Abstract
It is challenging to monitor the health of transplanted organs, particularly with respect to rejection by the host immune system. Because transplanted organs have genomes that are distinct from the recipient's genome, we used high throughput shotgun sequencing to develop a universal noninvasive approach to monitoring organ health. We analyzed cell-free DNA circulating in the blood of heart transplant recipients and observed significantly increased levels of cell-free DNA from the donor genome at times when an endomyocardial biopsy independently established the presence of acute cellular rejection in these heart transplant recipients. Our results demonstrate that cell-free DNA can be used to detect an organ-specific signature that correlates with rejection, and this measurement can be made on any combination of donor and recipient. This noninvasive test holds promise for replacing the endomyocardial biopsy in heart transplant recipients and may be applicable to other solid organ transplants.
Keywords: next-generation sequencing, noninvasive diagnosis, acute rejection
The surveillance of organ health, particularly to detect the onset of transplant rejection, is essential for the long-term survival of organ transplant recipients. For heart transplant recipients, the gold standard for diagnosis of rejection is the endomyocardial biopsy. However, the endomyocardial biopsy is an expensive and invasive procedure that is limited by sampling error and interobserver variability in grading. Furthermore, cardiac biopsies may cause patient discomfort and rare but serious complications, including arterial puncture, arrhythmias, conduction abnormalities, biopsy-induced tricuspid regurgitation, and even cardiac perforation (1–5).
There has been considerable effort to develop noninvasive techniques that might replace or reduce the need for endomyocardial biopsies, with much focus placed on monitoring the recipient's immune response to detect the onset of rejection. The expression profile of certain genes in peripheral blood mononuclear cells (PBMCs), assayed from patient blood samples, has been demonstrated to differ between quiescent patients and those with severe rejection episodes (6–8). The AlloMap molecular expression test (XDx) is the first FDA-approved test based on this research (9, 10). This test has a low positive predictive value; however, its use in conjunction with clinical observation and echocardiograms has been shown to safely reduce the number of biopsies performed without increasing risk of serious cardiovascular events (9).
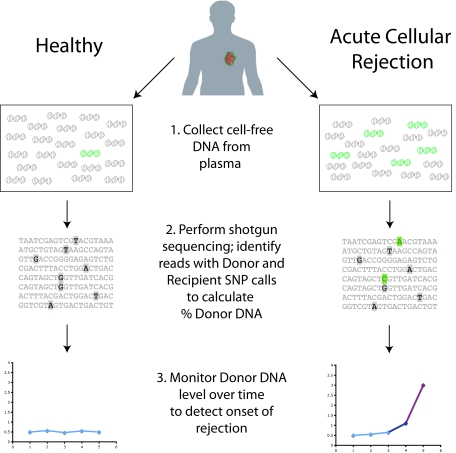
Instead of monitoring the recipient's immune response, we have developed an assay that directly interrogates the health of the donated organ. This technique involves measuring the signature of dying cells from the organ in the cell-free DNA circulating in the recipient's plasma (11). If a unique genomic signature of DNA from the donated organ (compared with the recipient's genome) can be identified, then the level of “donor DNA” from the transplanted organ can be monitored over time, and changes in organ health can be detected as changes in the donor DNA level. (Fig. 1) The rationale for this approach arises from the observation that both acute and chronic rejection processes are associated with apoptosis of specific cell types within the allograft (12, 13). Past research has attempted to identify cell-free DNA in sex-mismatched female recipients of male donor organs, where chromosome Y can serve as the donor genetic signature. This line of research, however, has yielded conflicting results on the existence of a donor-specific signature in the plasma of organ transplant recipients (14, 15). The clearest evidence has come from renal transplantation, where donor-specific chromosome Y has been detected in recipient urine and plasma (16–19). To date, most measurements of cell-free DNA in organ transplantation have been limited to the special case of women who receive male organs, which has prevented the widespread use of cell-free DNA as a diagnostic tool, because female recipients of male donor organs represent less than a quarter of all transplant procedures. HLA markers can be quantified to identify donor-derived DNA in pancreas–kidney transplant recipients (20), but the precision is low, making its utility to measure rejection unclear, and it is not applicable to cases when the donor and recipient are HLA matched.
Fig. 1.
General scheme for this study. Cell-free DNA collected in plasma contains a majority of molecules from the recipient (in gray) but may also include some from the transplanted organ (green). Due to increased cell death in the organ during a rejection episode, more donor molecules are expected to be present in the blood at these times. Shotgun sequencing of the purified DNA allows for counting recipient versus donor molecules by looking at single nucleotide polymorphisms (SNPs) that vary between donor and recipient. Very high levels of donor DNA, particularly changes from past measurements, will indicate the onset of rejection.
Here, we show that organ-specific donor DNA is detectable in the plasma of heart transplant recipients and that this genetic signature increases substantially before rejection events. We also demonstrate a universal, sex-independent strategy using shotgun sequencing to measure single nucleotide polymorphism (SNP) differences between individuals to quantify the donor DNA signal. This genome transplant dynamics (GTD) approach is applicable to any organ donor and any recipient, regardless of sex, by first genotyping the donor and recipient to establish a unique donor “genetic fingerprint,” which can be detected by high throughput sequencing of cell-free DNA in the recipient's blood following transplantation. The GTD assay provides a quantitative measure of organ health that can complement or possibly replace other approaches for posttransplant monitoring.
Results
Chromosome Y Detection in Sex-Mismatched Transplant Recipients.
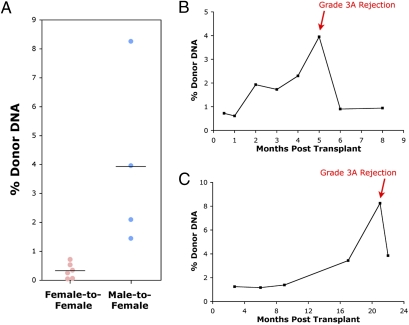
Because previous research has reported conflicting results on the possibility of detecting genetic signatures from transplanted organs using conventional PCR (14, 15), we first explored the use of the more sensitive technique of microfluidic digital PCR (21, 22) in sex-mismatched transplants where a female recipient has received a male donor heart. We purified DNA from the plasma of nine patients collected immediately before an endomyocardial biopsy that established a cellular rejection episode (≥grade 3A/2R). Six patients (identified as patients 1–6) were females who had received a heart from a female donor and three patients (identified as patients 7–9) were females who had received a heart from a male donor. Digital PCR was performed on the cell-free DNA using probes for chromosome (Chr) 1 and chromosome Y to establish the % Chr Y signal relative to Chr 1 (Fig. 2A). For the six female patients receiving organs from female donors, either no or a very low level of Chr Y was observed 0.32 ± 0.27% (SD). However, for the three female patients who received male organs, in four unique plasma samples (one patient had two documented rejection events), a greater than tenfold higher average level of Chr Y was observed, 3.93 ± 3.07% (SD), with a range from 1.4 to 8.2%. The signal from the male-donor samples is well separated from that of the female-donor controls (P = 0.018, Student's t test). These data establish that genetic material from the transplanted organ can be detected in the recipient's plasma during rejection episodes.
Fig. 2.
Donor DNA levels determined by digital PCR using a chromosome Y marker. (A) Ten plasma samples from patients with biopsy-determined rejection events with grade ≥3A–2R were analyzed, including six female patients (1–6) receiving female organs (six total events) and three female patients (7–9) receiving male organs (four total events). Group averages are marked by black lines. (B and C) Time-course graphs of chromosome Y levels (% donor DNA) in patients 7 (B) and 8 (C). Both patients had grade 3A–2R rejections as determined by biopsy at the indicated time point.
We then expanded our analysis to a larger set of 39 archived samples from female transplant patients who had received male hearts, some of whom had rejection events and some of whom did not. Each patient had been sampled at multiple time points and an endomyocardial biopsy was performed after each blood draw. Example time series are shown in Fig. 2 and Fig. S1; one can clearly see that donor DNA levels from the heart are increased at rejection events. Using all samples from all time points, we plotted a receiver operating curve (ROC) on the basis of different thresholds for the donor DNA level, with the endomyocardial biopsy results (grades ≥3A–2R) used as the indicator of true positive rejection events (Fig. S2). At a threshold of 2.0% donor DNA, we capture an 80% true positive rate with a 15% false positive rate. Our data suggest that values of donor DNA less than 1% and typically around 0.5% appear “normal” for heart transplant recipients; higher values are likely indicative of organ damage similar to that directly observed by endomyocardial biopsy showing myocyte damage that characterizes rejection grades ≥3A–2R, with a significant difference between the donor DNA levels in the five true positives and the other 34 samples analyzed (P = 0.0002, Student's t test).
For patient 10, there is a high level of chromosome Y at the first time point, 2 wk after transplantation. Examination of the clinical record revealed the presence of antibody-mediated rejection, due to presensitization before transplantation with high levels of circulating HLA antibodies directed against donor antigens. Following aggressive treatment for antibody-mediated rejection, the signal for this patient stabilized around 0.60 ± 0.41% (SD) for the other six plasma samples collected in the first year. This first data point is therefore treated as a true positive in our analysis. For patient 13, a high chromosome Y signal is seen both immediately following transplantation and at 8 mo after transplantation. These spikes in signal are not connected to any documented events in this patient's medical history, and in the absence of such records we treat them as false positives. For other patients without documented rejection events, there is no significant chromosome Y signal above 0.50% at any monitored time point (Fig. S1).
Samples after rejection episodes were also analyzed, where possible from the archived plasma record, to determine whether donor DNA levels decreased following treatment. For patient 7, samples both 1 and 3 mo following rejection were analyzed and revealed that the donor DNA levels had fallen significantly, to ∼0.9%. For patient 8, only one sample, 1 mo after treatment began, was available and it also showed a reduction in donor DNA from its peak at the time of rejection, albeit to a value that is still higher than what was seen before the rejection event. Details on each patient's medical status at the time of rejection and following treatment are given in Table S1.
Sequencing-Based Donor DNA Quantitation.
Although the above results demonstrate that rejection can be detected using chromosome Y as a genetic signature for the donor organ's DNA in the plasma, this type of assay can only be used in the minority of cases where a female recipient receives a male donor organ. We sought to demonstrate that using more detailed knowledge of the genomes of the donor and recipient—such as a large number of single nucleotide polymorphisms—could be used to monitor the genome transplant dynamics between arbitrary pairings of donor and recipient.
A frequent estimate for the variation between individuals is that approximately one base per thousand differs, for about 3 million total SNPs (23). However, not all of these sites will be useful for discriminating recipient and donor molecules in the plasma. Due to the overwhelming number of expected recipient DNA molecules in the plasma, the only usable locations are those where the recipient has a homozygous SNP with a single base present in both alleles. This leaves about 1.6 million positions to query by sequencing, of which about a quarter will be homozygous for a different allele in the donor's genome and the rest heterozygous with just one of the two bases differing.
For each usable SNP, we can identify the “recipient” base (i.e., A, from an AA SNP), the single base present in both alleles of that SNP in the recipient's genome. The donor base (i.e., T, from an AT or TT SNP) is the new base present in either one (heterozygous) or both (homozygous) alleles of the donor's genome that is not present in the recipient's genome. Any other possible base calls at the site (i.e., C or G) would be considered an “error” from PCR amplification and/or sequencing. Whenever a shotgun-sequencing read aligns over a site containing an identified usable SNP, we assign the read to one of three bins, depending on whether it shows the recipient base, the donor base, or something else at the SNP site. Using the total count of recipient and donor bases, we then calculate the ratio of donor DNA present. Although we may not observe reads from any particular SNP, by having hundreds of thousands of potential loci to discriminate donor and recipient molecules, there should be enough reads to establish an accurate donor DNA percentage. A diagram of the workflow used to assign SNPs into groups and to assign DNA sequences to donor or recipient is given in Fig. S3.
As proof of principle, we took genetic material from two HapMap cell lines (NA07348 and NA10830) that have been heavily genotyped, and treated one cell line as recipient and the other as donor (23). Approximately 660,000 usable homozygous recipient SNPs are characterized in these cell lines, with 160,000 of these being homozygous donor SNPs and the rest heterozygous donor SNPs. We mixed genomic DNA from NA10830, the mock “donor,” with DNA from NA07348, the mock recipient, at ratios between 1.5 and 7.5%, and prepared libraries for sequencing. A control library with just the recipient DNA—“0% donor”—was also created. Each library was sequenced in a single lane on an Illumina GAII, yielding between 3.6–10.8 million unique aligned sequences of which 30,000–100,000 contained SNP locations (Table 1). Recipient, donor, and error calls were counted from the sequenced bases. Whereas the majority of calls were from the recipient genome, as expected, an increasing number of donor calls was made as the proportion of donor DNA in the library increased. Illumina's quality scores were used to remove a majority of the sequencing errors, and the number of recipient and donor calls was much larger than the overall sequencing error rate.
Table 1.
Sequencing statistics for the control HapMap genomic libraries
| % donor | 0.0 | 1.5 | 2.0 | 2.5 | 4.0 | 5.0 | 7.5 |
| Total reads | 13,082,100 | 6,321,400 | 8,707,200 | 14,116,000 | 9,169,000 | 12,510,700 | 19,183,300 |
| Aligned | 9,019,118 | 4,581,260 | 6,629,726 | 10,885,018 | 6,278,041 | 7,552,199 | 14,145,769 |
| Unique | 8,747,074 | 3,651,817 | 5,145,156 | 7,076,860 | 5,077,904 | 5,003,458 | 10,824,332 |
| Total reads with SNPs | 77,201 | 30,421 | 46,892 | 63,884 | 45,708 | 39,047 | 98,384 |
| Heterozygous SNPs | |||||||
| Total reads | 58,047 | 22,812 | 35,410 | 48,293 | 34,541 | 29,348 | 74,099 |
| Recipient reads | 57,852 | 22,549 | 34,841 | 47,329 | 33,587 | 28,310 | 70,268 |
| Donor reads | 138 | 244 | 533 | 925 | 928 | 1,009 | 3,803 |
| Errors | 57 | 19 | 36 | 39 | 26 | 29 | 28 |
| Homozygous SNPs | |||||||
| Total reads | 19,154 | 7,609 | 11,482 | 15,591 | 11,167 | 9,699 | 24,285 |
| Recipient reads | 19,088 | 7,465 | 11,149 | 15,044 | 10,544 | 8,978 | 21,882 |
| Donor reads | 59 | 137 | 327 | 539 | 610 | 706 | 2,394 |
| Errors | 7 | 7 | 6 | 8 | 13 | 15 | 9 |
Reads from either heterozygous or homozygous donor SNPs are separated from the other calls. Only SNP base calls with a quality score (QS) ≥80 are used to minimize base-calling errors. The number of “error” calls is significantly less than the number of “donor” calls, even for the 0% donor library, which may result from errors in the established genotype with false homozygous calls. As only one in two reads for a heterozygous donor SNP will contain the donor base, the overall rate of such reads is about half the rate for homozygous donor SNPs.
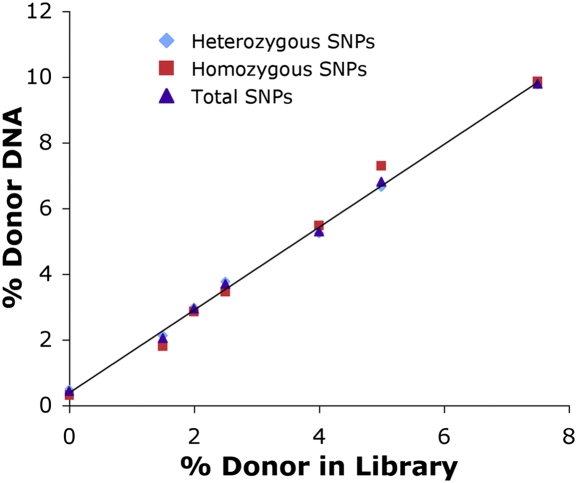
The raw counts were then used to calculate donor percentages, using just the homozygous SNPs, just the heterozygous SNPs, or total SNPs as shown in Fig. 3. Provided a correction is made for seeing only one out of every two donor molecules for the heterozygous SNPs, data from either heterozygous or homozygous donor SNPs give a reliable calculation of % donor DNA. The overall response was linear over the sampled range (R2 = 0.998) with sufficient sensitivity to measure transplant rejection, on the basis of the digital PCR results for donor percentage in the actual patient samples. These results establish a methodology to use SNPs to quantify the amount of one genome present in the background of another.
Fig. 3.
SNP-based detection of % donor DNA in control HapMap samples. There is a linear response (R2 = 0.998) of calculated % donor DNA compared with the % donor in the mock sequencing libraries. The trendline is given for all SNPs, including both heterozygous and homozygous donor SNPs, but the calculated percentages are similar in both subsets.
SNP Analysis of Patient Plasma Samples by Sequencing.
To demonstrate the feasibility of using SNPs as a marker for GTD in patient samples, we first needed to establish genotype information for the donor and recipient. We obtained whole blood from the recipient and banked splenocytes from the donor for patients 7 and 8, with rejection events, and patient 11, who had no such events. DNA was purified from these cells and was then genotyped at over a million loci using Illumina's Omni1-Quad Beadchip. Because our assay is particularly sensitive to false positive homozygous SNP calls for the recipient, we limited our focus to Beadchip SNPs with high GenCall and Cluster Separation scores, which yielded ∼150,000 usable loci in each case. Improvements in genotyping technology should eventually allow for many more of the potential 1.6 million usable SNPs to be identified, thereby improving the GTD assay's sensitivity even further.
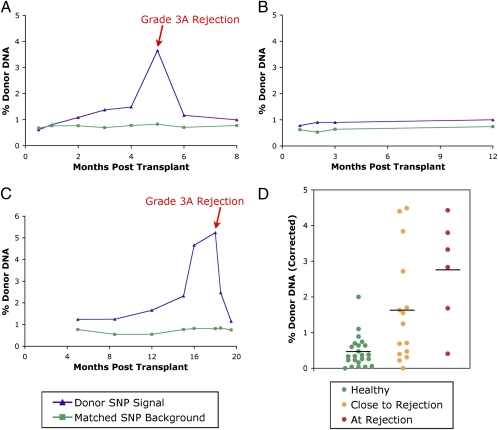
Sequencing libraries were prepared from the purified plasma DNA for each patient as previously described (24). Shotgun sequencing led to an average of 10–12 million unique aligning reads per sample, with ∼25,000 SNP-containing reads per time point (complete statistics are given in Table S2). Total donor DNA % was calculated using all SNPs (homozygous and heterozygous) and the results analyzed for each time point in each patient as before. Example time course graphs are shown in Fig. 4 and show similar trends to those observed by digital PCR for these same patients.
Fig. 4.
Donor DNA levels determined by sequencing. (A–C) Time-course graphs for patients 7 (A) and 11 (B), female patients receiving male hearts also analyzed by digital PCR, and for patient 14 (C), a male patient receiving a male heart. Patients 7 and 14 had grade 3A–2R rejections as determined by biopsy at the indicated time point, whereas patient 11 was a negative control with no rejection events. The calculated donor SNP signal is graphed in purple using both homozygous and heterozygous donor SNP positions. The matched SNP background is graphed in green, demonstrating the error for the assay arising from sequencing errors, genotyping errors, or sample contamination. The difference between the green and purple trend lines represents the changing level of donor-specific DNA. (D) Collected sequencing results for all patients analyzed by sequencing (7, 8, 11, 14–17) using the corrected % donor DNA values. Samples coincident with a biopsy determining an acute cellular rejection event are grouped together in red. Samples within 3 mo of a biopsy-determined rejection are shown in orange. Samples greater than 3 mo from any biopsy-determined rejection, or from a patient without any rejection events, are considered “healthy” normal readings and are shown in green. Group averages are marked by black lines.
Whereas our initial sequencing experiments were performed on samples from female recipients of male donor hearts to independently validate the results against the digital PCR measurements, we sought to demonstrate the universal nature of this approach by analyzing patients that could not be tracked using chromosome Y. We therefore performed our GTD assay on four male patients who received male organs (patients 14–17). A time course for one of these patients is shown in Fig. 4C.
The GTD assay allows for an internal control for the genetic signature of the donor organ compared with experimental background. In addition to considering all of the SNPs that differ between donor and recipient, the set of homozygous SNPs identified as the same for donor and recipient can be considered. Any nonrecipient signal observed at these sites will represent the assay background arising from sequencing errors, genotyping errors, or potential sample contamination with other human genetic material that would vary at some of these positions. The error in matched SNPs is plotted in green for these patients in Fig. 4 and is stable over all time points. The rise seen during biopsy-proven rejections is only seen in the donor-specific SNPs and therefore must be a specific signal from the donor organ that does not arise from changes in sequencing error rates or from sample contamination. The distance between the two curves is a reasonable value to report for observed donor DNA. Whereas several months before rejection this difference is negligible, this difference can rise to over 3–4%, a significant level of donor DNA, at the biopsy-proven rejection time points.
We generated an ROC curve for the collected sequencing data of all 44 patient samples (Fig. S4). Using biopsy grades as the indicator of “true positives” (rejection events), at a threshold of 1.70% donor DNA we can capture an 83% true positive rate with a 16% false positive rate. Comparing the donor DNA levels for the 6 true positives (2.75 ± 1.81%, SD) to the 38 other samples (0.92 ± 1.16%, SD) reveals a significant difference between the rejection samples and the other analyzed time points (P = 0.0013, Student's t test).
Although we have treated all time points in this study that were not coincident with a biopsy-proven rejection event as “negatives,” the observed trends by digital PCR and sequencing suggest that some of the time points, particularly those immediately before the rejection, may be elevated above baseline as an early indication of the onset of graft damage. For our sequencing results, we grouped together all time points not within 3 mo of an observed acute cellular rejection as “healthy.” Remaining time points were either grouped together as close to rejection or at rejection. The collected data from these groups are shown in Fig. 4D. Whereas some samples in the intermediate group have low values similar to the healthy time points, many have intermediate or high values similar to the rejection time points, suggesting that in some cases it may be possible to use this assay for earlier detection of rejection. Although more patient samples, particularly with biopsies graded at intermediate levels, will be needed to establish the significance of time points immediately before biopsy-determined rejections, our results clearly establish significant differences from normal once graft damage is severe, as determined by a grade ≥3A–2R biopsy.
Discussion
In this study we aimed to demonstrate that donor-derived cell-free DNA exists in the plasma of organ transplant recipients, and that elevated levels of donor DNA can be used as an indication of organ rejection. Although the existing cell-free DNA literature has presented conflicting reports on whether organ-specific signatures can be detected in plasma (14, 15), our data establish unambiguously that donor-specific DNA is present in the plasma of heart transplant recipients. By both methods of GTD demonstrated here, we establish a mean value below 1% as being normal for the level of donor-derived cell-free DNA when the patient is healthy. During organ rejection, however, the level of donor DNA signal rises in correlation with the endomyocardial biopsy results, with mean values increasing to 3–4% of the total cell-free DNA. Following treatment, the level of donor DNA tends to decline, and in several patients returns to baseline. Collectively these results establish that donor-derived DNA in the plasma is a promising biomarker for the onset of, and recovery from, heart transplant rejection. Whereas most earlier studies focused on the limited cases of females receiving male organs, here we have also demonstrated a generalizable strategy using single nucleotide polymorphisms that can be used for any possible donor and recipient pair.
In comparing GTD to noninvasive expression analysis tests, such as AlloMap, one observes some similarities and differences. It is not known whether AlloMap can detect rejection before biopsy, whereas we have shown evidence here that GTD is able to detect rejection before biopsy. Neither GTD nor AlloMap have been shown to distinguish between antibody-mediated rejection and cellular rejection. However, both conditions are treated with corticosteroids while awaiting a confirmatory test, so the fact that GTD can perform early detection may enable early intervention to prevent full blown rejection, whether cellular or antibody mediated. Because GTD measures the genetic signature of the donor organ, it should, like the endomyocardial biopsy, more directly report organ damage.
What could the health economic benefits of GTD be? Although it is difficult to calculate the precise value of early detection, there have been calculations of the benefit of using a noninvasive test to reduce the number of biopsies. On the basis of the results of the CARGO study, Evans et al. estimated that a noninvasive test with similar performance properties could save $12 million annually in health care costs in the United States (25). Because the GTD false positive rate is about half of that of the AlloMap test, the savings would be even greater, not including the benefits of early detection. Further studies, particularly of GTD in the clinic, will be required to determine the complete utility of this test as a replacement for the biopsy.
Because GTD and the AlloMap test look at different signals in the blood, and likely have different sources for false positives/negatives, a combination of the two approaches could be particularly powerful by reporting on both host immune response and graft injury. As GTD is not particularly dependent on physiology specific to the heart, it also has the potential to be used in the setting of other solid organ transplants (such as kidney, lung, and liver), where DNA from the transplanted organ may also exist in the recipient's plasma.
Materials and Methods
Posttransplant Monitoring and Clinical Sample Collection.
This study used stored plasma samples from a previously established cohort of 112 consecutive patients undergoing first heart transplantation between January 2002 and May 2005 at our institution. This cohort, funded by the National Institutes of Health (5P01AI050153-02), was assembled prospectively to study the relationship between cytomegalovirus (CMV) infection and the development of cardiac allograft vasculopathy. Age younger than 10 y, renal dysfunction requiring prolonged dialysis, and inability or unwillingness to provide signed informed consent represented exclusion criteria for study enrollment. All patients gave informed consent to the protocol approved by our institutional review board for studies in human subjects.
Posttransplant immunosuppression consisted of daclizumab (1 mg/kg i.v.) administered at the time of transplant surgery and on alternate weeks for a total of five doses, cyclosporine (3–5 mg/kg/d); prednisone initiated at 1 mg/kg/d and tapered to <0.1 mg/kg/d by the sixth postoperative month; and either mycophenolate mofetil 1,000–4,000 mg daily, or sirolimus 1–4 mg daily. All recipients received standard CMV prophylaxis consisting of 4 wk of i.v. ganciclovir. Those recipients who were CMV antibody negative and received a heart from a CMV antibody positive donor received an additional 3 mo course of CMV hyperimmune serum and up to 80 d of valganciclovir.
All study patients were monitored for acute cellular rejection by surveillance endomyocardial biopsies performed at scheduled intervals after transplant: weekly during the first month, biweekly until the third month, monthly until the sixth month, and then at months 9 and 12. Biopsies were graded according to the 1990 International Society for Heart and Lung Transplantation (ISHLT) classification system as 0, 1A, 1B, 2, 3A, 3B, and 4 (26). These grades are readily translatable to the ISHLT 2004 revised grading scale (0, 1R, 2R, and 3R) (27). Plasma samples were collected before performing the biopsy procedure and stored at the following time points posttransplant: day 14 and months 1–4, 6, 9, 12, 16, 20, 24, 38, 32, 36, 40, 44, 48, 52, 56, and 60.
Stored plasma samples were used for this study as follows: Serial plasma samples were retrieved for 13 patients with at least one episode of biopsy-proven acute cellular rejection (≥grade 3A–2R). Six of these 13 patients were females who had received hearts from female donors, 3 were females who had received hearts from male donors, and 4 were from males who had received hearts from male donors. Plasma was also retrieved for 4 female patients receiving hearts from male donors with no rejection episodes (all biopsies grade 0, or 1A–0, or 1R). For the recipients with male donors, plasma samples from as many as eight different time points, including any biopsy-proven rejection time points, were analyzed to determine a time course for the donor-specific DNA signature.
Plasma Purification and Digital PCR.
Plasma samples (1–2 mL total volume) were purified using the Nucleospin Plasma F kit (E&K Scientific Products). Digital PCR was performed on 12.765 digital array chips using the BioMark real-time PCR system (Fluidigm); FastStart TaqMan Probe Master Mix with Rox (Roche) was used with two probes targeted to a Chr 1 locus (EIF2C1) and a multicopy Chr Y locus (Dys14), as previously described (24). Control male and female genomic DNA (Promega) was used to calibrate the Chr 1 and Chr Y signals.
Control SNP Library Preparation.
Genomic DNA for the NA07348 and NA10830 HapMap lines was attained from the Coriell Institute for Medical Research (Camden, NJ). DNA was quantitated using the NanoDrop spectrophotometer (Thermo Scientific) and mixed at defined ratios. DNA was sheared to ∼200 to 300-bp fragments on a Covaris S2 (ABI) and purified on a Microcon YM-30 column (Millipore) before performing Illumina's single-end sequencing library preparation.
Patient Genotyping and Library Preparation.
Recipient DNA from whole blood and donor DNA from banked splenocytes was purified using the DNeasy blood and tissue kit (Qiagen). Where necessary, DNA was amplified by Repli-G Midi kit (Qiagen) before sending the samples to SA Biosciences for genotyping on the Omni1-Quad Beadchip (Illumina). To minimize false positive homozygous recipient SNP calls, only SNP loci with a GenCall score ≥0.70 and a Cluster Separation score of 1.00 were considered.
Sequencing libraries were prepared from the purified patient plasma DNA using the standard Illumina library preparation method with the exception of reduced adaptor concentration during ligation as previously described (24).
Sequencing.
Thirty-six–cycle single-end sequencing runs were performed for all DNA libraries. Each library was analyzed on a single lane of an Illumina GAII flowcell, with the exception of the second sample from patient 8, which combined data from two lanes due to poor sequencing yields. Reads were aligned to the reference human genome hg18 using ELAND, with an average of over 13 million aligned reads per lane and over 7 million unique aligning reads per lane. As nonunique reads at this low level of coverage most likely arise from the PCR amplification during library preparation, duplicated reads (reads that aligned to the same location) were removed before analysis to leave just a single aligning read at each site. All unique reads that span one of the SNP sites where the recipient has a homozygous allele that differs from the donor's genotype were then analyzed for the presence of a recipient, donor, or error (other) base call. Bases with reported quality scores lower than 80 were excluded from this analysis to minimize sequencing errors. The total donor DNA percentage was calculated by taking twice the number of donor heterozygous read calls plus the number of donor homozygous read calls over the total number of donor and recipient read calls, not including errors.
Supplementary Material
Acknowledgments
We thank Norma Neff and Gary Mantalas for assistance in carrying out sequencing runs. This work was supported by the National Institutes of Health Director's Pioneer Award and Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: Stanford University has applied for a patent relating to the method described in this study.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013924108/-/DCSupplemental.
References
- 1.Baraldi-Junkins C, et al. Complications of endomyocardial biopsy in heart transplant patients. J Heart Lung Transplant. 1993;12:63–67. [PubMed] [Google Scholar]
- 2.Williams MJ, et al. Biopsy-induced flail tricuspid leaflet and tricuspid regurgitation following orthotopic cardiac transplantation. Am J Cardiol. 1996;77:1339–1344. doi: 10.1016/s0002-9149(96)00202-0. [DOI] [PubMed] [Google Scholar]
- 3.Oldham N, Ott RA, Allen BA, Fopiano P, Dwyer M. Ventricular fibrillation complicating endomyocardial biopsy of a cardiac allograft. Cathet Cardiovasc Diagn. 1991;23:300–301. doi: 10.1002/ccd.1810230415. [DOI] [PubMed] [Google Scholar]
- 4.Bhat G, Burwig S, Walsh R. Morbidity of endomyocardial biopsy in cardiac transplant recipients. Am Heart J. 1993;125:1180–1181. doi: 10.1016/0002-8703(93)90138-y. [DOI] [PubMed] [Google Scholar]
- 5.Hamour IM, et al. Limited utility of endomyocardial biopsy in the first year after heart transplantation. Transplantation. 2008;85:969–974. doi: 10.1097/TP.0b013e318168d571. [DOI] [PubMed] [Google Scholar]
- 6.Lin D, et al. Biomarkers in Transplantation Team; NCE CECR Centre of Excellence for the Prevention of Organ Failure. Whole blood genomic biomarkers of acute cardiac allograft rejection. J Heart Lung Transplant. 2009;28:927–935. doi: 10.1016/j.healun.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Deng MC, et al. CARGO Investigators. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz PA, et al. Detection of cardiac allograft rejection and response to immunosuppressive therapy with peripheral blood gene expression. Circulation. 2004;110:3815–3821. doi: 10.1161/01.CIR.0000150539.72783.BF. [DOI] [PubMed] [Google Scholar]
- 9.Pham MX, et al. IMAGE Study Group. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 10.Starling RC, et al. Working Group on Molecular Testing in Cardiac Transplantation. Molecular testing in the management of cardiac transplant recipients: initial clinical experience. J Heart Lung Transplant. 2006;25:1389–1395. doi: 10.1016/j.healun.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta. 2006;363:187–196. doi: 10.1016/j.cccn.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 12.Chok R, et al. Apoptosis and expression of heme oxygenase-1 in heart transplant recipients during acute rejection episode. Transplant Proc. 2002;34:2815–2818. doi: 10.1016/s0041-1345(02)03526-1. [DOI] [PubMed] [Google Scholar]
- 13.Cailhier JF, Laplante P, Hébert MJ. Endothelial apoptosis and chronic transplant vasculopathy: Recent results, novel mechanisms. Am J Transplant. 2006;6:247–253. doi: 10.1111/j.1600-6143.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 14.Lo YM, et al. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351:1329–1330. doi: 10.1016/s0140-6736(05)79055-3. [DOI] [PubMed] [Google Scholar]
- 15.Lui YY, et al. Origin of plasma cell-free DNA after solid organ transplantation. Clin Chem. 2003;49:495–496. doi: 10.1373/49.3.495. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Ohkohchi N, Okazaki H, Guo Y. Use of PCR and PCR-SSP for detection of urinary donor-origin DNA in renal transplant recipients with acute rejection. Chin Med J (Engl) 2003;116:191–194. [PubMed] [Google Scholar]
- 17.Zhang J, et al. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: Urinary DNA chimerism. Clin Chem. 1999;45:1741–1746. [PubMed] [Google Scholar]
- 18.Zhong XY, et al. Cell-free DNA in urine: a marker for kidney graft rejection, but not for prenatal diagnosis? Ann N Y Acad Sci. 2001;945:250–257. [PubMed] [Google Scholar]
- 19.García Moreira V, Prieto García B, Baltar Martín JM, Ortega Suárez F, Alvarez FV. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem. 2009;55:1958–1966. doi: 10.1373/clinchem.2009.129072. [DOI] [PubMed] [Google Scholar]
- 20.Gadi VK, Nelson JL, Boespflug ND, Guthrie KA, Kuhr CS. Soluble donor DNA concentrations in recipient serum correlate with pancreas-kidney rejection. Clin Chem. 2006;52:379–382. doi: 10.1373/clinchem.2005.058974. [DOI] [PubMed] [Google Scholar]
- 21.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan HC, Blumenfeld YJ, El-Sayed YY, Chueh J, Quake SR. Microfluidic digital PCR enables rapid prenatal diagnosis of fetal aneuploidy. Am J Obstet Gynecol. 2009;200(5):543 e541–547. doi: 10.1016/j.ajog.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frazer KA, et al. International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans RW, et al. The economic implications of noninvasive molecular testing for cardiac allograft rejection. Am J Transplant. 2005;5:1553–1558. doi: 10.1111/j.1600-6143.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- 26.Billingham ME, et al. The International Society for Heart Transplantation. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. J Heart Transplant. 1990;9:587–593. [PubMed] [Google Scholar]
- 27.Stewart S, et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant. 2005;24:1710–1720. doi: 10.1016/j.healun.2005.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.