One of the least understood aspects of protein synthesis is communication between the ribosome and the nascent peptide. Chiba et al. (1) in this issue of PNAS reveal that, at least in some cases, these dialogues can be species-specific.
The ribosome assembles amino acids into proteins in the peptidyl transferase center (PTC) located at the interface side of the large ribosomal subunit. On its way out, the nascent peptide passes through the exit tunnel. This ∼100-Å-long and 10- to 20-Å-wide irregularly shaped vent spans the entire body of the subunit. The tunnel is not just a hole but rather a functionally important compartment where the structure of the nascent peptide is monitored and from which specific peptides can signal the ribosome to slow down its rate of elongation or even completely stop translation. The best-characterized manifestation of this feedback mechanism is nascent peptide-dependent ribosome stalling when the PTC becomes incapacitated after having polymerized the effector (stalling) domain of the regulatory nascent peptide. Such programmed translation arrest is sensitive to the cellular environment and therefore, can be used for control of gene expression. A number of genes regulated on the basis of nascent peptide recognition have been identified in bacteria and eukaryotes (2).
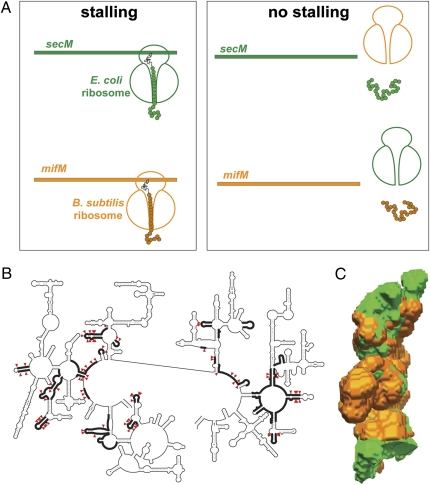
Despite the generally conserved nature of the ribosome, the fine structure of the exit tunnel exhibits considerable variations. Nearly 20% of rRNA nucleotides forming the exit tunnel differ between Gram-negative and Gram-positive bacteria (Fig. 1B). The variation in tunnel properties can be further influenced by the idiosyncratic posttranscriptional rRNA modifications and the divergence of tunnel proteins. These differences may affect the overall shape of the tunnel (Fig. 1C) as well as its chemical characteristics (polarity, pattern of hydrogen donors and acceptors, electrostatics, etc.) (3, 4), and thus, they may alter functional interactions of the ribosome with the protein being synthesized.
Fig. 1.
The variation in the ribosome tunnel structure between species may affect nascent peptide recognition. (A) The SecM and MifM stalling peptides arrest progression of only cognate ribosomes. (B) The tunnel nucleotide residues differing between Gram-negative E. coli and Gram-positive B. subtilis are shown by red arrowheads. Segments of the large ribosomal subunit rRNA that form the walls of the exit tunnel are indicated by thick lines (20). (C) Difference in the shape of the exit tunnels in ribosomes of Gram-negative (E. coli; green) and Gram-positive (Deinococcus radiodurans; orange) bacteria. The tunnel shape from the structures of the respective large ribosomal subunits was extracted as described in ref. 4.
Significant progress has been achieved in recent years in elucidating the mechanism of translation modulation by the nascent peptide. Sequences critical for stalling have been characterized for several regulatory peptides (5–8), a number of rRNA and protein residues at the walls of the exit tunnel that are involved in nascent peptide recognition and triggering of the ribosomal response have been mapped (6, 7, 9), and cryo-EM structures of stalled ribosome complexes are starting to emerge (10, 11). Nevertheless, very little is still known about the principles of nascent peptide–ribosome communication. Chiba et al. (1) illuminate an important aspect of the operation of this mechanism: the specificity of the ribosome to recognition of the stalling peptide.
Chiba et al. (1) examine programmed translation arrest induced by the regulatory proteins SecM and MifM found in Escherichia coli and Bacillus subtilis, respectively. In Gram-negative E. coli, nascent peptide-dependent ribosome stalling at the secM ORF activates translation of the downstream gene-encoding protein export chaperon SecA (7). Translation arrest at the mifM ORF in Gram-positive B. subtilis leads to activation of expression of an alternative membrane biogenesis factor YidC2 (12). Although SecM and MifM peptides elicit the same ribosomal response—programmed translation arrest—they do not share any obvious homology. By using a cell-free translation system driven by either E. coli or B. subtilis ribosomes, Chiba et al. (1) were able to show that the B. subtilis MifM nascent peptide prompts stalling of the B. subtilis ribosome but not of the foreign E. coli ribosome. Conversely, the SecM nascent peptide, which efficiently arrests progression of the homologous E. coli ribosome, fails to do so when translated by heterologous ribosomes from B. subtilis (Fig. 1A). Thus, it seems that SecM and MifM stalling domains have been evolutionarily optimized to operate specifically with their cognate ribosome. Translation arrest directed by SecM and MifM peptides is influenced by dissimilar factors—the activity of the secretion apparatus in the former case and membrane insertion in the latter case. However, Chiba et al. (1) were able to dissociate ribosome stalling from the peptide's sensory properties. By replacing the N terminus of MifM with a secreted signal sequence, Chiba et al. (1) converted MifM into a secretion sensor, thereby showing that the arrest domain operates independently of the peptide's sensory domain.
Stalling peptides, even those that operate in the same species, can be fine-tuned to exploit different sets of sensors in the exit tunnel (10, 13, 14). Even small alterations in the structure of the peptide or the ribosome can disrupt their functional communication. Not surprisingly, the closer-related that the species are, the higher the chance that the nascent peptide can overcome the language barrier in talking to a noncognate ribosome. Thus, the E. coli ribosome that does not understand the stalling message encoded in the MifM peptide from Gram-positive B. subtilis can nevertheless properly respond to stalling peptides from closely related Gram-negative bacteria (15, 16). Remarkably, however, some stalling peptides have apparently evolved to be truly multilingual and are able to communicate even with evolutionarily distant ribosomes. Vivid examples are the peptides that, through nascent peptide-dependent ribosome stalling, control inducible antibiotic resistance. Resistance to antibiotics is a trait that is often disseminated by horizontal gene transfer. A stalling peptide
Many proteins could have been optimized for proper translation specifically by their cognate ribosome.
that can properly direct translation arrest in a variety of species would have an evolutionary advantage over a species-specific regulator, because it would be more often retained in a new bacterial host in need of protection from the antibiotic. Indeed, nascent peptides that control inducible antibiotic resistance can efficiently stall ribosomes from evolutionarily distant Gram-positive and Gram-negative bacteria (6, 13, 17). The stalling nascent peptides found in eukaryotic cells also seem to be rather cosmopolitan in their nature: fungal arginine attenuator peptide and a regulatory peptide from human cytomegalovirus can efficiently stall plant ribosome from wheat (Triticum aestivum) (11, 18, 19). The intricate communication network within the ribosomal tunnel allows for nascent peptides to “choose” how private or universal their message to the ribosome is going to be.
Which structural features of the peptide and the ribosome account for their specific functional interactions? Although cryo-EM studies have provided important insights (10, 11), the resolution is not sufficient to confidently identify specific molecular and atomic contacts. Additionally, we are still awaiting high-resolution crystallographic structures of ribosome–nascent peptide complexes. However, the availability of the two specific ribosome–nascent peptide pairs uncovered by Chiba et al. (1) opens the attractive possibility of addressing this question through biochemistry and genetics. One can explore which of the MifM amino acid residues need to be changed to make the peptide recognizable by the E. coli ribosome and what changes need to be introduced in SecM so that it can stall the ribosome from B. subtilis. Conversely, it would be interesting to test what needs to be changed in the ribosome to allow recognition of a stalling domain in the foreign peptide.
Prolonged ribosome stalling is an extreme case of possibly many scenarios of retro-regulation of translation by the peptide from within the ribosome exit tunnel. Functionally meaningful interactions between the ribosome and nascent peptides may decrease the translation elongation rate to facilitate protein folding, interactions with chaperons, or secretion. Conversely, a nascent peptide-induced increased rate of elongation can help avoid unwanted interactions of incomplete translation products with the cellular environment. The findings of Chiba et al. (1) imply that many proteins could have been optimized for proper translation specifically by their cognate ribosome. We are only starting to understand the language that the ribosome and the nascent peptide speak to each other. Eavesdropping on their conversations can reveal many exciting mysteries.
Acknowledgments
The work on nascent peptide-dependent ribosome stalling in this laboratory is supported by National Science Foundation Grant MCB-0824739 (to N.V.-L. and A.S.M.) and National Institutes of Health (National Institute of Allergy and Infectious Disease) Grant AI083684 (to N.V.-L.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 6073.
References
- 1.Chiba S, et al. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci USA. 2011;108:6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 4.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Mayford M, Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989;206:69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 8.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Seidelt B, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhushan S, et al. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vázquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhushan S, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-Vera LR, Yang R, Yanofsky C. Tryptophan inhibits Proteus vulgaris TnaC leader peptide elongation, activating tna operon expression. J Bacteriol. 2009;191:7001–7006. doi: 10.1128/JB.01002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirsch DR, Lai MH. Regulation of a macrolide resistance-beta-galactosidase (ermC-lacZ) gene fusion in Escherichia coli. J Bacteriol. 1984;159:381–384. doi: 10.1128/jb.159.1.381-384.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang P, Wang Z, Sachs MS. Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J Biol Chem. 2000;275:26710–26719. doi: 10.1074/jbc.M003175200. [DOI] [PubMed] [Google Scholar]
- 19.Degnin CR, Schleiss MR, Cao J, Geballe AP. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]