Abstract
The human immunodeficiency virus envelope protein is the key element mediating entry into host cells. Conformational rearrangement of Env upon binding to the host CD4 receptor and chemokine coreceptor drives membrane fusion. We elucidated the quaternary arrangement of the soluble Env trimeric immunogen o-gp140ΔV2TV1, in both its native (unliganded) and CD4-induced (liganded) states by cryoelectron microscopy and molecular modeling. The liganded conformation was elicited by binding gp140 to the synthetic CD4-mimicking miniprotein CD4m. Upon CD4m binding, an outward domain shift of the three gp120 subunits diminishes gp120–gp41 interactions, whereas a “flat open” concave trimer apex is observed consequent to gp120 tilting away from threefold axis, likely juxtaposing the fusion peptide with the host membrane. Additional features observed in the liganded conformation include rotations of individual gp120 subunits that may release gp41 for N- and C-helix refolding and also may lead to optimal exposure of the elicited coreceptor binding site. Such quaternary arrangements of gp140 lead to the metastable liganded conformation, with putative locations of exposed epitopes contributing to a description of sequential events occurring prior to membrane fusion. Our observations imply a mechanism whereby a soluble Env trimeric construct, as opposed to trimers extracted from virions, may better expose crucial epitopes such as the CD4 binding site and V3, as well as epitopes in the vicinity of gp41, subsequent to conjugation with CD4m. Structural features gleaned from our studies should aid the design of Env-based immunogens for inducement of potent broadly neutralizing antibodies against exposed conformational epitopes.
Keywords: HIV-1, immunogen design, induced conformation
The human immunodeficiency virus envelope proteins gp120 and gp41 associate in a trimeric arrangement (1–3) and mediate membrane fusion through conformational rearrangement and fusion peptide insertion into the host membrane. The gp120 tertiary conformational change observed via X-ray crystallography has not been definitively characterized in the context of a quaternary structure. Several observations have been gleaned from tomography studies, though with inconclusive and sometimes contradictory results. Quaternary structural features reported previously include trimer morphology, with some groups reporting a mushroom-like shape and others a more rod-like arrangement, as well as the relative location of the primary epitope, hypervariable loops, and N- and C-termini, which have some variation among the different tomograms. Another point of conflict is the presence of a cavity at the threefold axis, capped by a density. One final feature is the arrangement of gp41, with some groups reporting a thin, stalk-like rod arrangement of gp41 and another group reporting a more splayed out, tripod-like conformation of gp41 proximal to the viral membrane. Taken together, these contrasting features portray a heterogeneous set of registers that seem to hinder a cohesive and thorough model of ligand-induced quaternary arrangement.
Reconstruction of density maps in combination with docking of X-ray crystallography atomic coordinates (4–6) serves to further characterize and build upon previous work. Structures of unliganded simian immunodeficiency virus (SIV) gp120 (7) and liganded HIV-1 gp120 (CD4-bound) (8) revealed a large conformational rearrangement in the gp120 inner domain, though its significance was then unclear in the quaternary context. As mentioned above, tomograms by various groups (9–11) have not reached consensus on several structural features. Both Kwong (8) and Chen (7) provided putative quaternary models based on their tertiary findings, and the subsequent tomograms either diverged from or converged to those models. Cryoelectron microscopy (cryo-EM) imaging and structure determination of gp140 as a soluble trimeric complex as opposed to imaging of envelopes purified from virions can provide evidence of quaternary conformational shifts identified upon ligand binding at higher resolution than previously achieved, and hopefully serve to address the discrepancies between previous observations, which currently hamper a consensus view on the crucial events preceding membrane fusion.
In this work we propose a model for events occurring prior to membrane fusion by generating and analyzing density maps of gp140 in an unliganded and also a CD4-induced liganded conformation. We directly assessed the impact of the gp120 tertiary conformational change onto the quaternary arrangement of the gp140 trimer by comparatively analyzing the density maps between the native gp140 structure and its liganded conformation. The gp140 construct utilized in this study had a partial V2 loop truncation, a modification that has resulted in altered immunogenic characteristics (12–17), and was derived from TV1, an R5-dependent subtype C isolate originating from South Africa. Previous work has determined that TV1 immunogens elicited neutralizing antibodies and provided breadth against isolates from subtypes B and C (18). Furthermore, the antibodies used in the immunochemical characterization of gp140 (12, 17) cover the most pertinent gp120 and gp41 epitopes, namely the CD4 binding site (CD4BS—IgGCD4 and sCD4), the coreceptor binding site (CrBS—17b and 447D), a neutralizing epitope on gp41 (2F5), and a glycan-dependent epitope (2G12), thus thoroughly covering the topology of gp140 so as to display its similarity to native Env trimers. The CD4 miniprotein (CD4m) used to elicit the liganded conformation displays nanomolar affinity to gp120 (19), and elicits a nearly identical gp120 conformation as does soluble CD4 (sCD4) (20). The small size of CD4m allowed the structural analysis in this work to focus on the quaternary changes in gp140 and circumvent possible steric aggregation induced by stoichiometric sCD4–gp140 binding. Locations of epitopes, hypervariable loops, and N- and C-termini prior to and following CD4 binding provide a rationale for epitope exposure, gp120/gp41 interface abolition, and quaternary rearrangement. Tying in the well-characterized tertiary shift in gp120 to the entire trimer also reveals how a change in conformation in one protein can be communicated to an adjacent, noncovalently associated protein, as well as to demonstrate the extent of epitope occlusion and exposure posttriggering by CD4. Such information, crucial to vaccine and drug development, can enhance understanding of Env and hopefully broaden the scope of understanding of this crucial protein complex.
Results
Unliganded and CD4m-Liganded gp140 Reconstructions.
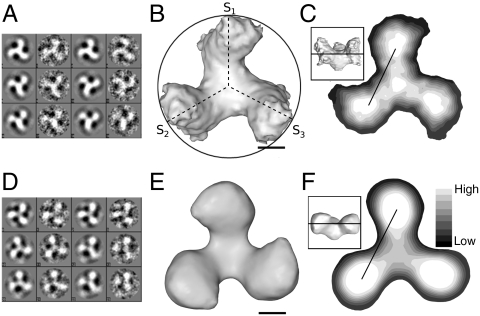
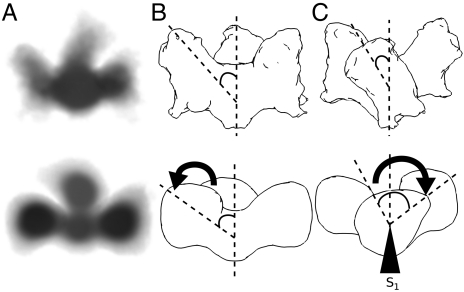
The density map of unliganded trimeric gp140 revealed a propeller blade motif, clockwise handedness and a rounded short stem at the gp41 site or hub of the propeller blade (Fig. 1B). The native trimer has a diameter of approximately 110 Å wide, and about 90 Å in height. The liganded density map has dimensions of 110 Å in diameter and 70 Å in height (Fig. 1E). One notable difference is the weakened density in the threefold interface, diminished after CD4m binding, which could be readily assessed by the distance of the density gravity center between the proximal and the distant peaks of high contour (approximately 5 σ above the average density value). Comparative analysis of the two density maps suggests that three events occur upon reaching the liganded state: an outward density shift, a rotation about axes normal to the threefold (z) axis in each gp120 subunit (rotation about axes S1, S2, and S3 at θ = 90°, 210° and 330° respectively, Fig. 1B) and a subunit tilt away from the z-axis that effectively flattens the density surrounding the threefold axis. Collectively, these events have significant impacts on a putative membrane fusion model.
Fig. 1.
Class averages of raw data showing top views of gp140. (A) Class averages of unliganded gp140. The resolution of this structure is 11 Å using the 0.143 Fourier shell correlation (FSC) cutoff criterion, and 19 Å using the 0.5 FSC cutoff criterion. The map shows good agreement between projections and class averages. (B) Top view of unliganded gp140. Note subunit axes S1, S2, and S3 at 90°, 210°, and 330°, respectively. (C) Segmentation of unliganded gp140 shows distinct gravity centers. Distance between gravity centers with highest intensity is approximately 58 Å. The Inset shows the plane at which density map was cut. (D) Class averages of liganded gp140. The resolution of this density map was low-pass filtered to 19 Å for effective comparison with the unliganded map. (E) Top view of liganded gp140. (F) Segmentation of liganded gp140, with distance between gravity centers of approximately 72 Å. The Inset shows the plane at which density map was cut. Scale bars: 25 nm for panels B, C, E, and F.
Density Shift Away from Threefold Axis.
Isosurface density analysis of unliganded gp140 timers at a contour value of 3σ above the mean value revealed that the trimer “fan blade” connection to the threefold density remained contiguous. The density of the trimer arms in the liganded state are more bulbous at the distal tip, observations best described as a shift in density away from the trimer core. Comparative segmentation of both unliganded and liganded trimers revealed a weakened density at the threefold axis of gp140 trimers in the liganded state, along with a prominent gravity center at each trimer arm locus (Fig. 1 C and F).
Segmentation analysis and center-to-center measurements of unliganded vs. liganded trimers were taken to measure the distances between dense regions in the trimer arm to assess the extent of density shift. The unliganded trimer contains a center of high density in the likely location of the inner domain (Fig. 1C). The distance from the threefold axis to the center of this dense domain is about 25 Å (Fig. 1C). In the liganded state there is no perceptible difference between domains in the trimer arm. The distance between the threefold axis and the dense domain in the liganded trimer was about 40 Å (Fig. 1F). The labile glycans do not appear to be as consistently represented by the data as the subunit core, because isosurface contour adjustment caused the density representative of the glycosylated face to disappear first.
Distinct Subunit Rotations Evident in the gp140 Liganded State.
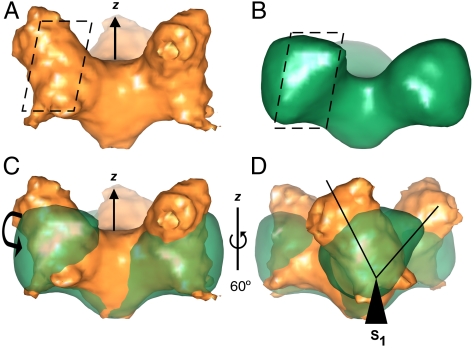
Evaluation of the side views of both unliganded and liganded density maps (Fig. 2 A and B) reveals a pair of subunit rotations in the liganded state. A subunit tilt away from the threefold (z) axis spans approximately 25°, which effectively flattens the trimer and may expose gp41 and the junction density among the subunits around the threefold axis (Fig. 2C), when compared to the depression at the threefold axis in the unliganded gp140 density map (Fig. 2 A and C). Juxtaposition of the threefold axis with the host membrane likely facilitates fusion peptide insertion, and this flattened quaternary state is likely stabilized by coreceptor binding. The subunit rotation about the axes normal to the z-axis was elucidated by overlapping both densities and viewing through the trimer arms (Fig. 2D). If one defines the axis perpendicular to the threefold axis, effectively looking down on the trimer in a top view (Fig. 1 B and E), as the z-axis, then one such observed subunit rotation is about the axes normal to the threefold (z) axis (axes S1, S2, and S3, as outlined above and in Fig. 1B) and another tilts away from the z-axis. The overlapped densities corresponding to the gp120 subunit appear to rotate clockwise by approximately 60° (Fig. 2D). Such a rotation may partly abrogate gp120–gp41 binding, free up the N-helix of gp41 to facilitate N- and C-helix binding, and translocate the CD4 binding site (CD4BS) on gp120 so that it faces the adjacent counterclockwise gp120 subunit, as well as rotate the location of the V3 loop such that it would be perpendicularly accessible and thus amenable to coreceptor binding.
Fig. 2.
Comparison of segmentation and side views of unliganded and liganded gp140. (A) Side view of unliganded gp140 (orange). The z-axis is also referred to as the threefold axis throughout the text. Slice refers to segmentation in Fig. 4A. (B) Side view of liganded gp140 (green). Slice refers to segmentation in Fig. 4B. (C) Side views of overlapped unliganded (orange) and liganded (green) gp140 densities reveal a tilt away from the z-axis observed upon CD4m binding. The decrease in the total height in the trimer is compensated by an extension in width. (D) Rotating by 60° about the z-axis reveals a subunit rotation of approximately 60° as measured through the S1 axis protruding out of the plane of the page.
Distances Between Epitopes Suggest Ligand Binding Angles.
The location of epitopes crucial to membrane fusion is suggestive of the receptor entry angles for binding. Because the CD4BS in the native, unliganded state appears to be located at the outermost trimer apex, about 60 Å apart, it appears that CD4 likely approaches the trimer from an angle perpendicular to the trimer plane (along the z-axis). As mentioned above, one change in gp120 subunit orientation upon CD4m binding is a clockwise rotation about axes normal to the threefold (z) axis (axes S1, S2, and S3), as observed by superimposition of trimer arms of both densities. The V3 loop in the unliganded trimer fitting was located on the outer edge of each trimer arm, and is oriented on the trimer surface in the liganded state fitting approximately 100 Å apart. Likely, the gp120 rotation results in maximal exposure of the V3 loop, such that both the V3 loop and the bridging sheet are perpendicularly accessible by the coreceptor. Subsequent binding of the coreceptor likely results in a more solvent-exposed gp41 locus at the threefold axis, possibly facilitating fusion peptide insertion into the host membrane.
Variable Loop Location and Subunit Rotations.
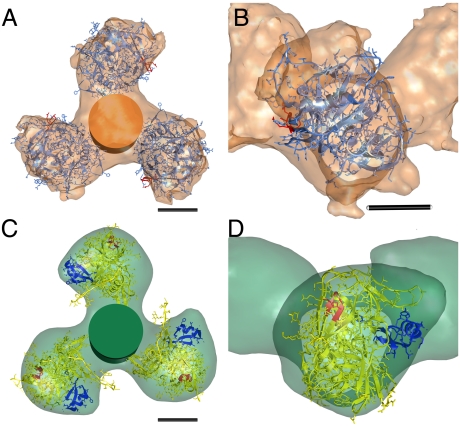
Docking of atomic coordinates resulted in good agreement between the density map and the X-ray coordinates utilized in fitting, with a correlation coefficient of 0.77. The native density map had unliganded SIV gp120 coordinates [Protein Data Bank (PDB) 2BF1] fitted into it (Fig. 3 A and B). In our unliganded fitting, residues involved in CD4 binding, such as Asp368 and Glu370, are approximately 60 Å apart on the trimer apex, dimensions relevant for the design of multivalent miniprotein-based inhibitors to compete for multiple CD4 binding sites simultaneously.
Fig. 3.
Docking of gp120 and gp41 coordinates into density maps. (A) Top view of native gp140 map with unliganded SIV gp120 coordinates (PDB: 2BF1, blue, with V3 colored red) and an orange cylinder representing gp41. CD4 binding sites are approximately 60 Å apart. (B) Close-up view of gp120 subunit showing coordinate location of V3 loop in red. (C) Top view of liganded gp140 map with CD4-bound HIV coordinates (PDB: 1GC1, yellow, with V3 colored red) and CD4m coordinates (PDB: 2I5Y, blue), with green cylinder representing gp41. (D) Close-up view of gp120 subunit, showing new position of V3 loop in red. Distance between V3 loop regions in trimer is approximately 100 Å. Scale bars: 25 Å for panels A, B, C, and D.
The density map of the trimer in the liganded state had HIV-1 CD4-bound (PDB 1GC1) and CD4m-bound (PDB 2I5Y) fitted into it, again with good agreement and with similar results (Fig. 3 C and D and Fig. S1). Coordinates for CD4m (PDB 2I5Y) were included in the fitting to ascertain the location of CD4m and thereby the CD4BS after binding (Fig. 3 C and D, in blue). In the unliganded density map, the region corresponding to the V3 loop as exhibited by the docking is located on the outer edge of each trimer fan blade (Fig. 3B), poised in position to become maximally exposed after the clockwise rotation that CD4m binding elicits. The coreceptor binding site, comprised of the newly formed bridging sheet and the V3 loop, is, in the liganded state, located proximal to the threefold axis (Fig. 3D), and is more accessible once gp120 and the CD4BS rotate about the axes normal to the threefold axis (S1, S2, and SM3). The location of the CD4BS suggests that CD4 binds perpendicular to the threefold plane, as opposed to obliquely parallel to this plane (Fig. 4A), and after subunit rotation, is oriented toward the adjacent counterclockwise subunit, resulting in a more accessible coreceptor binding site.
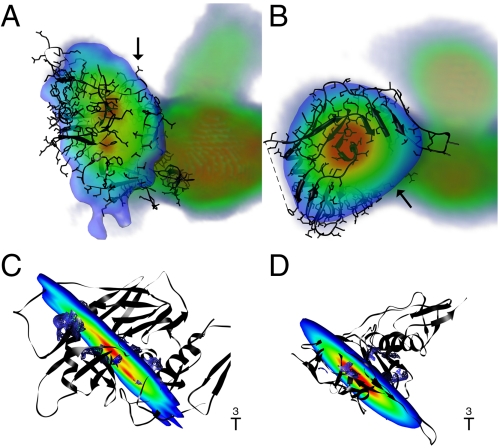
Fig. 4.
Segmentation of gp140 reveals density gap colocalizing with inner-outer domain boundary. (A) Volume rendering of unliganded gp140 docked with unliganded SIV gp120 in an oblique view with segmentation slice reveals areas of higher density (in red) compared to areas of lower density (in yellow, green, and blue). A clear separation between high density domains is observed while slicing through volume, which is likely indicative of the boundary between inner and outer domains. Arrow is pointing at the likely orientation of CD4 as it binds to the CD4-binding residues. (B) Volume rendering of CD4m-bound gp140 in an oblique view with segmentation slice reveals a uniform area of high density. As opposed to native, unliganded gp140, no discernable gap in high density areas is seen, suggesting closer association between the inner and outer domains of gp120. Arrow points to the location of the CD4 binding site subsequent to rotation of gp120 as described in Fig. 2. (C) Coordinates of unliganded SIV gp120 with slice from unliganded gp140 density map. Tilt reveals localization of the density gap at the inner-outer domain interface. (D) Coordinates of CD4-bound HIV-1 gp120 with slice from CD4m-bound gp140 density map. Tilt reveals lack of density separation, with corresponding closer association between inner and outer domains as observed in the coordinates.
The V2 loop, in the unliganded state, appears to be located proximal to the viral membrane on the leading edge of each trimer arm. Upon CD4m binding, the V2 loop rotates along the gp120–gp41 interface such that it points counterclockwise toward the adjacent gp120 subunit. Visual inspection of the docked coordinates in the unliganded density map also suggests that the V4 and V5 loops are situated at the trimer arm tip, along with some of the major glycosylation sites, rendering them fully solvent-exposed and thus allowing them to shield Env from the host immune system. The gp120 region at the gp120/gp41 interface is partly comprised of two β-strands (β2 and β3) that, in the liganded state, form half of the bridging sheet to provide part of the coreceptor epitope.
The conformation of gp41 in a native state is unknown. To address this uncertainty, as well as the lack of gp41 coordinates in an unliganded conformation, both structures were docked with a cylinder at the threefold axis (Fig. 3 A–D) with dimensions similar to the six-helix bundle conformation of gp41, the only available crystal structure conformation of gp41. The cylinder has a radius of 17 Å and a height of 48 Å. The use of this cylinder thus mimics the maximum space that gp41 could occupy, though it is more likely that the unbound gp41 conformation only has an ordered N-helix associated with the inner domain of gp120, which may keep it from associating with the gp41 C-helix. Superposition of both density maps reveals a decrease in gp41 height, though it is unclear whether this observation is suggestive of a gp41 conformational change.
Missing residues contributed to the small void volume at the threefold axis observed when docking the coordinates to our unliganded density map, whereas our density map of Env in the liganded state docked well with the HIV-1 CD4-bound gp120 crystal structure (PDB 1GC1) and the CD4m-containing gp120 crystal structure (PDB 2I5Y). In the docking of unliganded SIV gp120 crystal structure to our unliganded gp140 structure, the V1/V2 and V3 loops, along with 83 residues from the N-terminus and 9 from the C-terminus, were truncated. The HIV-1 gp120 coordinates in the liganded state also have truncated N- and C-termini, V1/V2, and V3 loops.
Inner-Outer Domain Interface on gp120.
Segmentation of the native gp140 trimer revealed a clear gap in density in the trimer arms (Fig. 4A and Fig. S2A). Inspection of the density map after docking of the SIV gp120 crystal structure colocalized the gap in density with the boundary between the inner and outer domains (Fig. 4C). Such a secondary structural element validated the gp120 orientation within the density map, with the inner domain oriented toward the threefold axis. The gap in density was not present in the density map of gp140 in the liganded state (Fig. 4 B and D and Fig. S2B), reflecting the closer association between the inner and outer domains in the liganded conformation when compared to the SIV unliganded conformation.
Discussion
The increased distance between the gp41 core and each gp120 subunit (Figs. 1 C and F) observed between the unliganded and the liganded states can be properly described as a density shift away from the threefold axis in our gp140 structure, but with a constrained extension of the outer domain at the tip of each trimeric blade to the central axis. Whereas the observed density shift of approximately 15 Å correlates well with the tertiary structural change previously measured by the displacement of the V2 loop stem, strands β2 and β3, elicited in the liganded state (7), such density weakening at the junction of gp120 subunits to the gp41 density at the threefold axis may not only sterically facilitate the formation of the gp41 prehairpin intermediate to expose the fusion peptide, but may also contribute to gp120 shedding as previously reported (21, 22). As part of the quaternary density movement in the gp140 trimer, the reported tertiary conformational change of gp120 could be operating in concert within the trimer to expose and elicit the conformational change of gp41. The locations of the domain movements with respect to the threefold axis are consistent in that the inner domain is juxtaposed to the threefold axis (Fig. 3 A and B), and the outer domain and glycosylated face is solvent-exposed. This counters one of the Zanetti models, in which the glycosylated face of the outer domain was oriented toward the threefold axis (9). The Zhu model revealed a trimer of a different morphology (10), with a gp120 orientation that was docked similar to our work, but with the distinction that there are likely many steric clashes at the threefold, because they did not allow sufficient space there to account for the missing residues in the crystal structure at the N- and C-termini. Moreover, whereas our gp120 subunit is oriented such that the inner domain is oriented toward the threefold axis, as speculated in the Chen model (7), our structure indicates the movement of the inner domain away from the threefold axis, instead of moving the outer domain and leaving the inner domain static, resulting in a more exposed threefold axis and a loosened gp120/gp41 interface. This may serve as intersubunit communication to promote conformational rearrangement of gp41, likely exposing the fusion peptide. Additionally, the position of the outer domain is further confirmed to be static, as the maximum diameter of gp140 (110 Å) was found to remain unchanged in the liganded density map. This observation is consistent with a tertiary shift in the gp120 inner domain proximal to the threefold axis, thus resulting in the diminished gp120/gp41 interface.
Our observed gp120 orientation accommodates CD4 entry and binding angle perpendicular to the threefold plane in agreement with the Chen model (Fig. 4A), and the gp120 orientation in the liganded state is in agreement with Kwong’s prediction regarding the gp120 trimeric arrangement (Fig. 4B) (8, 23). The orientations of gp120 docked in our two density maps supported the predicted model by Chen et al. (7) by placing the C-terminus next to the central gp41 stalk at the threefold axis of gp140 structure. The predominantly hydrophobic residues in the unliganded, inner domain β-strands of the bridging sheet (β2–3), which are at the gp120/gp41 junction in our docking, may stabilize the hydrophobic α-helices of gp41. Dissociation of these gp120 β-strands from gp41 thus may serve to enhance the accessibility of gp41 at the threefold axis for subsequent fusion peptide exposure and insertion, followed by genome release in endosomes as recently suggested (24). Rotation of gp120 subunits in the liganded state could remove part of the gp120/gp41 association and contribute to its weakening, possibly further promoting gp41 N- and C-helix association through exposure of hydrophobic surfaces, facilitating quaternary arrangement for the fusion peptide to insert into the host membrane. Comparison between our native and liganded gp140 structures corroborates the Liu model of subunit rotation (11). The gp120 subunit rotates, from having its CD4BS perpendicularly accessible prior to the binding, to a location laterally exposed from gp120, such that CD4 does not sterically occlude the coreceptor binding site, thus maximizing CCR5 or CXCR4 accessibility to the bridging sheet and V3 loop. Previous work has established that all three CD4 binding sites are exposed in the trimer construct (25), corroborating the exposed nature of the CD4BS as evidenced by our unliganded gp140 docking. However, the Liu model did not account for the observed tilt away from the z-axis, which flattens the trimer and exposes the threefold axis, the location where the N-terminal fusion peptide of gp41 would ostensibly protrude from and insert into the host membrane. Moreover, the location of the V2 loop in the Liu model is proximal to the threefold axis, a position that is likely a result of placing the b12-bound coordinates in their structure of the unbound state. In Liu’s ternary complex, the positioning of V2 does not populate the cap over the observed cavity, and such void volume does not colocalize to the N- and C-termini, either. The observed V2 location in the unliganded gp140 density map and its subsequent location in the liganded state suggest a drastic rearrangement such that the V2 stem is translocated vertically and rotated such that it is oriented toward the adjacent counterclockwise subunit (Fig. 4B). Also, the location of the V3 loop in our unliganded docking, proximal to the adjacent clockwise gp120 subunit (Fig. 3B), is in agreement with previous work demonstrating a joint V2/V3 antibody epitope (26), as well as with work suggesting gp41-independent intersubunit gp120 contacts mediated by V2 (27) and with the locations of V2 and V3 suggested by the Chen model (7). Furthermore, the gp140 construct utilized in this work contains mutations in both primary and secondary cleavage sites, leading to enhanced trimer stability (12). Whereas other studies have suggested altered quaternary profiles in cleavage-defective clade A gp140 constructs as opposed to cleavage-competent constructs, predominantly in the gp41 region (28), as well as a difference in immunogenicity between cleavage-defective and cleavage-competent clade B constructs (29), the enhanced stability reported for clade C cleavage-defective gp140, as well as the native-like immunochemical binding profile that clade C cleavage-defective gp140 displays (12), allowed for improved homogeneity crucial for data collection and averaging.
The discrepancy of a density-void cavity below the outermost surface of the trimer at the central threefold axis described by tomography density maps (9–11) is a noteworthy difference between the tomograms and the current work. Our density maps suggest that the upward swing of each gp120 subunit (Fig. 2 A and C) in the unliganded state might have contributed to the depression feature at the threefold axis, the concave nature of which may have been represented as an apparent cavity in the published tomography structures of lower resolution, where a density cap over the cavity results in a convex trimer apex. The tilt-induced depression would be raised in the liganded state, resulting in the aforementioned tilt away from the z-axis. Thus, the cavity may be an indication of a depression at the threefold axis, and the density over the cavity may well be a limitation of spatial resolution. However, having observed such a clear depression at the threefold axis, our density map concurs with the “cavity” observation, albeit without an overlapping cap at the surface. Another discrepancy of gp41 density is described between the Zanetti model and the Zhu model for having a thin, rod-like stalk and a tripod-like, broad gp41 conformation, respectively, at the threefold axis (9, 10). The lack of a rod-like thin stalk at the gp41 locus, as observed in the density map of unliganded gp140 (Fig. 2 A and C) and in the Zhu model (10), suggests extensive contacts between gp120 and gp41 at the interface. The gp140 construct contains the membrane-proximal external region, which likely contributes to the density protruding from the underside, viral membrane-proximal density of the trimer structure. The decreased gp120/gp41 interaction (Fig. 1C) elicited by ligand binding may serve as intersubunit communication required to elicit conformational changes in gp41. Exposure of the gp41 N-helix has been demonstrated to require CD4 binding (30). In line with recently published gp120 coordinates describing the gp41-associated seven-strand β-sandwich (31), the region at the gp120/gp41 interface comprising part of the gp120 inner domain may serve not only as an anchor for the three gp120 subunits to the threefold, but also as a trigger to sterically prevent premature association of the N- and C-helices of gp41. Our docking orientation of gp120, in which both of the termini are oriented toward the threefold axis, is in good agreement with the structure from Pancera et al. (31), as well as with earlier studies implicating N- and C-termini substitution mutants with abrogated gp120/gp41 association (32). However, there remains a need to further verify whether the density shift observed in this work can be appropriated to a tertiary inner domain shift or to a quaternary gp120 translation or rotation resulting in a weakened gp120–gp41 interface. Additionally, alterations (33) and truncations (34, 35) of the gp41 C-terminal cytoplasmic domain (CD) region result in CD4-independent membrane fusion, implicating likely cross-membrane allosteric cooperativity between gp120 and the gp41 CD. The altered gp140 was a chimeric construct derived from BaL gp120 elements and HXB2 gp41 elements, including the HXB2 gp41 CD (33). The mutated and truncated constructs included point mutations of transmembrane residues and truncations of cytoplasmic domains, resulting in increased fusion efficiency and exposure of CD4-induced epitopes (34, 35). Such constructs display an altered immunogenic profile than native Env, suggesting a pretriggered gp120 conformation exposing the coreceptor binding site. The similarity between binding profiles of native Env and gp140 to sCD4 and neutralizing antibodies (12), however, suggests that the native conformation of Env is retained in the gp140 construct, and that the initial point of contact of CD4 is very similar in both gp140 and native Env. Further downstream events including membrane fusion are observed to be impacted by the altered and truncated constructs. Furthermore, recent cryo-EM work based on detergent-solubilized trimers extracted from virions (36) portrays a cage-like morphology of Env with an accentuated cavity and a flat membrane proximal region, suggesting both minimal gp120–gp41 interactions and likely dissociated gp41 N-helices. Biochemical characterization of the extracted Env complex, as opposed to comparison with membrane-embedded Env in virus-like particles, would help to address whether a native quaternary arrangement is retained after extraction and solubilization, as done with the gp140 construct (12). The minimal gp120–gp41 contacts suggested by Wu’s density map would not agree with the abundant biochemical and mutagenesis data asserting close gp120–gp41 contacts (37–41).
The gp140 trimeric immunogens characterized in this work, along with the molecular models of conformational rearrangement suggested by them, will provide critical features of the designated gp140 immunogen. The three discrete events observed in the transition between the native, unliganded state and the liganded state, namely an outward density shift (Fig. 5A), a gp120 tilt away from the z-axis (Fig. 5B), and a gp120 rotation along the intramolecular axis (Fig. 5C), outline the major quaternary conformational shifts that derive from the gp120 and gp41 tertiary conformational alterations (Movie S1). Further work to quantitatively validate the quaternary epitope locations presented in this work will serve to corroborate our experimental observations. Analysis of additional deletion variants and full-length constructs will yield clues as to the quaternary impact of important gp120 regions such as the V2 loop, given the propensity of the partial V2 truncation in the construct utilized in this work to elicit an enhanced immune response. The quaternary structural arrangement of gp140 in a metastable state and the putative locations of its exposed epitopes contribute to the characterization of subsequent events occurring prior to membrane fusion, and utilization of gp140 as an immunogen in the unliganded and liganded forms with knowledge of epitope location and exposure could prove useful in elicitation of antibodies against both conserved epitopes, as opposed to the use of monomeric gp120 or of adenovirus-based cell-mediated immunity, thus providing a feasible direction for rational immunogen design (12, 42–44).
Fig. 5.
Model of quaternary changes induced by CD4 binding. Top panels show unliganded gp140, and bottom panels display CD4m-liganded gp140. (A) Density shift away from threefold axis results in weakened gp120/gp41 interface and closer association between gp120 inner and outer domain. (B) Tilt away from z-axis results in flatter trimer and exposed threefold axis, likely contributing to gp41 fusion peptide exposure and insertion into host membrane. (C) Subunit rotation along designated subunit axes (S1, S2, and S3, as previously described, protruding out of the plane of the page) further weakens gp120/gp41 interface, likely freeing up gp41 N-helix for association with the C-helix.
Materials and Methods
Sample Preparation and Cryoelectron Microscopy.
Samples of subtype C o-gp140ΔV2TV1 (hereinafter gp140) were biochemically characterized, purified, and imaged as previously described (6, 12, 45, 46) (see also Figs. S3, S4, and S5).
Image Processing and Docking of Subunit Tertiary Structures.
Deconvolution of contrast transfer function was done similarly as previously described (6, 45), where these particles were centered by autocorrelation, bandpass-filtered, and subjected to multivariate statistical analysis, namely correspondence analysis and hierarchical clustering, and submitted for refinement using EMAN (46). The density maps were validated by the agreement between the calculated reprojections and the class averages of micrographic images at each angular step (Fig. 1 A and D). Density maps were fitted to previously published X-ray coordinates of either unliganded SIV gp120 (7) or sCD4-bound HIV-1 gp120 (8) as previously described (6). To address the lack of unliganded gp41 coordinates, a cylinder with a radius of 17 Å and height of 48 Å, equivalent to the six-helix bundle, was placed at the threefold axis, thus conveying the maximum space that gp41 could ostensibly occupy.
Particle Distribution Across Euler Angles.
The distribution of the particles across Euler angles shows particles to have certain preferred orientations, as distinguished by inspection of class averages juxtaposed to projections of the trimer volume (Fig. 1 A and D). There are some Euler angle ranges that are not as well-represented by the data, particularly at values representative of trimer side views. Undersampling of side views is likely the result of side views being mischaracterized as monomeric breakdown products and being screened from the particle selection process. Whereas side view undersampling was likely a factor in both unliganded and liganded trimers, it was more accentuated in the unliganded reconstruction.
Supplementary Material
Acknowledgments.
We thank Fredrick Gibson for reviewing the manuscript, aiding in image processing and fruitful discussion. This work was sponsored by the National Institutes of Health (NIH) Human Immunodeficiency Virus Research and Development Grant 5 P01 AI48225-03 and Discovery program. C.G.M. was partially supported by an NIH Initiative for Maximizing Student Development Fellowship.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the Electron Microscopy Data Bank [accession nos. EMD-5263 (unliganded gp140) and EMD-5264 (CD4m-ligated gp140)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016113108/-/DCSupplemental.
References
- 1.Earl PL, Doms RW, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss CD, Levy JA, White JM. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J Virol. 1990;64:5674–5677. doi: 10.1128/jvi.64.11.5674-5677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinter A, et al. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng RH, et al. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: Three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Ref. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing L, et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 8.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanetti G, et al. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, et al. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava IK, et al. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2008;372:273–290. doi: 10.1016/j.virol.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyatt R, et al. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan N, et al. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson WE. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J Virol. 2002;76:2075–2086. doi: 10.1128/jvi.76.5.2075-2086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava IK. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol. 2003;77:11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian Y, et al. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J Virol. 2005;79:13338–13349. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Herrewege Y, et al. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J Antimicrob Chemother. 2008;61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- 20.Stricher F, et al. Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein. J Mol Biol. 2008;382:510–524. doi: 10.1016/j.jmb.2008.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore JP, et al. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 22.Sattentau QJ, Moore JP. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong PD, et al. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyauchi K, et al. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma VA, et al. Structural characteristics correlate with immune responses induced by HIV envelope glycoprotein vaccines. Virology. 2006;352:131–144. doi: 10.1016/j.virol.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 26.Kimura T, et al. Human monoclonal antibody 2909 binds to pseudovirions expressing trimers but not monomeric HIV-1 envelope proteins. Hum Antibodies. 2009;18:35–40. doi: 10.3233/HAB-2009-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Center RJ, et al. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J Virol. 2000;74:4448–4455. doi: 10.1128/jvi.74.10.4448-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dey AK, et al. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology. 2009;385:275–281. doi: 10.1016/j.virol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beddows S, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Furuta RA. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 31.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2009;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helseth E, et al. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BM, et al. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J Virol. 2008;82:5460–5471. doi: 10.1128/JVI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonavia A, et al. A single amino acid change and truncated TM are sufficient for simian immunodeficiency virus to enter cells using CCR5 in a CD4-independent pathway. Virology. 2005;341:12–23. doi: 10.1016/j.virol.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyss S, et al. Regulation of human immunodeficiency virus type 1 envelope glycoprotein fusion by a membrane-interactive domain in the gp41 cytoplasmic tail. J Virol. 2005;79:12231–12241. doi: 10.1128/JVI.79.19.12231-12241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs A, et al. Alanine scanning mutants of the HIV gp41 loop. J Biol Chem. 2005;280:27284–27288. doi: 10.1074/jbc.M414411200. [DOI] [PubMed] [Google Scholar]
- 38.Rits-Volloch S, et al. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 2006;25:5026–5035. doi: 10.1038/sj.emboj.7601358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen J, Jacobs A, Caffrey M. Role of the HIV gp120 conserved domain 5 in processing and viral entry. Biochemistry. 2008;47:7788–7795. doi: 10.1021/bi800227z. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, et al. Role of the HIV gp120 conserved domain 1 in processing and viral entry. J Biol Chem. 2008;283(47):32644–32649. doi: 10.1074/jbc.M806099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.York J, Nunberg JH. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J Virol. 2004;78:4921–4926. doi: 10.1128/JVI.78.9.4921-4926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnett SW, et al. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001;75:5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 44.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawano MA, et al. Calcium bridge triggers capsid disassembly in the cell entry process of simian virus 40. J Biol Chem. 2009;284:34703–34712. doi: 10.1074/jbc.M109.015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.