Abstract
Many proteins reside at the cell poles in rod-shaped bacteria. Several hypotheses have drawn a connection between protein localization and the large cell-wall curvature at the poles. One hypothesis has centered on the formation of microdomains of the lipid cardiolipin (CL), its localization to regions of high membrane curvature, and its interaction with membrane-associated proteins. A lack of experimental techniques has left this hypothesis unanswered. This paper describes a microtechnology-based technique for manipulating bacterial membrane curvature and quantitatively measuring its effect on the localization of CL and proteins in cells. We confined Escherichia coli spheroplasts in microchambers with defined shapes that were embossed into a layer of polymer and observed that the shape of the membrane deformed predictably to accommodate the walls of the microchambers. Combining this technique with epifluorescence microscopy and quantitative image analyses, we characterized the localization of CL microdomains in response to E. coli membrane curvature. CL microdomains localized to regions of high intrinsic negative curvature imposed by microchambers. We expressed a chimera of yellow fluorescent protein fused to the N-terminal region of MinD—a spatial determinant of E. coli division plane assembly—in spheroplasts and observed its colocalization with CL to regions of large, negative membrane curvature. Interestingly, the distribution of MinD was similar in spheroplasts derived from a CL synthase knockout strain. These studies demonstrate the curvature dependence of CL in membranes and test whether these structures participate in the localization of MinD to regions of negative curvature in cells.
Keywords: lipid microdomains, microfabrication, lipid rafts, hydrogel, anionic phospholipids
A central question in cell biology is how the spatial organization of proteins and lipids is established, maintained, and replicated and how it fluctuates in response to external stimuli. Eukaryotic cells use several mechanisms to accomplish this task, including a dynamic cytoskeleton that controls the spatial and temporal position of proteins, nucleic acid, and organelles (1). The formation of lipid microdomains in the membrane also is involved in the localization of integral membrane proteins (2). These mechanisms play a critical role in cell physiology and behavior.
Bacteria also use mechanisms for controlling the intracellular location of proteins, lipids, and nucleic acid. The persistent historical view of bacterial cells as lacking spatial control over their intracellular components inhibited the field. Bacteria are sophisticated organisms that use tightly regulated physiological mechanisms similar to many of those used by eukaryotic cells, including controlling shape, regulating growth and division, transporting intracellular components (e.g., proteins, plasmids, DNA, and RNA), and polarizing the cell (3, 4). These organisms have several remarkable structural characteristics, but one of the most fundamental questions in this area of microbiology—how cells produce, maintain, and replicate their spatial organization—still is not entirely understood (5).
Many different families of bacterial proteins are positioned at the cell poles (6, 7). TipN and TipF in Caulobacter crescentus and SpoIIB in Bacillus subtilis are recruited to the midcell before division and are positioned at the poles after septation is complete (8–10). Other proteins appear to respond to cell morphology and become localized at the poles and septa (e.g., the cell division sites) of rod-shaped bacteria where they participate in a variety of physiological processes, including division, spore formation, chemotaxis, and cell-wall remodeling (5). The lack of available techniques for predictably controlling the shape of the bacterial cell wall and membrane makes it difficult to study the mechanisms by which biomolecules localize in response to cell shape.
The poles and the septa are regions of the cell that have the largest curvature. [Curvature is defined by 1/r along the cylindrical region of the cell wall and 2/r at the pole, where r is the radius (11).] In Escherichia coli cells, r is ∼0.5 μm, and curvature is ∼2 μm−1 at the pole and and ∼10 μm−1 at the septa, as determined from electron microscopy images (12). Several observations suggest that cell curvature influences the distribution of cytoplasmic, amphiphilic proteins in bacteria. For example, in the rod-shaped bacterium B. subtilis, membrane curvature affects the localization of the sporulation factor protein SpoVM (via positive curvature) and multimers of the division protein DivIVA (via negative curvature) (13–15). The mismatch in length scales between a single molecule of DivIVA or SpoVM and the curvature of the membranes with which these proteins interact makes it unclear how individual proteins sense curvature. Huang and Ramamurthi (11) carefully estimated the physical requirements for curvature sensing and suggest that oligomers of DivIVA or SpoVM may have physical dimensions that are compatible for sensing membrane curvature.
Negative curvature also may influence the distribution of lipids within leaflets of bacterial membranes. E. coli membranes consist of ∼5% cardiolipin (CL, also referred to as “diphosphatidylglycerol”), 20–25% phosphatidylglycerol (PG), and 70–80% phosphatidylethanolamine (PE) (16). CL is distributed between the two leaflets of the bilayers and is located preferentially at the poles and septa in rod-shaped cells of E. coli, B. subtilis, and Pseudomonas putida (17–19). The polar positioning of the proline transporter ProP and the mechanosensitive ion channel of small conductance MscS in E. coli is dependent on CL (20). These observations suggest that cell shape may influence the localization of CL and proteins. One hypothesis for polar protein localization in bacteria is that large cell curvature reorganizes the membranes and creates ordered lipid domains that position protein at specific regions of cells.
Thermodynamic models of inner membrane organization in bacteria support this hypothesis. Wingreen and coworkers (21, 22) used numerical simulations to study CL microdomain formation at the poles of bacterial cells. CL has an intrinsic curvature (∼1–5 nm−1) because the cross-sectional area of the head-to-tail groups makes it conical in shape (11). The shape of CL makes it favorable for this lipid to be positioned at negatively curved regions of the inner leaflet of bacterial membranes. Short-range interactions between CL molecules in bacterial membranes are thermodynamically favorable and may lead to the formation of microdomains, which localize to the poles of cell membranes in silico; in contrast, individual molecules remain uniformly distributed in the membrane (21, 22). CL microdomains that exceed a critical size overcome entropy and localize at the poles. Osmotic forces that pin the membrane against the peptidoglycan layer of the cell wall regulate the size of the microdomains. These simulations are supported by recent experiments that suggest CL domains may be responsible for polar protein localization in E. coli cells (20).
In this paper we present a microtechnology-based technique for studying the intracellular organization of bacterial cells. The technique uses polymer microchambers to control the curvature of intact bacterial membranes. We created giant E. coli spheroplasts and confined them in microchambers with a user-defined shape. The microchambers were embossed in a layer of agarose and had a volume that matched the spheroplasts. The shape of the confined spheroplast membrane conformed to the geometry of the microchamber walls and provided a mechanism for controlling membrane curvature precisely.
We imaged the localization of CL microdomains in geometrically constrained spheroplasts confined in polymer microchambers using the CL-specific fluorophore nonyl acridine orange (NAO) (18, 23) and epifluorescence microscopy and analyzed the data using image analysis tools. We found that CL microdomains localized to regions of large, negative spheroplast curvature. Furthermore, by expressing YFP fused to the N-terminal domain of the cytoplasmic division protein MinD (MinD-YFP), we demonstrate the dependence of negative membrane curvature on MinD localization in spheroplasts and its colocalization with CL microdomains. These experiments provide a quantitative measure of the relationship between cell curvature in bacteria, CL microdomain localization at the poles, and the positioning of amphiphilic, cytoplasmic proteins. Lipid microdomains in bacteria recently have been described as “lipid rafts” because they may have striking similarities to the structure and function of lipid-ordered domains in eukaryotic membranes (24, 25).
Results
Controlling Membrane Curvature in Bacteria.
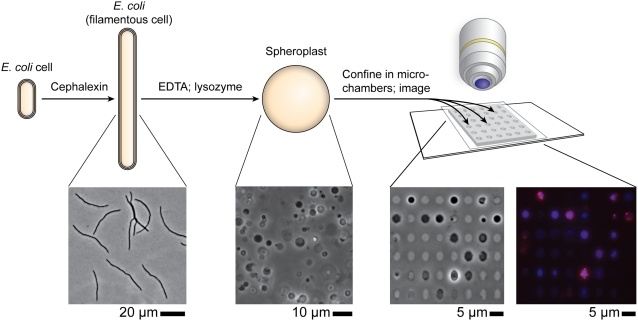
There are very few techniques available for manipulating the shape and curvature of intact bacterial membranes in vivo (26, 27). To control the curvature of bacterial membranes quantitatively, we developed the microtechnology-based procedure summarized in Fig. 1. We used cephalexin to grow cells of E. coli strain MG1655 into filaments that had an average length of ∼50 μm (Fig. S1). Stripping away the outer cell membrane followed by hydrolysis of the peptidoglycan with lysozyme produced giant spheroplasts with an average diameter of ∼3–4 μm and a volume of ∼20–25 μm3 (28) (Fig. 1 and Fig. S1). We found that the removal of the outer membrane and the peptidoglycan had no effect on the integrity of the bacterial membrane: Labeling the lipids with the membrane dye FM 4–64 (Invitrogen) and imaging the spheroplasts using epifluorescence microscopy confirmed that the membranes were spherical, continuous, and intact (Fig. 1 and Fig. S2). The majority of spheroplasts retained their cellular machinery and DNA, and the membrane contained randomly distributed domains of CL (Fig. 2C). In native cells CL microdomains are observed commonly at the cell poles (Fig. 2A).
Fig. 1.
A schematic representation of the experimental procedure: E. coli cells are grown into filaments with a mean length of 50 μm. Removing the outer membrane and the peptidoglycan converts the filaments into spheroplasts. Spheroplasts are labeled with NAO (for CL, false-colored red) and DAPI (for DNA, false-colored blue), confined in individual microchambers with varying curvature, and imaged using phase-contrast and epifluorescence microscopy.
Fig. 2.
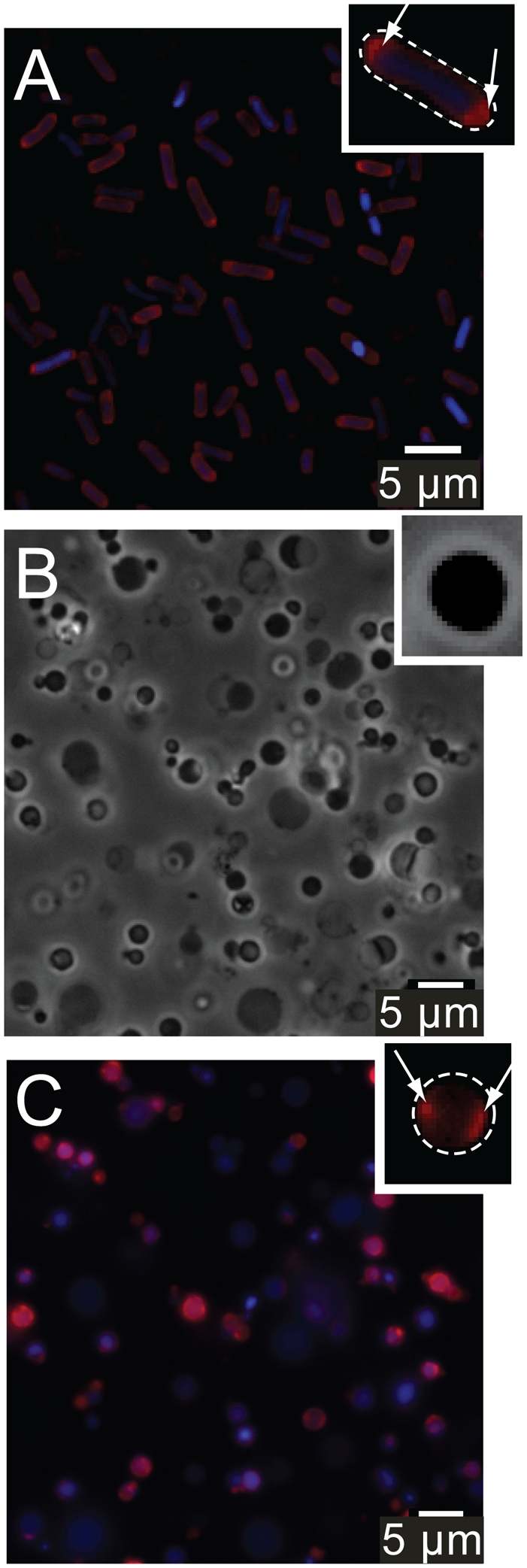
(A) Epifluorescence microscopy images of E. coli cells with labeled CL (NAO, red) and DNA (DAPI, blue). (B) Phase-contrast microscopy images of freestanding spheroplasts. (C) Epifluorescence microscopy images of freestanding spheroplasts with labeled CL (red) and DNA (blue). (Insets) White dashed lines indicate the perimeter of the cell (A) and spheroplast (C). White arrows (C) highlight CL microdomains.
We used microfabrication and soft lithographic techniques to emboss the surface of a layer of agarose with an array of microchambers containing patterns of six distinct features that progressively morphed from spherical to rod-shaped with hemispherical ends (Fig. S3) (29, 30). The curvature of the series of compartments was 0.67, 0.73, 1.03, 1.25, 1.56, and 2.08 μm−1, and the volume was approximately constant at 22 μm3 (see Fig. S4 and Table S1 for microchamber dimensions). The microchambers were ∼3 μm tall with a rectangular cross-section. We measured the Young's modulus, E, of the agarose using atomic force microscopy and found it to be ∼0.5 MPa. In contrast, an E. coli cell has an E of ∼10–20 MPa, and a lipid bilayer has a much lower elasticity than either of these materials and is deformable (31).
We used capillary pressure to draw spheroplasts into microchambers so that ∼40% of the microchambers were filled with spheroplasts. The spherical shape of the spheroplasts deformed to accommodate the geometry of the microchambers, and the membrane followed the contour line of the agarose walls (Fig. 1). The fabrication of thousands of arrays of microchambers in parallel in one layer of agarose enabled us to identify and image ∼2,500 spheroplasts that were isolated in microchambers and free of defects (e.g., were not folded, bent, or bulging out of the microchamber) (Fig. S5 and Movies S1 and S2).
CL Localizes to Regions of Curved E. coli Membranes.
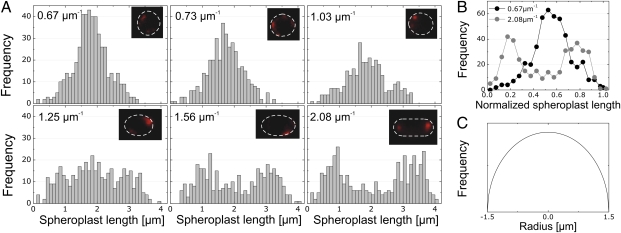
We prepared E. coli spheroplasts, labeled CL with NAO and DNA with DAPI, and imaged the spheroplasts using epifluorescence microscopy. To visualize NAO bound specifically to CL microdomains, we used an appropriate filter set to detect the far red-shifted emission of the dye (λ = 620 nm) that arises from π–π stacking of two molecules bound to CL (32). The Stokes shift makes it possible to distinguish between NAO bound to CL and NAO associated with PG, because the latter complex emits at λ = 535 nm. We found that the majority of the CL in spheroplasts was localized in microdomains that were distributed randomly throughout the membrane; the majority of the spheroplasts were intact and contained DNA. We analyzed the relationship between the position of CL microdomains in the membrane and spheroplast curvature. In microchambers that imposed spheroplast curvature of 0.67 and 0.73 μm−1, the CL microdomains displayed little preference for their position in the membranes of confined spheroplasts (Fig. 3A). The CL distribution started to broaden for a spheroplast curvature of 1.03 μm−1.
Fig. 3.
(A) An analysis of the position of CL microdomains in spheroplasts confined in microchambers with different curvatures. CL was visualized by labeling spheroplasts with NAO. Each panel contains a representative image of CL localization in one confined spheroplast with the curvature indicated; the dashed line indicates the perimeter of the spheroplast. (B) Comparison of the CL microdomain distribution in spherical (curvature = 0.67 μm−1, dark circles) and rod-shaped (curvature = 2.08 μm−1, light circles) spheroplasts plotted against the normalized length of spheroplasts. We analyzed ≥400 spheroplasts for each microchamber curvature. (C) The predicted localization of CL microdomains in a spheroplast confined in a spherical microchamber.
CL microdomains displayed a significant preference for localization to the regions of confined spheroplast membranes that mimicked the curvature of native E. coli cell poles (Fig. 3A). In membranes in which the spheroplast curvature was 1.56 and 2.08 μm−1, the distribution of CL microdomains was bimodal, and the mean location was centered close to the pseudopoles of the rod-shaped microchambers. Note, that the actual position/polarity (0 or 1) of CL microdomains is arbitrary and is a product of the analysis. The localization of CL in these structures is different from the distribution in spheroplasts that either are unconfined or are confined in spherical microchambers (Figs. 2C and 3A). This divergence in CL localization is notable when comparing the CL microdomain frequency versus position for spheroplast curvatures of 0.67 and 2.08 μm−1 (Fig. 3B). We calculated the CL microdomain distribution in the membrane of a spheroplast confined in a spherical microchamber. We assumed a uniform CL distribution in a sphere with dimensions z = −r to z = r, where r is the radius. We sectioned the spheroplast into infinitesimally small intervals (dz) and calculated the surface area (dA) as 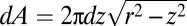 . The predicted distribution of CL for this spheroplast is parabolic (Fig. 3C).
. The predicted distribution of CL for this spheroplast is parabolic (Fig. 3C).
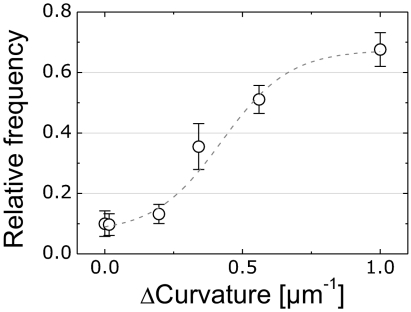
A plot of the relationship between the relative frequency of CL microdomains and the difference in microchamber curvature, Δcurvature, is depicted in Fig. 4. We calculated Δcurvature for each spheroplast by subtracting the region of lowest curvature from the region of highest curvature. To determine the relationship between membrane curvature and CL microdomain frequency, we divided epifluorescence microscopy images of confined spheroplasts into three segments consisting of two opposing poles and the midcell. Using these segments, we calculated the relative frequency of CL microdomains at the poles of spheroplasts as a function of Δcurvature from the CL data depicted in Fig. 3A. The resulting plot demonstrates the transition from nonpolar to polar localization of CL domains with increasing values of Δcurvature (Fig. 4). A plot of the data versus the elastic energy difference for a single CL molecule at different regions of membrane curvature is shown in Fig. S6. Smaller values of Δcurvature resulted in membranes in which CL was randomly or homogeneously distributed. These results quantitatively demonstrate that CL microdomains respond to differences in membrane curvature and display a statistically significant preference for localization at a spheroplast curvature >2 μm−1, which is comparable to the curvature of the poles in native E. coli cells.
Fig. 4.
The relative frequency of CL microdomains at the polar regions in spheroplasts plotted against the difference in the highest and lowest curvature regions of the membrane (Δcurvature). The data (open circles) were fitted to a sigmoidal function (dashed line) as a visual guide.
CL microdomains appeared to be <500 nm in diameter. However, the spatial resolution of this technique was limited by the diffraction limit of optical microscopy, which made it impossible to determine the size of the CL structures quantitatively. The limited temporal resolution of the technique made it difficult to observe the movement of CL microdomains in spheroplast membranes. In a typical experiment, the time that elapsed between confining the labeled spheroplasts into microchambers and the start of epifluorescence microscopy was several minutes. By the time we started imaging, the CL microdomains were localized in the membrane, and we were unable to observe the movement of CL microdomains by time-resolved microscopy.
MinD Localizes to High-Curvature Regions of Membranes.
Many E. coli proteins, including ProP, MscS, MscL, LacY, MinC, and MinD, have been reported to localize to the poles and septa (14, 15, 20, 33, 34). We used MinD as a model system to test the effect of membrane curvature on protein localization using the technique of confining spheroplasts in microchambers. The Min family of proteins—MinC, MinD, and MinE—form a pole-to-pole oscillator in E. coli cells that regulates the formation of the division plane (35). The N-terminal region of MinC binds to FtsZ and inhibits its assembly into the Z-ring at the future site of cell division. The C-terminal region of MinC binds MinD, which functions as a topological determinant for formation of the MinCDE complex. MinD is an ATPase and has an amphiphilic domain for binding the membrane (36). Varma et al. (34) demonstrated that ectopic poles play an important role in the oscillation of the Min system and that geometrical cues may play a role in the selection of the interaction site.
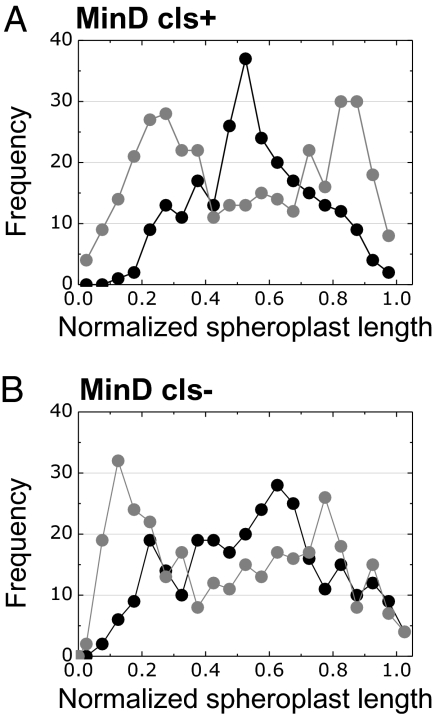
We imaged the localization of MinD in the confined membranes and observed that the effect of spheroplast curvature on the localization of MinD closely resembles the data for CL microdomains (Fig. 5A). These observations support earlier studies suggesting that MinD responds to geometric constraints of the cell, which direct it to the poles (34). The data provide quantitative support for the hypothesis that membrane curvature influences the positioning of some E. coli proteins.
Fig. 5.
Plots depicting the relationship between the frequency of MinD localization and the normalized position in the confined spheroplast. Distributions of (A) MinD-YFP in CL synthase-positive (cls+) (MG1655 pFX40) and (B) CL synthase-negative (cls−) (JW1241 Δcls pFX40) E. coli strains. Dark circles represent data for spheroplasts confined in spherical microchambers (curvature = 0.67 μm−1). Light circles represent data for rod-shaped microchambers (largest curvature = 2.08 μm−1).
MinD and CL Colocalize to High-Curvature Regions in E. coli Membranes.
Our data on CL and MinD localization led us to the hypothesis that CL microdomains may be landmarks for recruiting the protein to the polar regions of E. coli cells. Several pieces of indirect evidence support this hypothesis. (i) Amphiphilic proteins have a high interaction potential with lipids (37). (ii) The amphiphilic, positively charged helix of MinD has a preference for association with negatively charged phospholipids (36, 38–40). (iii) CL increases the binding efficiency of MinD to liposomes in vitro (41). (iv) CL influences the polar localization of ProP and MscS in E. coli cells in vivo (20).
To study the interaction between MinD and CL microdomains in vivo, we fluorescently labeled CL and DNA in spheroplasts of an E. coli strain expressing MinD-YFP and confined the spheroplasts in microchambers. MinD colocalized with CL in ∼80% of the spheroplasts that we analyzed (Fig. 6). Collecting photons emitted at λ = 620 nm made it possible to image the location of NAO at a wavelength that was far from YFP (λexcitation = 500 nm; λemission = 535 nm) (18). There is a caveat, however, because NAO bound to PG has λexcitation = 488 nm and λ emission = 535 nm. Consequently some of the intensity that we attribute to YFP may be caused by NAO bound to PG that colocalizes with MinD-YFP.
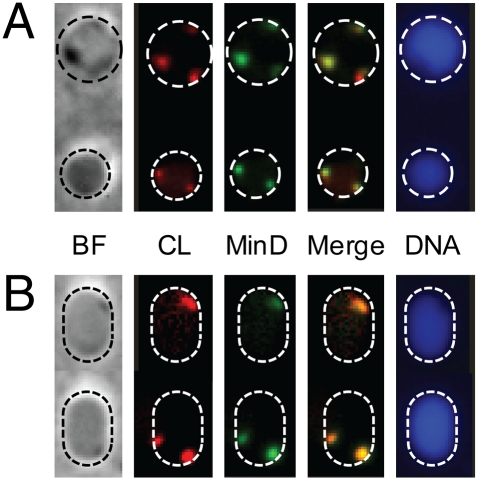
Fig. 6.
MinD and CL colocalization in E. coli MG1655 (+pFX40) spheroplasts confined in microchambers. (A) Microscopy images of spheroplasts confined in spherical microchambers. (B) Images of spheroplasts confined in rod-shaped microchambers. The microscopy images depict bright field (BF; phase contrast); CL labeled with NAO (CL); MinD-YFP (MinD); the overlay of the CL and MinD images (Merge); and DNA labeled with DAPI (DNA). Dashed circles indicate the perimeter of the microchambers and were added to the images for clarity.
Dependence of CL on MinD Localization.
To determine if CL microdomains are required to position MinD at the E. coli cell poles, we studied protein localization in spheroplasts of the CL synthase knockout JW1241 Δcls pFX40 (Fig. 5B and Figs. S7–S9) (42). We also explored the PG synthase knockout strain UE54 pFX40 (43); however, we were unable to find adequate conditions to filament cells of this strain. We observed that MinD-YFP localization in spheroplasts of the Δcls strain was dependent on membrane curvature and that the distribution was similar to MinD-YFP in spheroplasts from wild-type E. coli cells (Fig. 5A). The result suggests that MinD positioning is dependent on curvature, as suggested earlier (11, 34), and may be independent of CL. Although we were unable to detect CL microdomains in strain JW1241 Δcls using NAO, E. coli Δcls cells have been reported to contain ∼10% of the CL content of native cells (44). We cannot exclude the possibility that small amounts of CL present might participate in MinD localization. Furthermore, to compensate for the depletion of CL, cells of the Δcls strain may synthesize acidic phospholipids with physicochemical properties that are similar to CL. In vitro studies of proteins with synthetic lipid membranes may play a key role in establishing the role of CL in protein localization.
Discussion
In this paper we present a technique for controlling the curvature of membranes in giant E. coli spheroplasts. By removing the outer cell membrane and the layer of peptidoglycan, we created the functional equivalent of a bacterial cell that is “soft” and can be deformed using mechanical forces. The simplicity of the technique will enable the analysis of a large number of proteins that have been described as responding to curvature or that have been observed at the poles or septa of bacterial cells, including DivIVA, MinE, ProP, LacY, and MscS.
The approach enables the study of the effect of membrane curvature on lipid and protein organization, which typically are explored in vitro using micropipettes to aspirate giant unilamellar vesicles (45). Confining spheroplasts in microchambers has two advantages over micropipette aspiration: (i) The maximum curvature of the membrane is confined in one focal plane, which simplifies microscopy and image analysis; and (ii) aspiration is a serial technique, whereas spheroplast confinement makes it possible to study large numbers of samples simultaneously and in parallel.
Our experiments support observations of the localization of CL microdomains at the poles of rod-shaped bacteria in vivo and provide evidence for the hypothesis that this spatial preference is a consequence of geometric and physical constraints of the cell. The polar localization of CL at regions of negative curvature in spheroplast membranes confirms observations by several groups studying CL–protein interactions in E. coli (18, 20) and lipid domain organization in B. subtilis (17, 46). The data provide in vivo experimental support for the models described by Wingreen and coworkers (21, 22). Importantly, this research extends these models to include all the components and complexities of intact cells, including proteins, the chromosome, and protein–lipid interactions in membranes consisting of CL, PG, and PE molecules with a wide variety of tail groups. Refining models using localization data from an in vivo assay avoids technical simplifications that may arise from in vitro experiments. Although no data are available yet on curvature-induced assembly and positioning of CL domains in bacterial membranes in vitro, studies have been carried out with mixtures of eukaryotic lipids. For example, Subramaniam et al. (47) demonstrated that soft polymer surfaces displaying hemispherical protrusions localize lipids in synthetic eukaryotic membranes. Parthasarathy et al. (48) determined that a critical curvature of ∼0.8 μm−1 was required to influence the spatial organization of lipids on topographically patterned inorganic surfaces.
Our measurements of MinD localization in response to curvature confirm experimental and theoretical data by Varma et al. (34). The preferential localization of MinD to the highly curved regions of cells has been explained by a mechanism in which protein multimers sensed the geometry of membranes (34). Another hypothesis is that MinD associates with CL microdomains at curved regions of the cell. In this mechanism, MinD binds to membranes via hydrophobic matching between the acyl chains of CL and hydrophobic domains on the protein (36, 40). The diversity of its four acyl chains may influence CL packing in microdomains and its interaction with proteins (49). Barák et al. (38) described MinD as an amphiphilic protein with a membrane-associated peptide helix and a region of positive charge in contact with the membrane. The amphiphilic and cationic regions of the C terminus of MinD were reported to create hydrophobic and electrostatic interactions that interacted with anionic lipids (36).
In this paper we provide data exploring a mechanism in which CL microdomains function as landmarks to recruit MinD to curved regions of the membrane. Experiments with spheroplast membranes containing reduced amounts of CL relative to wild-type membranes did not alter the localization pattern of MinD significantly. We were unable to detect CL microdomains in Δcls membranes with epifluorescence microscopy; however, the presence of residual CL makes it difficult to rule out this mechanism.
Interactions between CL and MinD may form sites of initiation or nucleation for the MinCDE complex that are consistent with recent experiments on supported lipid bilayer membranes (50). These interactions may present a mechanism for inhibiting polar cell division in vivo. In confined spheroplasts, the Min complex may be associated with CL microdomains at regions of large, negative membrane curvature. We did not observe the dynamic oscillation of MinD-YFP in our experiments with spheroplasts. This result may be explained by the lack of division termination in spheroplasts, arising from stable interactions between CL and the proposed helix-binding domain of MinD, which inhibits its diffusion. Another possibility is that the amount of ATP present is insufficient.
The present study describes a systematic approach to test a mechanism for controlling spatial organization in bacterial cells and supports the hypothesis that geometrical cues influence the position of biomolecules in bacterial cells. Complementing this approach with in vitro techniques will guide the study of protein–lipid interactions in bacteria and play a fundamental role in identifying mechanisms and targets for the development of the next generation of antimicrobial compounds.
Materials and Methods
Spheroplasts were formed by growing E. coli MG1655 (± pFX40) and JW1241 Δcls pFX40 to an OD of ∼0.5–0.7 in modified LB medium, followed by filamentation with 60 μg/mL cephalexin for 3–4 h until an average cell length of 50 μm was reached. Filamented cells were treated with 1.3 mM EDTA and 20 μg/mL lysozyme for 5–10 min to remove the outer membrane and the peptidoglycan layer (28). Spheroplasts were labeled with 400 nM NAO (to visualize CL) for 55 min and 100 nM DAPI (to visualize DNA) for 5 min. The microchambers were created using soft lithography (29). A 3-μL solution of spheroplasts was incubated on top of a 4% agarose slab containing the features for 2 min and then was sealed with a glass coverslip and imaged by fluorescence microscopy. Details of the methods, materials, and procedures used in this study can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Julius Adler, Nick Abbott, Dushko Kuzmanovski, and K. C. Huang for helpful discussions and comments on the manuscript, Sarah Crittenden and Judith Kimble for the use of their Zeiss laser-scanning confocal microscope, Larry Rothfield and K. C. Huang for providing E. coli strain pFX40 (YFP-MinD), and Bill Dowhan and Hiroshi Hara for providing E. coli strain UE54. This work was supported by National Science Foundation Grant DMR-0520527, Grant RGY0069/2008-C from the Human Frontiers Science Program, Grant WIS01366 from the US Department of Agriculture, and by the Searle Scholars Program, 3M, and DuPont.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015757108/-/DCSupplemental.
References
- 1.Théry M, Bornens M. Cell shape and cell division. Curr Opin Cell Biol. 2006;18:648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 3.Montero Llopis P, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebersbach G, Jacobs-Wagner C. Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol. 2007;15:101–108. doi: 10.1016/j.tim.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai EM, Nair U, Phadke ND, Maddock JR. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol Microbiol. 2004;52:1029–1044. doi: 10.1111/j.1365-2958.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 7.Bardy SL, Maddock JR. Polar explorations: Recent insights into the polarity of bacterial proteins. Curr Opin Microbiol. 2007;10:617–623. doi: 10.1016/j.mib.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 10.Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang KC, Ramamurthi KS. Macromolecules that prefer their membranes curvy. Mol Microbiol. 2010;76:822–832. doi: 10.1111/j.1365-2958.2010.07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tocheva EI, Li Z, Jensen GJ. Electron cryotomography. Cold Spring Harb Perspect Biol. 2010;2:a003442. doi: 10.1101/cshperspect.a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowhan W. Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 17.Kawai F, et al. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernal P, Muñoz-Rojas J, Hurtado A, Ramos JL, Segura A. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ Microbiol. 2007;9:1135–1145. doi: 10.1111/j.1462-2920.2006.01236.x. [DOI] [PubMed] [Google Scholar]
- 20.Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM. Protein localization in Escherichia coli cells: Comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J Bacteriol. 2010;192:912–924. doi: 10.1128/JB.00967-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLOS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petit JM, Maftah A, Ratinaud MH, Julien R. 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur J Biochem. 1992;209:267–273. doi: 10.1111/j.1432-1033.1992.tb17285.x. [DOI] [PubMed] [Google Scholar]
- 24.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010;24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabeen MT, et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–1823. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinac B, Buechner M, Delcour AH, Adler J, Kung C. Pressure-sensitive ion channel in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2297–2301. doi: 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia YN, Whitesides GM. Soft lithography. Angew Chem Int Ed. 1998;37:551–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 30.Weibel DB, Diluzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci USA. 2010;107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mileykovskaya E, et al. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001;507:187–190. doi: 10.1016/s0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 33.Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varma A, Huang KC, Young KD. The Min system as a general cell geometry detection mechanism: Branch lengths in Y-shaped Escherichia coli cells affect Min oscillation patterns and division dynamics. J Bacteriol. 2008;190:2106–2117. doi: 10.1128/JB.00720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bramkamp M, van Baarle S. Division site selection in rod-shaped bacteria. Curr Opin Microbiol. 2009;12:683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burn P. Amphitropic proteins: A new class of membrane proteins. Trends Biochem Sci. 1988;13:79–83. doi: 10.1016/0968-0004(88)90043-6. [DOI] [PubMed] [Google Scholar]
- 38.Barák I, Muchová K, Wilkinson AJ, O'Toole PJ, Pavlendová N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol. 2008;68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szeto TH, Rowland SL, Habrukowich CL, King GF. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem. 2003;278:40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- 41.Mileykovskaya E, et al. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 42.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiba Y, et al. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J Bacteriol. 2004;186:6526–6535. doi: 10.1128/JB.186.19.6526-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romantsov T, et al. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- 45.Heinrich M, Tian A, Esposito C, Baumgart T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc Natl Acad Sci USA. 2010;107:7208–7213. doi: 10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 47.Subramaniam AB, Lecuyer S, Ramamurthi KS, Losick R, Stone HA. Particle/Fluid interface replication as a means of producing topographically patterned polydimethylsiloxane surfaces for deposition of lipid bilayers. Adv Mater (Deerfield Beach Fla) 2010;22:2142–2147. doi: 10.1002/adma.200903625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parthasarathy R, Yu CH, Groves JT. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 49.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci USA. 2010;107:8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.