assignment analysis using equal numbers (n = 11) of age-matched individuals across the three species (Fig. S1). Equalizing sample size across the three species allowed us to assign metabolic differences to the human and the chimpanzee evolutionary linage with equal confidence. To avoid artifacts caused by differences in lifespan duration, we used the following two approaches to match individuals’ age across species: (i) stage-of-life approach: scaling ages linearly within each species to the same maximal lifespan, using 120 y for humans, 60 for chimpanzees, and 40 for rhesus macaques as a reference (19); and (ii) chronological approach: using calendar age directly. Using both approaches, we again found an excess of human-specific metabolic changes in PFC and not in CBC (Fig. 1E, Fig. S9, and Table S5). Thus, an acceleration of metabolic changes in the PFC is specific to the human evolutionary linage and is not observed in CBC.
Protein Expression and Metabolic Pathway Analyses.
To further test the validity of the identified human-specific metabolic changes and to assess their functional significance, we analyzed the expression of the corresponding metabolic enzymes in 12 human, 12 chimpanzee, and 12 rhesus macaque PFC samples using label-free quantitative mass spectrometry.
In humans, chimpanzees and rhesus macaques, we identified a total of 1301042, 1004360 and 1151143 peptides, which were mapped on 10769, 7945, and 9339 IPI peptide sequences, respectively. Requiring at least 10 peptide counts per protein, we detected 2,747 proteins in humans, 2,343 proteins in chimpanzees, and 2,842 proteins in rhesus macaques. The 1,951 genes detected in all three species (proteinwise FDR < 5%) were used in further analyses.
Using these data, we first tested whether metabolic changes with age found in the human PFC correlate with expression changes of enzymes directly interacting with these metabolites (i.e., enzymes connected to metabolites by a single edge in the KEGG pathway annotation). In total, we found 47 such metabolite-enzyme pairs, 21 of them showing significant correlation between metabolite and enzyme concentration changes with age (Pearson correlation, P < 0.0001, |r|>0.5). Finding such a high proportion of correlated changes is unexpected (P = 0.001), as estimated by randomly substituting expression profiles of the enzymes by those of other proteins 1,000 times. The result was also robust at different correlation coefficient cut-offs (Fig. S11).
Second, we tested whether enzymes directly associated with metabolites with human-specific concentration profiles (for simplicity: human-specific metabolites) show more expression differences between human and chimpanzee PFC than enzymes directly associated with metabolites that change with age in the human PFC but are not human specific. We found a greater expression divergence for 13 such enzymes associated with human-specific metabolites (one-sided Wilcoxon test, P = 0.003) compared with 24 enzymes associated with non–human-specific metabolites (Fig. S12A). Finally, we tested whether enzymes directly associated with human-specific metabolites show more human-specific expression than enzymes directly associated with non–human-specific metabolites, by estimating the ratio between their human–macaque and chimpanzee–macaque expression divergence. We found that for enzymes associated with human-specific metabolites, this ratio was significantly greater (one-sided Wilcoxon test, P = 0.004) (Fig. S12B). Thus, we observed significant agreement between human-specific metabolic profiles in the PFC and expression of the corresponding enzymes in terms of both ontogenetic changes and evolutionary divergence.
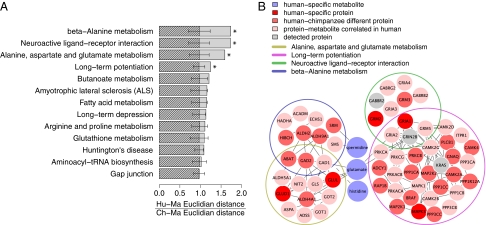
To determine which pathways are associated with human-specific metabolic changes, we first mapped the detected proteins onto the KEGG pathways containing at least one human-specific metabolite and more than 10 proteins. This resulted in 13 KEGG pathways that include five metabolites (glutamate, histidine, spermidine, oxoproline, and hexadecanoic acid) and 209 proteins. Four pathways had greater than expected human-specific protein expression divergence: long-term potentiation (P = 0.039), neuroactive ligand-receptor interaction (P = 0.015), alanine, aspartate, and glutamate metabolism (P = 0.022), and β-alanine metabolism” (P = 0.012) (Fig. 3A and Fig. S13). For all four pathways, Q values were <0.02, indicating significance of results given the number of pathways tested.
Fig. 3.
Pathway analysis of human-specific metabolites. (A) Shaded bars show median value of the human-specific protein expression divergence calculated as ratio of Euclidian distances between human–macaque and chimpanzee–macaque expression profiles of protein detected in the 13 KEGG pathways. Each pathway contains at least one human-specific metabolite and more than 10 detected proteins. Hatched bars show median human-specific divergence values expected by chance. Chance expectations were calculated by randomly assigning the same number of detected proteins to a given pathway, 1,000 times. Error bars show SD of the chance human-specific divergence estimates. (B) Network diagram showing four KEGG pathways with significantly greater human-specific protein expression divergence and three associated human-specific metabolites: glutamate, histidine, and spermidine. Colors indicate the following: bright red, proteins with significant human-specific expression; light red, proteins with significant human-chimpanzee expression divergence; pink, proteins with significant correlation to the corresponding metabolite profile; gray, detected proteins.
These pathways are not independent, but share common genes and a common metabolite, i.e., glutamate, which is present at lower concentrations in humans than in chimpanzees or macaques, particularly at the first years of life (Figs. 2B and 3B). Besides glutamate, two other metabolites with human-specific profiles in PFC, histidine, and spermidine, are involved in the β-alanine metabolism pathway.
Discussion
Our results show that human, chimpanzee, and macaque brain metabolomes change substantially with age: Specifically, 88% of the metabolites change their concentration significantly over the course of lifespan in at least one species or one brain region (Table S6). Many of these metabolic changes with age differ significantly among the three species (53% and 47% in PFC and CBC, respectively). Considering the two brain regions together, 77% of metabolic changes with age differed among species (Table S6). Large numbers of significant metabolic changes with age and differences among species are in line with substantial proportion of the total metabolic variance explained by these factors (17% and 49%, respectively; Fig. S5).
It must be noted, however, that most of the identified metabolic differences between species are not due to a difference in the pattern of concentration changes with age. Instead, 67% of metabolic changes in PFC and 82% in CBC reflect differences in the rate of age-related changes and/or differences in the mean metabolite concentration among species (Table S9). Thus, the majority of metabolic changes with age, especially in CBC, follow the same trajectory in all three species.
Our main result is the discovery of an approximately fourfold excess of metabolic changes in the PFC on the human evolutionary lineage. By contrast, CBC shows no such acceleration. This result is robust to choice of the normalization procedure, as well as chronological and stage-of-life age-matching subsets. Furthermore, we observed significant agreement between concentration profiles of metabolites in the human PFC and expression profiles of the corresponding metabolic enzymes. As protein expression measurements were carried out using independently dissected tissue samples and different methodologies, significant correspondence between protein and metabolic changes underscores the validity of the results.
Importantly, PFC and CBC samples used in our study come from the same individuals. This fact largely excludes individual demographic variables, such as postmortem delay, cause of death, medication, and so on, as causative factors for the difference in numbers of human-specific metabolic changes between PFC and CBC. Still, if environmental factors had a different effect on PFC and CBC metabolism, this could result in observed differences between the two brain regions.
Some aspects of environment and lifestyle, such as diet and life in captivity, could be argued to be more similar between chimpanzees and macaques, consequently making humans more distinct. To estimate the extent to which the species-specific metabolic changes might be caused by these environmental factors, we collected 37 metabolites reported to be affected by diet and 71 by exercise (Table S7). Using Fisher's exact test, we did not find significant overrepresentation of diet- or exercise-affected metabolites among metabolites with species-specific concentration profiles (P > 0.2 in all tests). Furthermore, reported diet and exercise effects were not compatible with the species-specific metabolic pattern that we observed (SI Appendix). Thus, our results provide no indication that excess of human-specific metabolic changes found in PFC could be explained by the influence of environmental factors.
Glutamate is one of the human-specific metabolites found in the PFC in this study. Glutamate release and recycling is a major metabolic pathway consuming 60–80% of the energy supplied by the glucose oxidation in the human cerebral cortex (32). Importantly, in the human brain, glutamate pathways play a central role in both energy metabolism and neurotransmission. Evolutionary changes in glutamate metabolism were previously indicated by the finding that glutamate dehydrogenase, an enzyme central to the glutamate and energy metabolism of the cell, has undergone duplication and subfunctionalization in the common ancestor of apes and humans, resulting in appearance of an ape- and human-specific GLUD2 gene (33). Our findings show that additional changes in glutamate metabolism took place in PFC, but not in CBC, on the human evolutionary lineage.
Specifically, we find that glutamate concentrations were lower in humans, and particularly in human newborns, compared with chimpanzees and macaques. Previous studies indicated that GLUD2 is optimized to rapidly metabolize brain glutamate (34, 35). It is therefore appealing to speculate that low glutamate concentrations observed in human infants might represent an evolutionary continuation of the rapid glutamate turnover trend. Indeed, based on previously published mRNA data (30), we observed higher expression levels of GLUD2 in human newborns compared with chimpanzee newborns (Fig. S14). This observation supports our notion that low glutamate levels in human newborns are caused by higher glutamate turnover. Functional interpretation of this change, however, is meaningful only in the context of the large metabolic flux of glutamate release and recycling taking place in glia and neurons (36, 37). Furthermore, it must be noted that glutamate concentrations in tissues other than brain were previously shown to be affected by diet and exercise (38, 39). Although such an environmental effect cannot be fully excluded in our study, human-specific changes in glutamate metabolism are intriguing, and might be an interesting and important subject of further research.
We must note, however, that even though our analysis covers more metabolites than other studies conducted to date, it is far from being comprehensive. Thus, although our findings indicate that changes in brain metabolism have contributed to the evolution of human cognition, many important metabolic pathways might have been missed in our analysis. Further studies are needed to elucidate the entire scope of metabolic changes that took place on the human evolutionary lineage.
Materials and Methods
Details on materials and methods are provided in SI Appendix. Briefly, samples were dissected from the frozen postmortem tissue from healthy individuals. PFC dissections were made from the frontal part of the superior frontal gyrus. CBC dissections were taken from the lateral part of the cerebellar hemispheres. For metabolite data, metabolites were extracted from ∼100 mg tissue, using GC-MS measurements as detailed elsewhere (40). All sample replicates were processed in a completely randomized order to minimize any possible batch effects. For protein data, proteins were extracted from 100 mg tissue, using label-free quantitative mass spectrometry according to a previously published method (41). Analysis was performed on an LTQ mass spectrometer equipped with a metal needle electrospray interface mass spectrometer (ThermoFinnigan) in a data-dependent collection model (each full scan followed by 10 MS/MS scans of the most intense ions).
Supplementary Material
Acknowledgments
We thank the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders, the Netherlands Brain Bank, the Chinese Brain Bank Center, and Dr. H. R. Zielke, Dr. J. P. Dai, and Dr. Y. Sun in particular for providing the human samples; the Yerkes Primate Center, the Biomedical Primate Research Centre, the Anthropological Institute, and Museum of the University of Zurich for chimpanzee samples, and Dr. R. Martin and Dr. W. Scheffran in particular for providing the chimpanzee samples; Suzhou Drug Safety Evaluation and Research Center, and C. Lian, H. Cai, and X. Zheng in particular for providing the macaque samples; J. Dent for editing the manuscript; E. Lizano and F. Xue for assistance; and all members of the Comparative Biology Group in Shanghai for helpful discussions and suggestions. We thank the following for financial support: the Ministry of Science and Technology of the People's Republic of China (Grants 2007CB947004, 2006CB910700, and 2011CB910200), the Max Planck Project “Age-related changes in human-chimpanzee and rhesus macaque brain metabolism,”the Chinese Academy of Sciences (Grants KSCX2-YW-R-094 and KSCX2-YW-R-251), the Shanghai Institutes for Biological Sciences (Grant 2008KIT104), the European Union Proteomage program, the Max Planck Society, and the Bundesministerum fuer Bildung und Forschung.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019164108/-/DCSupplemental.
References
- 1.Aiello L, Wheeler P. The expensive-tissue hypothesis: The brain and the digestive system in human and primate evolution. Curr Anthropol. 1995;36:199–221. [Google Scholar]
- 2.Mink JW, Blumenschine RJ, Adams DB. Ratio of central nervous system to body metabolism in vertebrates: Its constancy and functional basis. Am J Physiol. 1981;241:R203–R212. doi: 10.1152/ajpregu.1981.241.3.R203. [DOI] [PubMed] [Google Scholar]
- 3.Caceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uddin M, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci USA. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman LI, Wildman DE, Schmidt TR, Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Khaitovich P, et al. Positive selection on gene expression in the human brain. Curr Biol. 2006;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- 7.Sherwood CC, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci USA. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott C, Bustillo J. What have we learned from proton magnetic resonance spectroscopy about schizophrenia? A critical update. Curr Opin Psychiatry. 2006;19:135–139. doi: 10.1097/01.yco.0000214337.29378.cd. [DOI] [PubMed] [Google Scholar]
- 9.Dwyer DS, Bradley RJ, Kablinger AS, Freeman AM., 3rd Glucose metabolism in relation to schizophrenia and antipsychotic drug treatment. Ann Clin Psychiatry. 2001;13:103–113. doi: 10.1023/a:1016671725396. [DOI] [PubMed] [Google Scholar]
- 10.Khaitovich P, et al. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 2008;9:R124. doi: 10.1186/gb-2008-9-8-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jellum E, Bjornson I, Nesbakken R, Johansson E, Wold S. Classification of human cancer cells by means of capillary gas chromatography and pattern recognition analysis. J Chromatogr A. 1981;217:231–237. doi: 10.1016/s0021-9673(00)88077-2. [DOI] [PubMed] [Google Scholar]
- 12.Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD. Analytical and statistical approaches to metabolomics research. J Sep Sci. 2009;32:2183–2199. doi: 10.1002/jssc.200900152. [DOI] [PubMed] [Google Scholar]
- 13.Barbey AK, Krueger F, Grafman J. An evolutionarily adaptive neural architecture for social reasoning. Trends Neurosci. 2009;32:603–610. doi: 10.1016/j.tins.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan J, et al. A neural basis for general intelligence. Science. 2000;289:457–460. doi: 10.1126/science.289.5478.457. [DOI] [PubMed] [Google Scholar]
- 15.Winterer G, Goldman D. Genetics of human prefrontal function. Brain Res Brain Res Rev. 2003;43:134–163. doi: 10.1016/s0165-0173(03)00205-4. [DOI] [PubMed] [Google Scholar]
- 16.Clark DA, Mitra PP, Wang SS. Scalable architecture in mammalian brains. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- 17.MacLeod CE, Zilles K, Schleicher A, Rilling JK, Gibson KR. Expansion of the neocerebellum in Hominoidea. J Hum Evol. 2003;44:401–429. doi: 10.1016/s0047-2484(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 18.Weaver AH. Reciprocal evolution of the cerebellum and neocortex in fossil humans. Proc Natl Acad Sci USA. 2005;102:3576–3580. doi: 10.1073/pnas.0500692102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Magalhaes JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol. 2009;22:1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 20.Leigh SR. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol. 2004;62:139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- 21.Opstad KS, Bell BA, Griffiths JR, Howe FA. An assessment of the effects of sample ischaemia and spinning time on the metabolic profile of brain tumour biopsy specimens as determined by high-resolution magic angle spinning (1)H NMR. NMR Biomed. 2008;21:1138–1147. doi: 10.1002/nbm.1296. [DOI] [PubMed] [Google Scholar]
- 22.Opstad KS, Wright AJ, Bell BA, Griffiths JR, Howe FA. Correlations between in vivo (1)H MRS and ex vivo (1)H HRMAS metabolite measurements in adult human gliomas. J Magn Reson Imaging. 2010;31:289–297. doi: 10.1002/jmri.22039. [DOI] [PubMed] [Google Scholar]
- 23.Perry TL, Hansen S, Gandham SS. Postmortem changes of amino compounds in human and rat brain. J Neurochem. 1981;36:406–410. doi: 10.1111/j.1471-4159.1981.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 24.Shank RP, Aprison MH. Post mortem changes in the content and specific radioactivity of several amino acids in four areas of the rat brain. J Neurobiol. 1971;2:145–151. doi: 10.1002/neu.480020207. [DOI] [PubMed] [Google Scholar]
- 25.Kontur PJ, al-Tikriti M, Innis RB, Roth RH. Postmortem stability of monoamines, their metabolites, and receptor binding in rat brain regions. J Neurochem. 1994;62:282–290. doi: 10.1046/j.1471-4159.1994.62010282.x. [DOI] [PubMed] [Google Scholar]
- 26.Michaelis T, Helms G, Frahm J. Metabolic alterations in brain autopsies: Proton NMR identification of free glycerol. NMR Biomed. 1996;9:121–124. doi: 10.1002/(SICI)1099-1492(199605)9:3<121::AID-NBM409>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Petroff OA, Ogino T, Alger JR. High-resolution proton magnetic resonance spectroscopy of rabbit brain: Regional metabolite levels and postmortem changes. J Neurochem. 1988;51:163–171. doi: 10.1111/j.1471-4159.1988.tb04850.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen GG, Turecki G, Mamer OA. A quantitative GC-MS method for three major polyamines in postmortem brain cortex. J Mass Spectrom. 2009;44:1203–1210. doi: 10.1002/jms.1597. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Bushel PR, Chu T-M, Wolfinger RD. Principal variance components analysis: Estimating batch effects in microarray gene expression data. In: Scherer A, editor. Batch Effects and Noise in Microarray Experiments: Sources and Solutions. Wiley, Chichester: 2009. pp. 141–154. [Google Scholar]
- 30.Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturman JA, Gaull GE. Taurine in the brain and liver of the developing human and monkey. J Neurochem. 1975;25:831–835. doi: 10.1111/j.1471-4159.1975.tb04414.x. [DOI] [PubMed] [Google Scholar]
- 32.Rothman DL, Behar KL, Hyder F, Shulman RG. In vivo NMR studies of the glutamate neurotransmitter flux and neuroenergetics: Implications for brain function. Annu Rev Physiol. 2003;65:401–427. doi: 10.1146/annurev.physiol.65.092101.142131. [DOI] [PubMed] [Google Scholar]
- 33.Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- 34.Plaitakis A, Spanaki C, Mastorodemos V, Zaganas I. Study of structure-function relationships in human glutamate dehydrogenases reveals novel molecular mechanisms for the regulation of the nerve tissue-specific (GLUD2) isoenzyme. Neurochem Int. 2003;43:401–410. doi: 10.1016/s0197-0186(03)00028-7. [DOI] [PubMed] [Google Scholar]
- 35.Shashidharan P, et al. Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J Biol Chem. 1994;269:16971–16976. [PubMed] [Google Scholar]
- 36.Benjamin AM, Quastel JH. Locations of amino acids in brain slices from the rat. Tetrodotoxin-sensitive release of amino acids. Biochem J. 1972;128:631–646. doi: 10.1042/bj1280631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Berg CJ, Garfinkel D. A stimulation study of brain compartments. Metabolism of glutamate and related substances in mouse brain. Biochem J. 1971;123:211–218. doi: 10.1042/bj1230211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howarth KR, LeBlanc PJ, Heigenhauser GJ, Gibala MJ. Effect of endurance training on muscle TCA cycle metabolism during exercise in humans. J Appl Physiol. 2004;97:579–584. doi: 10.1152/japplphysiol.01344.2003. [DOI] [PubMed] [Google Scholar]
- 39.Li LO, et al. Early hepatic insulin resistance in mice: A metabolomics analysis. Mol Endocrinol. 2010;24:657–666. doi: 10.1210/me.2009-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 41.Fu X, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.