Abstract
The mammalian liver regenerates upon tissue loss, which induces quiescent hepatocytes to enter the cell cycle and undergo limited replication under the control of multiple hormones, growth factors, and cytokines. Endocannabinoids acting via cannabinoid type 1 receptors (CB1R) promote neural progenitor cell proliferation, and in the liver they promote lipogenesis. These findings suggest the involvement of CB1R in the control of liver regeneration. Here we report that mice lacking CB1R globally or in hepatocytes only and wild-type mice treated with a CB1R antagonist have a delayed proliferative response to two-thirds partial hepatectomy (PHX). In wild-type mice, PHX leads to increased hepatic expression of CB1R and hyperactivation of the biosynthesis of the endocannabinoid anandamide in the liver via an in vivo pathway involving conjugation of arachidonic acid and ethanolamine by fatty-acid amide hydrolase. In wild-type but not CB1R−/− mice, PHX induces robust up-regulation of key cell-cycle proteins involved in mitotic progression, including cyclin-dependent kinase 1 (Cdk1), cyclin B2, and their transcriptional regulator forkhead box protein M1 (FoxM1), as revealed by ultrahigh-throughput RNA sequencing and pathway analysis and confirmed by real-time PCR and Western blot analyses. Treatment of wild-type mice with anandamide induces similar changes mediated via activation of the PI3K/Akt pathway. We conclude that activation of hepatic CB1R by newly synthesized anandamide promotes liver regeneration by controlling the expression of cell-cycle regulators that drive M phase progression.
The mammalian liver has a unique ability to regenerate fully after tissue loss or injury, thus ensuring that the critical functions of the liver are maintained. Liver regeneration is tightly regulated by multiple growth factors, cytokines, and hormones that induce quiescent hepatocytes to enter the cell cycle and undergo limited replication to restore liver mass (1, 2). Loss of liver tissue can occur in response to toxic chemicals, such as alcohol or CCl4, or viral infection, in which case the regenerative process is superimposed on an inflammatory response aimed at removing dead cells or the infectious agent. There is no inflammatory response during the regenerative response triggered by surgical removal of part of the liver, as in the case of liver tumors or liver transplantation. Two-thirds partial hepatectomy (PHX) is an experimental model for liver regeneration uncomplicated by an inflammatory response that frequently is used to gain insight into humoral factors that modulate hepatocyte proliferation. In mice, the peak proliferative response occurs 40 h after PHX.
Endocannabinoids, such as arachidonoyl ethanolamide (anandamide, AEA) and 2-arachidonoylglycerol (2-AG), are lipid mediators that promote neural progenitor cell proliferation via activation of cannabinoid type 1 receptors (CB1R) in the brain (3, 4). CB1R and endocannabinoids also are present in the liver (5) where they promote de novo lipogenesis (5–7), a process also activated during regeneration (8–10). The ability of endocannabinoids to promote cell proliferation and to stimulate hepatic lipogenesis suggests that they may contribute to the early stages of the regenerative response of the liver. In the present study, we tested this hypothesis by examining the effects of pharmacological and genetic manipulation of CB1R on the proliferative response to PHX in mice and on the PHX-induced changes in the hepatic gene-expression profile, using ultrahigh-throughput RNA sequencing followed by pathway analysis. The results indicate that PHX induces hyperactivation of the hepatic AEA/CB1R system that contributes to the proliferative response via induction of cell-cycle proteins that drive M-phase progression.
Results
Effect of Genetic or Pharmacological Ablation of CB1R on the Proliferative Response to PHX.
To investigate the effect of CB1R signaling on the regenerative response to PHX, we quantified proliferating hepatocytes by BrdU staining 40 h after PHX or sham operation in wild-type mice and in mice lacking CB1R globally (CB1R−/−) or in hepatocytes only (LCB1R−/−). After PHX, the high proportion of BrdU+ cells in the wild-type liver was reduced dramatically in CB1R−/− and LCB1R−/− livers as well as in livers of wild-type mice treated with the CB1R antagonist rimonabant, 10 mg·kg−1·d−1 for 3 d before sacrifice (Fig. S1). Accordingly, a small, but significant post-PHX increase in liver weight was evident at 40 h in wild-type but not in CB1R−/− mice; however, the percent recovery of liver weight by the sixth day was similar in the two strains, indicating catch-up growth in CB1R−/− mice (Fig. S2). This result suggests a prominent role for hepatic CB1R in the early proliferative response.
Up-Regulation of CB1R and Hyperactivation of AEA Synthesis in the Posthepatectomy Remnant Liver.
In wild-type mice, PHX up-regulated the hepatic endocannaboid/CB1R system, as indicated by a 7.5-fold increase in CB1R mRNA, a more modest increase in CB1R protein levels (Fig. 1A), and a dramatic increase in the tissue levels of AEA (Fig. 1B) but not 2-AG (2.6 ± 0.5 pmol/mg in 0-h samples vs. 1.3 ± 0.4 pmol/mg in 40 h samples; P > 0.1). The increase in hepatic AEA was short-lived, because levels were back to control levels by the sixth day posthepatectomy (Fig. S3). The unprecedented, >100-fold increase in hepatic AEA content, which exceeds brain levels by two orders of magnitude, led us to explore the underlying mechanism. The steady-state levels of AEA reflect the balance between its biosynthesis and degradation, the latter catalyzed by fatty-acid amide hydrolase (FAAH), yielding arachidonic acid and ethanolamine (11). To assess AEA biosynthetic activity, mice subjected to PHX were treated with the FAAH inhibitor URB597 (1 mg/kg) 1 h before sacrifice to prevent AEA degradation. Unexpectedly, URB597 blunted rather than potentiated the PHX-induced increase in hepatic AEA (Fig. 1C). PHX similarly failed to increase hepatic AEA in FAAH−/− mice (Fig. 1D). This result strongly suggests that in the post-PHX liver, AEA synthesis is catalyzed by FAAH operating in reverse, resulting in the conjugation of arachidonic acid and ethanolamine. Accordingly, trace amounts of [2H4]ethanolamine added to liver extracts were incorporated into [2H4]AEA in 40-h post-PHX tissue but not in 0-h samples from wild-type mice, nor in either 0-h or 40-h samples from FAAH−/− or CB1R−/− mice (Fig. 1E). The levels of another FAAH substrate, oleoylethanolamide (OEA), also increased in 40-h vs. 0- h liver samples (573 ± 187 vs. 134 ± 14 fmol/mg; n = 3), and this increase similarly was absent in FAAH−/− mice (40 h: 202 ± 45 fmol/mg vs. 0 h: 175 ± 14 fmol/mg; n = 3). The increase in the AEA synthase activity of FAAH in 40-h vs. 0-h samples was paralleled by a decrease in its amidase activity, as assayed by the degradation of [3H]AEA (Fig. 1F), presumably because of the inhibition by the end products.
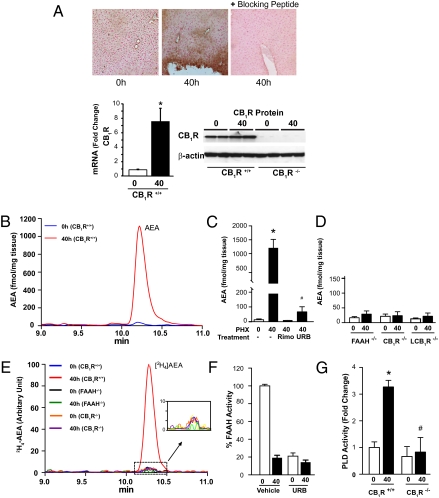
Fig. 1.
PHX up-regulates hepatic CB1R and hyperactivates hepatic AEA synthesis via the conjugation pathway. (A) Expression of CB1R in the liver is induced 40 h following PHX, as documented by immunohistochemistry (Upper), Western blotting (Lower Right), and real-time PCR (Lower Left). Specificity of CB1R immunostaining (brown color, Upper Center) is indicated by its elimination by a blocking peptide. *P < 0.05 relative to 0-h control value; n = 5. Note that for Western blots, CB1R first were immunoprecipitated using N-terminal antibody and then were gel-fractionated and blotted with C-terminal antibody, as described in Materials and Methods. (B) Hepatic AEA content is measured by LC-MS/MS. Representative ion chromatograms from the post-PHX remnant livers (40 h) and the resected livers (0 h) from the same mice are shown. (C) Hepatic levels of AEA in 0-h and 40-h post-PHX samples of untreated, rimonabant (Rimo)-pretreated (10 mg·kg−1·d−1 i.p. for 3 d) and URB597 (URB)-treated mice (1 mg/kg i.p. at 38 h post-PHX). Results shown are mean ± SE; n = 6 mice per group. *P < 0.005. In the same samples, 2-AG levels remained unchanged (0 h: 225 ± 42 fmol/mg vs. 40 h: 310 ± 85 fmol/mg). (D) PHX fails to induce AEA biosynthesis in FAAH−/−, CB1R−/−, and LCB1R−/− mice. (E) [2H4]ethanolamine is converted into [2H4]AEA in remnant but not in control liver from wild-type but not from CB1R−/− or FAAH−/− mice. Liver homogenates were incubated with 50 μM [2H4]ethanolamine at 37 °C for 30 min and then were extracted for LC-MS/MS analysis of [2H4]AEA. (F) The amidase activity of FAAH is lost in the post-PHX liver. FAAH activity was assayed as the release of [3H]ethanolamine from [3H]AEA, subject to inhibition by in vivo pretreatment with 1 mg/kg URB597. (G) PLD activity is induced by PHX in CB1R+/+ but not in CB1R−/− mouse liver. PLD activity in liver homogenates was measured as described in Materials and Methods. *P < 0.05 relative to 0-h, #P < 0.05 relative to 40-h value in wild-type mice.
Reversal of the reaction catalyzed by FAAH may occur only at high substrate concentrations (12). Indeed, PHX increased hepatic arachidonic acid from 121 ± 48 μM at 0 h to 798 ± 189 μM at 40 h (n = 5 P < 0.01), and a parallel increase in hepatic ethanolamine levels from 0.32 to 1.47 mM at 48 h post-PHX was reported earlier (13). To explore further which of the two reaction products inhibits the amidase activity of FAAH, the FAAH activity was measured in extracts of control livers supplemented with either ethanolamine or arachidonic acid. Although the addition of ethanolamine up to a final concentration of 1.2 mM did not affect FAAH activity, the addition of arachidonic acid caused concentration-dependent inhibition in the 10- to 100-μM range (Fig. S4). In contrast, the AEA synthase activity of FAAH was increased strongly by excess arachidonic acid (100 μM), as measured by the incorporation of [2H4]ethanolamine into AEA in extracts of normal mouse liver (Fig. S4).
The PHX-induced increase in hepatic AEA was blunted in CB1R−/− and LCB1R−/− mice (Fig. 1D) and in rimonabant-treated wild-type mice (Fig. 1C). This result suggests that AEA can induce its own biosynthesis via activation of CB1R, and this effect likely involves activation of phospholipase D (PLD), analogous to the CB1R-mediated increase in PLD activity in HEK293 cells stably transfected with CB1R and PLD2 (14). Indeed, hepatic PLD activity was increased threefold in the 40-h vs. 0-h samples from wild-type mice, whereas baseline PLD activity was lower and did not change after PHX in CB1R−/− mice (Fig. 1G), a finding that is compatible with CB1R-mediated activation of hepatic PLD in the regenerating liver.
The post-PHX increase in AEA apparently is limited to liver, because AEA levels remained unaltered by PHX in the heart (4.6 vs. 4.4 fmol/mg tissue), lungs (5.6 vs. 4.0 fmol/mg tissue), and spleen (4.3 vs. 5.2 fmol/mg tissue) at 0 vs. 40 h (P > 0.2 for all three tissues).
CB1R Induction of Cell-Cycle Proteins Involved in Mitotic Progression.
To identify downstream targets of CB1R in the regenerating liver, genome-wide hepatic gene expression was profiled in control and post-PHX liver from wild-type and CB1R−/− mice, using ultrahigh-throughput RNA sequencing and pathway analysis software. Arbitrary cutoffs were set at a greater than fivefold difference in gene expression between the 0-h and 40-h samples from wild-type mice, coupled with a less than fivefold difference for the corresponding values in CB1R−/− mice. A distinct set of cell-cycle proteins with a key role in mitotic progression displayed robust up-regulation in the post-PHX liver in wild-type but not in CB1R−/− mice (Fig. 2). These proteins include the transcription factor forkhead box protein M1 (FoxM1) and its targets cyclin B2 (Ccnb2) (15) and cyclin-dependent kinase 1 (CDK1), with additional proteins involved in DNA replication and centrosome organization, including kinesin-like 1 (Knsl1) (16), proliferating nuclear cell antigen (PCNA) (17), and Rad21 (18). In addition, the lipogenic transcription factor sterol regularity element-binding protein 1c (SREBP-1c) showed the same pattern of activation, in agreement with evidence that its hepatic expression is regulated by CB1R (5) and that lipogenesis is increased in the regenerating liver (8–10). These targets were validated at the mRNA and protein levels by real-time PCR and Western blotting, respectively (Figs. 3 and 4 A–C). Furthermore, acute in vivo treatment of mice with 10 mg/kg AEA resulted in rapid up-regulation of Foxm1, Ccnb2, Cdk1, Rad21, Pcna, and Knsl1 in the liver of wild-type, but not CB1R−/−, mice, as detected by real-time PCR (Fig. S5). Similar treatment with 2-AG caused a much smaller and delayed response (Fig. S6), perhaps in part because exogenous 2-AG is degraded much more rapidly than AEA in vivo (19).
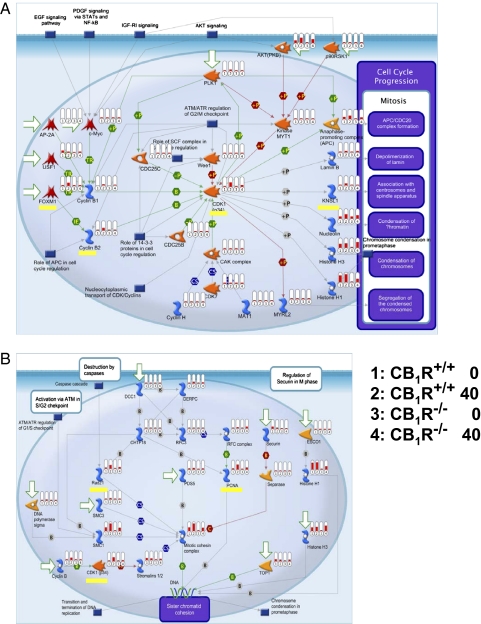
Fig. 2.
Global transcriptome analysis reveals CB1R-dependent, PHX-induced up-regulation of key cell-cycle proteins. Transcript levels of cell-cycle proteins involved in mitosis progression (A) or spindle separation (B) were determined using ultrahigh-throughput RNA sequencing and grouped on the basis of pathway analyses. Genes whose expression is increased by PHX in wild-type but not in CB1−/− mouse liver are highlighted in yellow and include Foxm1 and its targets Ccnb2, Cdk1, and Knsl1 (A) as well as Rad21 and Pcna (B). Relative mRNA levels are displayed as vertical red bars, with the four treatment groups juxtaposed in the indicated order for easy visual analysis.
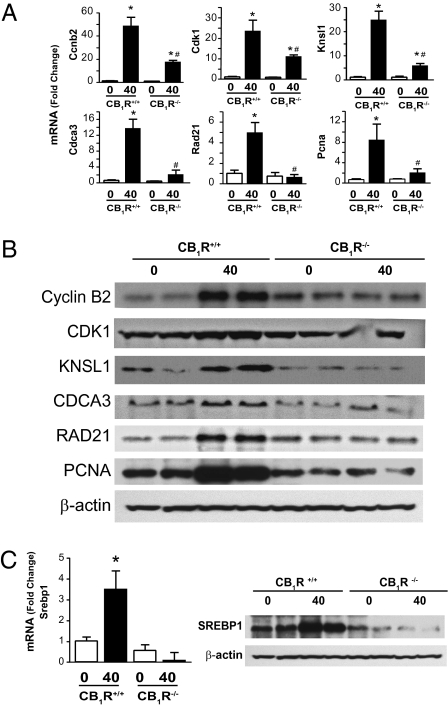
Fig. 3.
PHX-induced hepatic expression of cell-cycle proteins and the lipogenic transcription factor SREBP1c is blunted in CB1R−/− mice. (A) Cell-cycle protein mRNA quantified by real-time PCR. *P < 0.05 and **P < 0.01 from corresponding 0-h value; n = 4–6 mice per group. (B) Protein expression quantified by Western blotting. (C) SREBP1c mRNA and nuclear protein expression determined as above. *P < 0.05 from corresponding 0-h value.
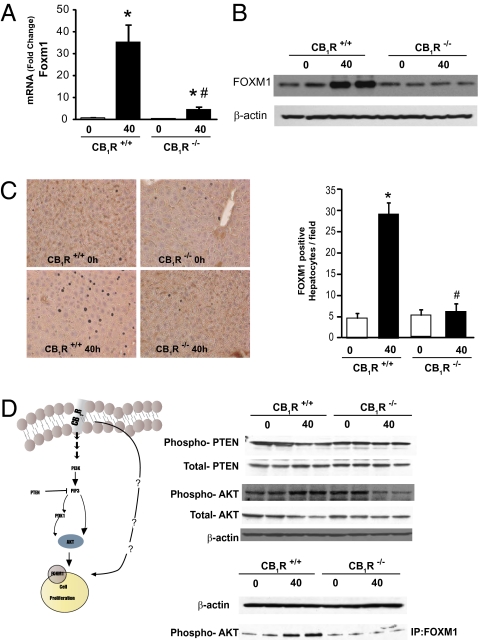
Fig. 4.
PHX-induced hepatic expression of FoxM1 is mediated by CB1R via the Akt/PTEN pathway. Induction of Foxm1 mRNA assessed by real-time PCR (A) and FoxM1 protein documented by Western blotting (B) and immunohistochemistry (C) is blunted in the liver of CB1R−/− mice. (D) PHX increases p-Akt, decreases phosphorylated PTEN (p-PTEN), and increases the association of p-Akt with FoxM1 in livers from wild-type but not from CB1R−/− mice. p-Akt/FoxM1 complexes (bottom lane) were identified by immunoprecipitation with a FoxM1 antibody followed by Western blotting with a p-Akt antibody.
CB1R Induces Foxm1 Expression via the PI3K/Akt Signaling Pathway.
CB1R couple to multiple signaling pathways, including the PI3K/Akt cell-survival pathway, which mediates its effect on neural progenitor cell proliferation (3, 4). Akt and its negative regulator, phosphatase and tensin homolog (PTEN), also are involved in regulating the expression of Foxm1 (20, 21). Accordingly, Akt phosphorylation was increased and PTEN phosphorylation was decreased in 40-h post-PHX vs. 0-h liver samples from wild-type mice, but no such changes were noted in corresponding tissue samples from CB1R−/− mice (Fig. 4D). The association of Akt with FoxM1 was demonstrated further by immunoprecipitating the complex using a FoxM1 antibody and then visualizing the gel-fractionated complex by an antibody against phosphorylated Akt (p-Akt) (Fig. 4D). This result indicates that the CB1R-mediated induction of Foxm1 occurs via the Akt/PTEN signaling pathway.
Discussion
This study demonstrates that AEA acting via hepatic CB1 receptors plays a key role in liver regeneration by controlling the expression of cell-cycle regulators that drive M-phase progression. It also provides evidence for hyperactivation of the biosynthesis of AEA in the regenerating liver via the enzymatic conjugation of arachidonic acid and ethanolamine, a pathway that previously had been thought to operate only under artificial, in vitro conditions.
The in vivo biosynthesis of AEA originally was proposed to occur via the conjugation of arachidonic acid and ethanolamine (22–24). However, the Ca2+-dependent biosynthesis of AEA from larger membrane phospholipid precursors now is widely considered the mechanism of AEA biosynthesis in vivo, via either the originally proposed transacylation/phosphodiesterase pathway (25) or alternative, parallel pathways (26, 27). As a result, the conjugation pathway has become viewed as an artifact of the high in vitro concentrations of arachidonic acid and ethanolamine used in the assay (12), which may occur only in postmortem tissue (28). The present findings present evidence for AEA biosynthesis via the conjugation pathway under well-defined in vivo conditions. At the exceptionally high, micromolar levels of AEA generated via the conjugation pathway, hepatic CB1R would be maximally occupied and activated in the regenerating liver, as also is suggested by the decrease observed in the early proliferative response with rimonabant treatment or in the CB1−/− liver.
It has been speculated that coincident activation of phospholipase A2 (PLA2) and PLD could trigger the conjugation pathway via the release of arachidonic acid and ethanolamine, respectively (23), thus reversing the direction of the enzymatic reaction because of the end-product inhibition of the amidase activity of FAAH. Indeed, we found that the addition of free arachidonic acid, but not ethanolamine, caused concentration-dependent inhibition of the amidase activity and a reciprocal dramatic increase in the AEA synthase activity of FAAH in extracts of normal liver (Fig. S4). The relative importance of arachidonic acid versus ethanolamine may be tissue dependent, because in extracts of peritoneal macrophages (29) or rat testis membranes (30), it was the addition of 0.1–1 mM ethanolamine that reportedly increased AEA synthesis by the conjugation pathway, whereas arachidonic acid mobilization by calcium-induced PLA2 activation had no such effect (29).
The transacylase/phosphodiesterase pathway is not specific for arachidonic acid-containing N-acylethanolamides (25), but the conjugation pathway is (23, 30), and this specificity may explain its greater capacity to generate AEA (>100-fold increase) than other acylethanolamides, such as OEA, levels of which were increased only fourfold.
The robust activation of proteins involved in cell-cycle progression during liver regeneration is expected and in some cases has been documented by targeting individual genes (31, 32) or using gene-array methodology (33). By sequencing of the entire transcriptome of liver tissue at a critical early phase of regeneration and by comparing transcriptional changes in wild-type and CB1R-deficient mice which have a delayed regenerative response, this study presents an unbiased genome-wide view of the transcriptome change during liver regeneration. By pathway analysis, the whole-genome sequencing implicated previously known networks involved in cell-cycle progression and also detected a key role for endocannabinoids in regulating the activity of those networks. The finding that the set of genes induced by PHX in the wild-type liver is not induced in the CB1R−/− liver strongly implicates AEA acting via CB1R as the mediator involved. This activity is supported further by the observation that acute in vivo treatment of mice with AEA induced the expression of the same genes in the liver of wild-type but not of CB1R−/− mice.
The FoxM1 transcription factor plays a central role in controlling the expression of a network of genes involved in mitotic progression (34) and also has been implicated in hepatocellular carcinoma (35). Mice with hepatocyte-specific deletion of Foxm1 have reduced DNA replication and complete inhibition of mitotic activity in hepatocytes after PHX (31) and also are highly resistant to chemically induced hepatocellular carcinoma (35). The finding that PHX or AEA up-regulates Foxm1 gene expression as well as the level of FoxM1 protein in wild-type but not CB1R−/− mice clearly indicates that endogenous AEA acting via CB1R plays a key role in this effect. This role is confirmed further by the ability of in vivo treatment with AEA to induce Foxm1 expression in the liver of wild-type but not CB1R−/− mice.
In CCl4-induced liver injury, increased CB2R activity in nonparenchymal liver cells and infiltrating leukocytes has been reported to reduce tissue damage and promote hepatocyte proliferation via a paracrine mechanism involving IL-6 and TNF-α (36). These effects probably are mediated by 2-AG, because CCl4 has been shown to cause a selective increase in hepatic 2-AG content (37). However, CB2R are unlikely to contribute to the PHX-induced changes in gene expression described here, as indicated by the absence of such changes in the liver of CB1R−/− mice (Figs. 2 and 3).
The present findings add endocannabinoids and CB1 receptors to the list of established promitogenic factors, including hepatocyte growth factor (HGF) and its receptor cMET, the epidermal growth factor receptor (EGFR), and the cytokine TNF-α, whose mitogenic action is mediated via the Akt-dependent activation of NFκB (2). The redundancy of hepatic mitogens is the most likely reason regeneration is delayed, but not prevented, in the absence of CB1. Whether endocannabinoids may interact with other hepatic mitogens is not yet clear, although there is reason to suspect that they do. In certain paradigms, AEA synthesis is triggered via NFκB and Akt activation (38), and in turn, CB1R can activate Akt (39) and transactivate the EGFR (40). Unlike HGF, whose levels are increased by PHX both in liver and in distant organs, suggesting the involvement of a hormonal factor in its induction (41), the PHX-induced increase in AEA is limited to the remnant liver, compatible with its action as an autocrine or paracrine-acting mitogen.
The present findings reveal a prominent role for endogenous AEA acting via CB1R in regulating the expression of key cell-cycle proteins required for mitotic progression in both regenerating liver and in liver carcinoma. Increased activity of the hepatic endocannabinoid/CB1R system also has been implicated in the pathology of liver cirrhosis, where it contributes to increased fibrogenesis (42) and hemodynamic abnormalities leading to ascites formation and hemorrhage (43). Indeed, reversal of these effects by CB1R blockade may have therapeutic value in cirrhosis (42, 43). Although CB1R blockade inhibits the early stages of regeneration, this effect is transient and would be more than offset by the benefit from suppressing the expression of cell-cycle proteins implicated in hepatocellular carcinoma, the incidence of which is known to be increased in cirrhotic individuals (44).
Materials and Methods
Animals.
All procedures were approved by the Institutional Animal Care and Use Committee and were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. C57BL/6J mice were from Jackson Laboratories. Male mice 10–12 wk of age were used in all experiments. CB1R−/− and CB1R+/+ littermates were obtained by breeding heterozygotes that had been backcrossed to a C57BL/6J background, as described (45). Mice with hepatocyte-specific knockout of CB1R (LCB1R−/− mice) were generated as described (46). FAAH−/− mice generated and maintained as described (47) had been backcrossed to a C57Bl6/J background.
Partial Hepatectomy.
Two-thirds PHX was performed as described (48). The removed liver tissue was snap-frozen in liquid nitrogen and used for analyses as 0-h control samples. The animals were killed by decapitation 40 h or 6 d after surgery, and the remnant liver was collected and snap-frozen in liquid nitrogen.
Endocannabinoid Measurements.
The tissue levels of endocannabinoids were measured by stable isotope dilution liquid chromatography/tandem mass spectrometry (LC-MS/MS), as detailed in SI Materials and Methods.
Enzymatic Formation of [2H4]AEA from [2H4]Ethanolamine.
Liver was homogenized in 10 mM Tris buffer, pH 7.6, containing 1 mM EDTA in a volume of 2.5 mL/g wet tissue weight. The homogenate was centrifuged at 1,000 × g for 5 min, and 200-μL aliquots of the supernatant containing 70 μg protein were incubated with 50 μM [2H4]ethanolamine (Sigma-Aldrich) for 30 min at 37 °C. The reaction was stopped by the addition of 2 mL of CHCl3/MeOH (2:1, vol/vol), and the mixture was extracted as described above. [2H4]AEA in the extracted sample was measured by LC-MS/MS.
FAAH Activity.
FAAH activity in liver homogenates was assayed by the release of [3H]ethanolamine from [3H]AEA labeled on the ethanolamine moiety, as described (49).
RNA Isolation.
Total RNA was isolated from liver using TRIzol reagents (Invitrogen) according to the manufacturer's instructions. The isolated RNA was treated with RNase-free DNase (Ambion) to remove any traces of genomic DNA contamination. RNA was purified further using the Qiagen RNA Clean-Up Kit.
RNA Sequencing by the SOLiD System.
Total RNA was isolated as described above from liver samples pooled from six CB1R+/+ and six CB1R−/− mice at the time of PHX (0 h) and at 40 h post-PHX. rRNA was depleted by two rounds of passage of 10 μg of total liver RNA through a RIBOMINUS concentration module (Ambion). rRNA-depleted RNA, 100 ng for each sample, was treated with RNase III to generate 100- to 200-nt fragments, hybridized, ligated with Adaptor Mix B, reverse-transcribed, size-selected, amplified, and purified using the Applied Biosystems SOLiD WTAK kit. Amplified libraries were ligated to beads using the Applied Biosystems SOLiD 3 System Templated Bead Preparation Guide. Approximately 6 × 107 templated beads for each sample were sequenced on the Applied Biosystems SOLiD Version 3.0 instrument using 50-nt chemistry.
Analyses of RNA Sequencing Data.
Short sequences of 50 bases from SOLiD v3.0 were aligned to the reference genome (UCSC mm9) using IMAP-0.2.5.1 (Applied Biosystems). The uniquely mapped reads were parsed with in-house Perl scripts to generate coverage of each nucleotide. The expression level of NCBI mouse refSeq genes (Build 37) was estimated by high50Ave, which is the average RNA sequence read coverage for the 50-bp window with the highest count and corresponding to a single exon. The raw data were log2 transformed and quantile normalized using the Limma package from Bioconductor (50). The total uniquely mapped reads were 5,948,676 (wild-type, 0 h); 6,354,410 (wild-type, 40 h); 2,357,632 (CB1R−/−, 0 h); and 6,966,732 (CB1R−/−, 40 h).
Pathway Analyses of RNA Sequencing Data.
Normalized values of high50Ave for each gene were analyzed using MetaCore (GeneGo) pathways software.
CB1R Immunoprecipitation.
For Western blotting, cell extracts were immunoprecipitated using a CB1R N-terminal antibody. The proteins were size-fractionated by gel electrophoresis and visualized using a C-terminal CB1R antibody, as described (51).
PLD Assay.
PLD activity in liver tissue was assayed using the Amplex Red Phospholipase D Assay Kit (Invitrogen). In this enzyme-coupled assay, PLD cleaves phosphatidylcholine to yield choline and phosphatidic acid. Choline then is oxidized by choline oxidase to betaine and H2O2. Finally, H2O2, in the presence of HRP reacts with Amplex Red reagent to generate the fluorescent product, resorufin, which is monitored at 571 and 585 nm.
Statistical Analyses.
Results are reported as mean ± SE. Statistical significance among groups was determined by one-way ANOVA followed by post hoc Newman–Keuls analysis using GraphPad Prism 4.3 software. Probability values of P < 0.05 were considered significant. Statistical significance between two groups was determined by the two-tailed unpaired Student t test.
For details of real-time PCR and Western blot analyses, coimmunoprecipitation of Akt with FoxM1, and immunohistochemistry, see SI Materials and Methods. Primers used for real-time PCR are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Ken Mackie for the CB1 antibodies and Beat Lutz and Giovanni Marsicano for the CB1 floxed mice used to generate LCB1−/− mice. This work was funded by the intramural program of the National Institute on Alcohol Abuse and Alcoholism.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017689108/-/DCSupplemental.
References
- 1.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2) Suppl 1:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos GK. Liver regeneration after partial hepatectomy: Critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina-Holgado E, et al. Cannabinoids promote oligodendrocyte progenitor survival: Involvement of cannabinoid receptors and phosphatidylinositol-3 kinase/Akt signaling. J Neurosci. 2002;22:9742–9753. doi: 10.1523/JNEUROSCI.22-22-09742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trazzi S, Steger M, Mitrugno VM, Bartesaghi R, Ciani E. CB1 cannabinoid receptors increase neuronal precursor proliferation through AKT/glycogen synthase kinase-3beta/beta-catenin signaling. J Biol Chem. 2010;285:10098–10109. doi: 10.1074/jbc.M109.043711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osei-Hyiaman D, et al. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J Clin Invest. 2005;115:1298–1305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osei-Hyiaman D, et al. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J Clin Invest. 2008;118:3160–3169. doi: 10.1172/JCI34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jourdan T, et al. CB1 antagonism exerts specific molecular effects on visceral and subcutaneous fat and reverses liver steatosis in diet-induced obese mice. Diabetes. 2010;59:926–934. doi: 10.2337/db09-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schofield PS, Sugden MC, Corstorphine CG, Zammit VA. Altered interactions between lipogenesis and fatty acid oxidation in regenerating rat liver. Biochem J. 1987;241:469–474. doi: 10.1042/bj2410469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gove CD, Hems DA. Fatty acid synthesis in the regenerating liver of the rat. Biochem J. 1978;170:1–8. doi: 10.1042/bj1700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tijburg LB, Nyathi CB, Meijer GW, Geelen MJ. Biosynthesis and secretion of triacylglycerol in rat liver after partial hepatectomy. Biochem J. 1991;277:723–728. doi: 10.1042/bj2770723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cravatt BF, et al. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 12.Arreaza G, et al. The cloned rat hydrolytic enzyme responsible for the breakdown of anandamide also catalyzes its formation via the condensation of arachidonic acid and ethanolamine. Neurosci Lett. 1997;234:59–62. doi: 10.1016/s0304-3940(97)00673-3. [DOI] [PubMed] [Google Scholar]
- 13.Houweling M, Tijburg LB, Vaartjes WJ, van Golde LM. Phosphatidylethanolamine metabolism in rat liver after partial hepatectomy. Control of biosynthesis of phosphatidylethanolamine by the availability of ethanolamine. Biochem J. 1992;283:55–61. doi: 10.1042/bj2830055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch T, Wu DF, Yang LQ, Brandenburg LO, Höllt V. Role of phospholipase D2 in the agonist-induced and constitutive endocytosis of G-protein coupled receptors. J Neurochem. 2006;97:365–372. doi: 10.1111/j.1471-4159.2006.03736.x. [DOI] [PubMed] [Google Scholar]
- 15.Brandeis M, et al. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc Natl Acad Sci USA. 1998;95:4344–4349. doi: 10.1073/pnas.95.8.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irelan JT, et al. A role for IkappaB kinase 2 in bipolar spindle assembly. Proc Natl Acad Sci USA. 2007;104:16940–16945. doi: 10.1073/pnas.0706493104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acharya N, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beauchene NA, et al. Rad21 is required for centrosome integrity in human cells independently of its role in chromosome cohesion. Cell Cycle. 2010;9:1774–1780. doi: 10.4161/cc.9.9.11524. [DOI] [PubMed] [Google Scholar]
- 19.Járai Z, et al. Cardiovascular effects of 2-arachidonoyl glycerol in anesthetized mice. Hypertension. 2000;35:679–684. doi: 10.1161/01.hyp.35.2.679. [DOI] [PubMed] [Google Scholar]
- 20.Dai B, et al. FoxM1B regulates NEDD4-1 expression, leading to cellular transformation and full malignant phenotype in immortalized human astrocytes. Cancer Res. 2010;70:2951–2961. doi: 10.1158/0008-5472.CAN-09-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, et al. EPS8 upregulates FOXM1 expression, enhancing cell growth and motility. Carcinogenesis. 2010;31:1132–1141. doi: 10.1093/carcin/bgq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deutsch DG, Chin SA. Enzymatic synthesis and degradation of anandamide, a cannabinoid receptor agonist. Biochem Pharmacol. 1993;46:791–796. doi: 10.1016/0006-2952(93)90486-g. [DOI] [PubMed] [Google Scholar]
- 23.Devane WA, Axelrod J. Enzymatic synthesis of anandamide, an endogenous ligand for the cannabinoid receptor, by brain membranes. Proc Natl Acad Sci USA. 1994;91:6698–6701. doi: 10.1073/pnas.91.14.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruszka KK, Gross RW. The ATP- and CoA-independent synthesis of arachidonoylethanolamide. A novel mechanism underlying the synthesis of the endogenous ligand of the cannabinoid receptor. J Biol Chem. 1994;269:14345–14348. [PubMed] [Google Scholar]
- 25.Di Marzo V, et al. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, et al. A biosynthetic pathway for anandamide. Proc Natl Acad Sci USA. 2006;103:13345–13350. doi: 10.1073/pnas.0601832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon GM, Cravatt BF. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- 28.Patel S, et al. The postmortal accumulation of brain N-arachidonylethanolamine (anandamide) is dependent upon fatty acid amide hydrolase activity. J Lipid Res. 2005;46:342–349. doi: 10.1194/jlr.M400377-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Kuwae T, Shiota Y, Schmid PC, Krebsbach R, Schmid HH. Biosynthesis and turnover of anandamide and other N-acylethanolamines in peritoneal macrophages. FEBS Lett. 1999;459:123–127. doi: 10.1016/s0014-5793(99)01226-0. [DOI] [PubMed] [Google Scholar]
- 30.Schmid PC, Schwindenhammer D, Krebsbach RJ, Schmid HH. Alternative pathways of anandamide biosynthesis in rat testes. Chem Phys Lipids. 1998;92:27–35. doi: 10.1016/s0009-3084(97)00109-6. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trembley JH, Ebbert JO, Kren BT, Steer CJ. Differential regulation of cyclin B1 RNA and protein expression during hepatocyte growth in vivo. Cell Growth Differ. 1996;7:903–916. [PubMed] [Google Scholar]
- 33.Li J, et al. Relationships between deficits in tissue mass and transcriptional programs after partial hepatectomy in mice. Am J Pathol. 2009;175:947–957. doi: 10.2353/ajpath.2009.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang IC, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalinichenko VV, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira-Clerc F, et al. Beneficial paracrine effects of cannabinoid receptor 2 on liver injury and regeneration. Hepatology. 2010;52:1046–1059. doi: 10.1002/hep.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegmund SV, et al. The endocannabinoid 2-arachidonoyl glycerol induces death of hepatic stellate cells via mitochondrial reactive oxygen species. FASEB J. 2007;21:2798–2806. doi: 10.1096/fj.06-7717com. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, et al. Lipopolysaccharide induces anandamide synthesis in macrophages via CD14/MAPK/phosphoinositide 3-kinase/NF-kappaB independently of platelet-activating factor. J Biol Chem. 2003;278:45034–45039. doi: 10.1074/jbc.M306062200. [DOI] [PubMed] [Google Scholar]
- 39.Galve-Roperh I, Rueda D, Gómez del Pulgar T, Velasco G, Guzmán M. Mechanism of extracellular signal-regulated kinase activation by the CB(1) cannabinoid receptor. Mol Pharmacol. 2002;62:1385–1392. doi: 10.1124/mol.62.6.1385. [DOI] [PubMed] [Google Scholar]
- 40.Yang H, et al. Epidermal growth factor receptor transactivation by the cannabinoid receptor (CB1) and transient receptor potential vanilloid 1 (TRPV1) induces differential responses in corneal epithelial cells. Exp Eye Res. 2010;91:462–471. doi: 10.1016/j.exer.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kono S, Nagaike M, Matsumoto K, Nakamura T. Marked induction of hepatocyte growth factor mRNA in intact kidney and spleen in response to injury of distant organs. Biochem Biophys Res Commun. 1992;186:991–998. doi: 10.1016/0006-291x(92)90844-b. [DOI] [PubMed] [Google Scholar]
- 42.Teixeira-Clerc F, et al. CB1 cannabinoid receptor antagonism: A new strategy for the treatment of liver fibrosis. Nat Med. 2006;12:671–676. doi: 10.1038/nm1421. [DOI] [PubMed] [Google Scholar]
- 43.Bátkai S, et al. Endocannabinoids acting at vascular CB1 receptors mediate the vasodilated state in advanced liver cirrhosis. Nat Med. 2001;7:827–832. doi: 10.1038/89953. [DOI] [PubMed] [Google Scholar]
- 44.Anthony PP. Hepatocellular carcinoma: An overview. Histopathology. 2001;39:109–118. doi: 10.1046/j.1365-2559.2001.01188.x. [DOI] [PubMed] [Google Scholar]
- 45.Zimmer A, Zimmer AM, Hohmann AG, Herkenham M, Bonner TI. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc Natl Acad Sci USA. 1999;96:5780–5785. doi: 10.1073/pnas.96.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong WI, et al. Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008;7:227–235. doi: 10.1016/j.cmet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Cravatt BF, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 49.Fowler CJ, Tiger G, Stenström A. Ibuprofen inhibits rat brain deamidation of anandamide at pharmacologically relevant concentrations. Mode of inhibition and structure-activity relationship. J Pharmacol Exp Ther. 1997;283:729–734. [PubMed] [Google Scholar]
- 50.Smyth GK, editor. Limma: Linear Models for Microarray Data. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 51.Mukhopadhyay B, et al. Transcriptional regulation of cannabinoid receptor-1 expression in the liver by retinoic acid acting via retinoic acid receptor-gamma. J Biol Chem. 2010;285:19002–19011. doi: 10.1074/jbc.M109.068460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.