Abstract
Natural killer (NK) cells are a group of innate immune cells that carry out continuous surveillance for the presence of virally infected or cancerous cells. The natural cytotoxicity receptor (NCR) NKp30 is critical for the elimination of a large group of tumor cell types. Although several ligands have been proposed for NKp30, the lack of a conserved structural feature among these ligands and their uncertain physiological relevance has contributed to confusion in the field and hampered a full understanding of the receptor. To gain insights into NKp30 ligand recognition, we have determined the crystal structure of the extracellular domain of human NKp30. The structure displays an I-type Ig-like fold structurally distinct from the other natural cytotoxicity receptors NKp44 and NKp46. Using cytolytic killing assays against a range of tumor cell lines and subsequent peptide epitope mapping of a NKp30 blocking antibody, we have identified a critical ligand binding region on NKp30 involving its F strand. Using different solution binding studies, we show that the N-terminal domain of B7-H6 is sufficient for NKp30 recognition. Mutations on NKp30 further confirm that residues in the vicinity of the F strand, including part of the C strand and the CD loop, affect binding to B7-H6. The structural comparison of NKp30 with CD28 family receptor and ligand complexes also supports the identified ligand binding site. This study provides insights into NKp30 ligand recognition and a framework for a potential family of unidentified ligands.
Keywords: X-ray crystallography, BIAcore binding, NKp30 mutants, antibody epitope mapping
Natural killer (NK) cells are a group of innate immune cells that recognize and destroy transformed cells via a combination of both activating and inhibitory receptors. This combination of positive and negative signaling within NK cells ensures the differentiation between healthy and transformed cells. Once NK cell-mediated cytotoxicity is triggered, perforin and granzyme B are released, resulting in target cell death. Activated NK cells also release chemokines and cytokines, which modulate both the innate and adaptive immune system, leading to a large scale response and control of an infection or tumor. NK cell receptors are made up of molecules from both the Ig superfamily and the C-type lectin family. To date, the most widely studied NK cell receptors are the major histocompatibility complex (MHC) class I recognizing killer Ig-like receptors (KIR) (1–5). In addition, among the major activating receptors, the structure of NKG2D in complex with several ligands, including MHC class I-related chains A and UL 16-binding protein, have allowed a detailed understanding of ligand specificity and NK cell activation (6, 7). In contrast, only limited structural information is available for the natural cytotoxicity receptors NKp30, NKp46, and NKp44 despite their pivotal role in NK cell control of cancerous and virally infected cells (8). In particular, the 3D structure of NKp30 is currently unknown.
NKp30 has been shown to be the dominant activating receptor responsible for the lysis of a number of tumor cell types (9). In addition, NKp30 has been shown to cause the activation and expansion of resting NK cells upon interaction with dendritic cells (DCs) and to cause the death of immature DCs (10). NKp30 has a single extracellular Ig-like domain with a short stalk region (∼5 aa), and a transmembrane domain that associates with CD3ζ homodimers through a transmembrane charged interaction (9). The physiological ligands for NKp30 remain a controversial issue as a large number of NKp30 interacting molecules have been proposed. These include a human cytomegalovirus tegument protein pp65 (11), Duffy-binding-like (DBL)-1α of Plasmodium falciparum erythrocyte membrane protein-1 (PfEMP-1) (12), leukocyte antigen-B-associated transcript 3 (BAT3) (13, 14), and a group of heparan sulfate/heparin molecules (15). Most recently, NKp30 was shown to recognize a B7 family homolog (B7-H6) as a ligand (16). Unlike the other protein ligands proposed, B7-H6 is expressed on a number of tumor cell lines, such as K562 and Raji, as well as on primary cancer cells. However, the lack of structural information for NKp30 makes it difficult to parse out the most important ligand involved in cancer destruction from the proposed ligands.
To facilitate our understanding of NKp30 ligand recognition, we determined the crystal structure of the extracellular domain of human NKp30. Using a structure-based peptide epitope mapping method of a functional blocking antibody of NKp30, we have identified a surface region of the receptor critical for ligand binding and have confirmed the importance of this region with mutational analyses and binding studies with B7-H6.
Results and Discussion
Structure of NKp30.
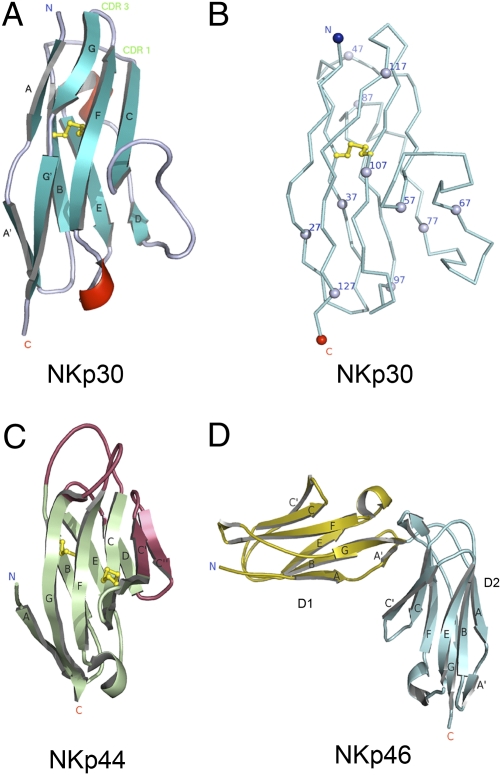
The extracellular domain of human NKp30 was crystallized and its structure was determined by molecular replacement to 1.85 Å resolution (Fig. 1 A and B). Data collection and refinement statistics are shown in Table 1. The final refined structure encompasses residues 18–130 of NKp30, an N-terminal 6xHis tag and six residues from a Tev digestion site (residues ELYFLG). No interpretable electron density was observed for the 6xHis residues. There are two molecules in the asymmetric unit which are highly similar with a root-mean-square deviation (RMSD) of 0.73 Å over 112 NKp30 Cα atoms. The Tev region differs between the two molecules and forms crystal contacts likely aiding crystallization. NKp30 displays an I-type Ig-like topology consisting of eight β-strands forming two antiparallel β-sheets made up of ABED and A′GFC strands, respectively. Compared with that of a V-type Ig structure, it lacks the C′ and C′′ strands that forms the canonical Ig CDR3 region (Fig. 1C) (17, 18). In addition, a short α-helix exists between the D and E β-strands and a short 310 helix bisects the loop between the E and F strands.
Fig. 1.
Structures of natural cytotoxicity receptors. (A) Structure of NKp30 adopts an I-type Ig-fold with β strands labeled and disulphide bonds shown in stick representation colored yellow. The common Ig-fold disulfide bond is formed between Cys-39 and Cys-108. (B) C-alpha ribbon representation of NKp30 with every 10th residue indicated by a sphere. (C) Structure of NKp44 displays a typical V-type Ig-fold with distinct regions different from NKp30 colored in red; disulphide bonds are shown in stick representation colored yellow. (D) Cartoon representation of NKp46 with β strands labeled.
Table 1.
Crystallographic data and refinement statistics
| Space group | P21 |
| Cell dimensions | |
| a | 45.53 |
| b | 53.04 |
| c | 51.85 |
| β | 107.7 |
| No. of observations | |
| Total | 132,338 |
| Unique | 19,666 |
| Completeness | 97.9% (95.6%) |
| I/σI | 150.8 (2.0) |
| Rsym | 0.105 |
| Rcryst | 0.1742 |
| Rfree | 0.2000 |
| Average B factor (Å) | 28.6 |
| RMSD bond lengths (Å) | 0.008 |
| RMSD bond angles (o) | 1.236 |
All data 50–1.85 Å. Last shell data are 1.88–1.85 Å.
Structural Comparison with Ig-Like Immunoreceptors.
Among the three activating natural cytotoxicity receptors, the structure of NKp30 is more similar to the V-type Ig-like NKp44 structure than to the two domain C2-type Ig-like NKp46 structure (Fig. 1) (19–21). In particular, the core region of NKp30 can be superimposed with NKp44 giving an RMSD of 1.68 Å over 77 Cα atoms. The main structural differences between NKp30 and NKp44 are the presence of interactions between the A strand and the G strand of NKp30 (an additional six H bonds) and the presence of a helix between the D and E strands seen in NKp30 but not observed in NKp44. Conversely, the presence of the C′ and C′′ strands and an atypical disulfide bond between the C and C′ strands are seen in NKp44 but not observed in NKp30. In addition, the structure of NKp30 exhibits distinct conformations in the BC and FG loops as well as a dissimilar surface electrostatic potential distribution from that seen in NKp44 (Fig. S1). For example, NKp30 lacks an extensive positively charged groove observed in the region around the C, C′, F, and G strands of NKp44.
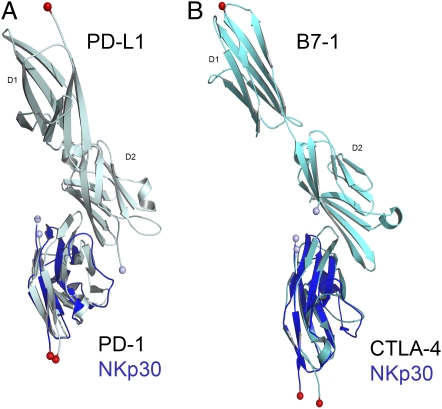
Despite less than 30% sequence similarity, NKp30 displays significant structural homology with programmed cell death 1 Ligand-1 (PDL-1) (RMSD 1.8 Å; PDB:3BIK), programmed cell death-1 (PD-1) (RMSD 2.1 Å; PDB:3BP6), myelin oligodendrocyte glycoprotein (MOG; RMSD 2.2 Å; PDB:1PKQ), coxsackie virus and adenovirus receptor (CAR; RMSD:2.2 Å; PDB:2W9L), T-cell coreceptors CTLA-4 (RMSD 2.6 Å; PDB:1I8L) and CD28 (RMSD 2.6 Å; PDB:1YJD), and also sialoadhesin (RMSD 3.0 Å, PDB:1QFO) (Fig. S2).
NKp30 Dimeric Structure.
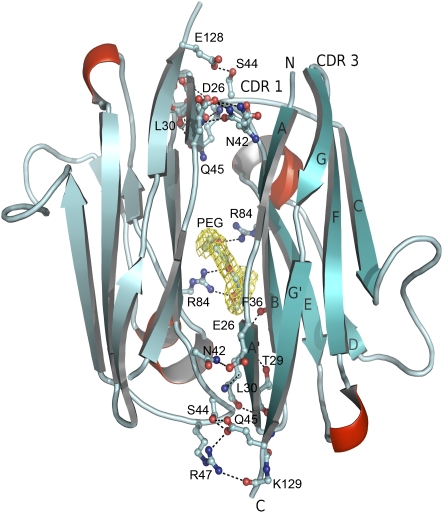
Within the NKp30 crystal, there are three intermolecular contacts observed between NKp30 and adjacent molecules; two of these contacts cover small areas, whereas the third contact area spans 975 Å2 of surface area per monomer as determined by Protein Interfaces Surfaces and Assemblies (PISA) (22) with a surface complementarity of 0.731 (23). This interaction is made up of β strands A, B, and E from both molecules in a twofold symmetrical fashion (Fig. 2). The two molecules dimerize in a head-to-tail orientation with over 20 residues from both molecules interacting via both hydrogen bonds and van der Waals interactions between the two molecules as determined using PDBsum (24). A polyethylene glycol (PEG) molecule is observed at the interface forming hydrogen bonds between Arg84 from two separate NKp30 molecules and six van der Waals contacts with Asp88, Gln90, and Phe36 from molecule 1 and Gln90, Gln92, and Phe36 from molecule 2. The interaction of NKp30 with the PEG molecule may mimic its interaction with the cell membrane. The head-to-tail form of dimerization has been previously observed in a number of protein molecules involved in cell–cell interactions such as the integrin binding domain of ICAM-1 (17). This domain, which is directly involved in integrin binding, is also of the I-set Ig-like structural fold and dimerizes using residues from β strands B, D, and E. It is intriguing to postulate dimerization may occur following ligand binding and thus initiate the intracellular signaling cascade.
Fig. 2.
NKp30 dimer as observed in the crystal, shown in cartoon representation. Interacting residues are displayed in ball and stick representation with hydrogen bonds displayed as dashed lines. The bound PEG molecule is colored in yellow surrounded by 2Fo-Fc electron density map at 1 σ level.
Identification of a Functional Ligand Binding Site on NKp30.
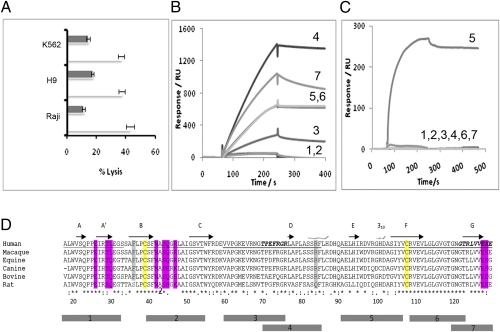
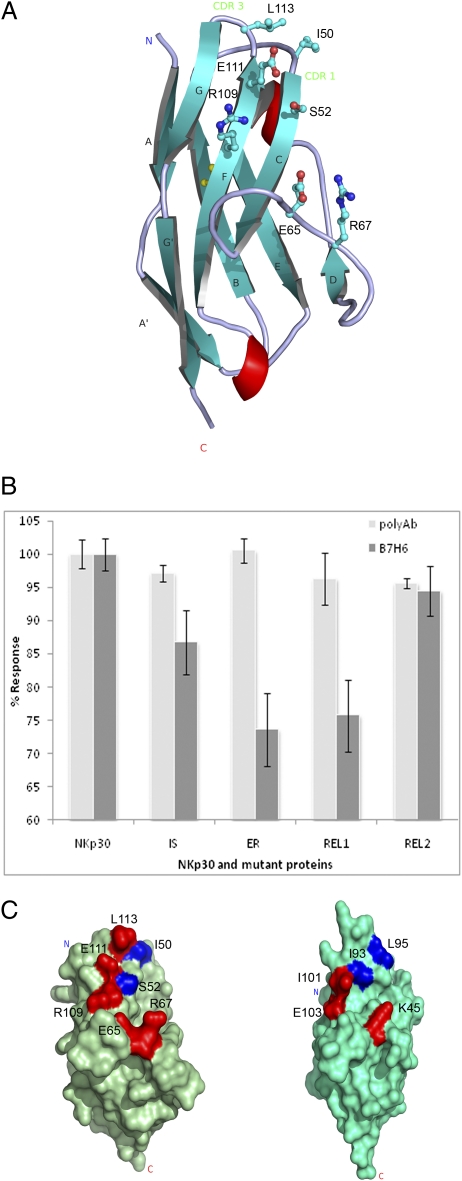
Several ligands have been identified for NKp30 (11–16). Although the physiological significance of these ligands remains to be investigated, the ligands do not share any sequence homology nor a predicted structural fold. The diversity of NKp30 ligands provides little insight into how and where these molecules interact with the receptor. The current structure enabled us to probe its potential ligand binding site on the basis of an assumption that functional blocking antibodies recognize a receptor epitope either overlapping or in the vicinity of the ligand binding site. Among the available NKp30 antibodies, one (clone 210845 from R&D Systems) significantly blocked NK cell lysis of H9, K562, and Raji cells (Fig. 3A). Normally, peptide-based epitope mapping requires the synthesis of hundreds of overlapping short peptides to span the amino acid sequence of the antigen (25). However, the crystal structure of NKp30 provided a clear definition of its secondary structure and surface elements and allowed us to map the receptor surface with a small number of peptides. Specifically, seven NKp30 peptides of 14–17 amino acids in length were synthesized to overlap the unique secondary structural elements of the receptor (Fig. 3D). These peptides span 98 of the total 114 amino acids in the extracellular domain of NKp30. Together, they map the entire solvent accessible surface of the receptor. All peptides, including a polyalanine control peptide, were synthesized with an N-terminal biotinylation modification and the peptide–antibody interactions were examined via surface plasmon resonance solution binding assays using immobilized streptavidin sensor chips. Antibody at 0.4 μM concentration was used as analyte with a flow rate of 20 μL/min. The results showed that the individual peptides were recognized to various degrees (ranging in response from 50 to 1,400 RU) by a goat anti-NKp30 polyclonal antibody, suggesting conformational mimicry exists between the peptides and the receptor (Fig. 3B). Peptide 5 (AELHIRDVRGHDASIYV) was the only peptide that was bound by the monoclonal blocking antibody at any level, resulting in an affinity of 38 nM, with no response observed for the remaining peptides (Fig. 3C). The high affinity, specific recognition of peptide 5 by the monoclonal antibody suggests that the antibody epitope resides in this peptide, residues 91–107, comprising the E strand (residues 91–97), EF loop (residues 98–102), and part of the F strand (103–107) (Fig. 3). The EF-loop region also contains a 310 α-helix (99–102). Interestingly, this region contains seven charged residues and thus is electrostatic rich. Although additional mutational analysis on peptide 5 is needed to further map the blocking antibody epitope within the secondary structure elements of the peptide, the location of peptide 5 is in the vicinity of a common ligand binding site observed in structurally homologous CD28 family receptors, such as CD28, CTLA-4, and PD-1 that all recognize the B7 family of proteins (Table 2) (26–28). Given that the blocking antibody epitope ensures the proximity of the ligand binding site, the involvement of the receptor F-strand region in ligand recognition among the CD28 family receptors suggests NKp30 uses a similar region for its ligand recognition.
Fig. 3.
(A) NK cell cytotoxicity assays against K562, H9, and Raji target cell lines in the presence (black) and absence (white) of an anti-NKp30 antibody, clone 210845 from R&D systems. The E:T ratios are 2.5:1 for K562 and H9, and 5:1 for Raji cells. (B and C) Surface plasmon resonance binding response curves between individually immobilized peptides 1 through 7 and a polyclonal (B) or monoclonal (C) anti-NKp30 antibody. (D) Alignment of human NKp30 protein homologs from multiple species. Typical Ig-fold cysteine residues are shown in yellow, whereas residues involved in dimerization are shown in magenta and Arg84, which interacts with a PEG molecule in the crystal structure shown in gray. The regions corresponding to peptides 1–7 are indicated below the sequences.
Table 2.
Comparison in ligand recognition between NKp30 and CD28 family of receptors
| Receptor | Ig fold | Ligand | Ig fold | Ligand binding site |
| NKp30 | V like | B7-H6 | V + C2 | F–G, C, C′ strands |
| PD-1 | V like | B7-H1/PD-L1 | V + C2 | F–G, C, C′ strands |
| B7-DC/PD-L2 | V + C2 | |||
| CTLA-4 | V like | B7-1 | V + C2 | F–G, C, C′ strands |
| CD28 | V like | B7-2 | V + C2 |
NKp30 Recognizes the N-Terminal IgV Domain of B7-H6.
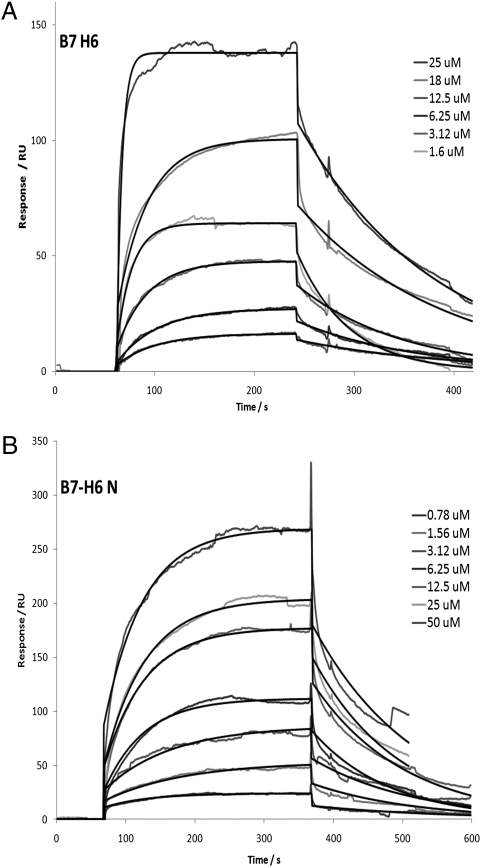
Because B7-H6 is expressed on both K562 and Raji cells used in the blocking assays (16), it is tempting to speculate that the blocking antibody interfered with NKp30 recognition of B7-H6 on K562 and Raji cells. The extracellular region of B7-H6 is made up of two domains, a V-type and a C-type Ig-like domain. To investigate which domain of B7-H6 is involved in NKp30 binding, we expressed both the two domain extracellular B7-H6 receptor and a truncated receptor with only the N-terminal IgV-like domain. Both the IgV only and the two-domain B7-H6 bound to soluble NKp30 with comparable affinities of 2.5 ± 0.7 μM and 3.5 ± 1.3 μM, respectively (Fig. 4), indicating that the N-terminal V domain of B7-H6 is sufficient for NKp30 recognition. This is also similar to CTLA-4 and PD-1; both recognize the N-terminal domain of their respective B7 family ligands.
Fig. 4.
Solution binding between recombinant NKp30 and immobilized B7-H6 molecules. The recombinant B7-H6 protein analyzed was either (A) a single N-terminal V-type Ig-like domain (B7H6 N) or (B) an intact extracellular two-domain construct (B7H6) with the protein concentration for NKp30 shown. Fitting curves determined by BIA evaluation are shown as the black line over the blank adjusted data curves.
Mutations of NKp30 Disrupt B7-H6 Binding.
The resemblance of the NKp30 ligand binding region mapped by the blocking antibody to that of the CD28 receptors and that both recognize the N-terminal V domain of their respective B7 family ligands suggest NKp30 recognizes B7-H6 in a similar mode as CD28 receptor–ligand complexes. To further investigate the ligand binding site on NKp30, we mutated three clusters of residues on the surface of NKp30 in the region of the F strands, C strands, and the CD loop (Fig. 5A). The alanine or serine mutations were designed to alter either surface-charged residues or hydrophobic residues on the receptor. They include two double mutations, I50A/S52A (IS) and E65S/R67S (ER) and two triple mutations, R109S/E111S/L113A (REL1) and R109E/E111R/L113A (REL2). The soluble mutant receptors were expressed and refolded similar to the wild-type NKp30 receptor. To ensure no gross folding defects introduced by the mutagenesis, each mutant protein was assessed by binding to a nonblocking polyclonal anti-NKp30 antibody and all gave highly similar binding. Their binding to immobilized B7-H6, however, varied compared with the wild-type binding data (Fig. 5B). Specifically, charged residues found on the F strand appear to be important for NKp30 binding to B7-H6 as E65S/R67S (ER) and R109S/E111S/L113A (REL1) mutants bound less effectively to B7-H6 than the R109E/E111R/L113A (REL2) mutation, which maintains the overall receptor charge in the F-strand region. Mutations in the C-strand region I50A/S52A (IS) did lower binding as did mutations in the CD-loop region E65S/R67S (ER), illustrating the role of these regions in contributing to B7-H6 binding. Moreover, the region of NKp30 important for B7-H6 binding illustrated by mutagenesis is highly similar to the region of PD-1, important for PD-L1 and PD-L2 binding as judged by similar mutagenesis experiments (Fig. 5C) (29), further demonstrating that NKp30 uses a similar ligand binding site as that observed within the CD28 family of receptors. Interestingly, the F, G, and C strands also form the ligand binding sites in other I-type Ig-like receptors, such as ICAM and Siglec receptors (17, 30, 31). On the basis of these binding and mutational studies of NKp30 and B7-H6, their structural homology with PD-1 and CTLA-4 receptor ligand pairs, and the presence of a conserved structural recognition mechanism among members of the CD28 family receptors and their B7 ligands, we hypothesize that NKp30 may recognize other unidentified homologs of B7-H6 in a similar way to that seen in PD-1 and CTLA-4 ligand recognition (Fig. 6).
Fig. 5.
(A) Structure of NKp30 with mutated residues shown in ball and stick representation. (B) Binding studies of NKp30 and mutant proteins to B7-H6 protein and polyclonal NKp30 antibody (binding was carried out on multiple CM5 chips with replicates measured in each case, n = 5). The NKp30 mutants are designated as I50A/S52A (IS), E65S/R67S (ER), R109S/E111S/L113A (REL1), and R109E/E111R/L113A (REL2). (C) Surface representation of NKp30 and PD-1 (29) with residues highlighted by mutational analyses to have the greatest (colored red) or moderate (blue) effects on ligand binding; the orientation of NKp30 is identical to that shown in A.
Fig. 6.
(A) Structure of PD-L1 in complex with PD-1 with NKp30 overlaid. (B) Structure of B7-1 in complex with CTLA-4 with NKp30 overlaid. N and C termini of each molecule are illustrated by a light blue and red sphere, respectively.
In summary, we have determined the crystal structure of the extracellular domain of human NKp30 at 1.85 Ǻ resolution. The receptor exhibits an I-type Ig-like fold that is similar to the CD28 family receptors. Combining the peptide epitope mapping and mutational analysis, we have identified a ligand binding site near the F-strand face of the receptor. We have also shown that the N-terminal V domain of B7-H6 is sufficient for NKp30 recognition, and we speculate that NKp30 may recognize other B7-H6 homolog proteins in a similar manner to the ligand promiscuity seen within the CD28 family receptors.
Materials and Methods
Purification of Recombinant Proteins.
The extracellular portion of NKp30, residues 18–130, were cloned into a pET 30a vector (New England Biolabs) with an additional N-terminal 6xHis tag and a Tev cleavage site (ENLYFQ). The protein was expressed as inclusion bodies in BL21 (DE3) star Escherichia coli cells (Invitrogen). Inclusion bodies containing 80 mg of NKp30 were resolubilized in 70 mL of 6M guanidine-HCl, 1 mM DTT, and 100 mM Tris-HCl at pH 7.5 and added dropwise into a 4-L solution containing 0.4 M l-arginine, 1 mM oxidized glutathione, and 5 mM reduced glutathione and 0.1 M Tris HCl, pH 8 at 4 °C with vigorous mixing. Following refolding for 2 d, the sample was dialyzed extensively against H2O, loaded onto a 15 mL Ni-NTA affinity column (GE) at a flowrate of 2 mL/min, and eluted over a 300 mM imidazole gradient. The fractions containing NKp30 were concentrated and further purified by gel filtration using a Superdex-200 column (GE) with a running buffer of 50 mM NaCl, 20 mM Tris-HCl at pH 9. NKp30 eluted at a volume indicative of a monomer species. Mutagenesis was carried out using a single oligonucleotide primer encompassing multiple base mutations and using the Quick change procedure. A total of four cluster mutants were made; two of these are double amino acid mutations, I50A/S52A (IS) and E65S/R67S (ER), whereas two are triple amino acid mutations, R109E/E111R/L113A (REL1) and R109S/E111S/L113A (REL2). The mutant NKp30 receptor proteins were expressed and refolded as described for the wild type.
DNA encoding residues 31–144, and 31–317, corresponding to the extracellular V and V + C domains of B7-H6, respectively, were synthesized by Genscript with an additional His6 and Tev cleavage site at the 5′ end of the coding region. Both of these constructs were then cloned into a pET30a vector and protein was expressed using E. coli BL21 (DE3) star cells (Invitrogen). Refolding was carried out as for NKp30 by the rapid-dilution procedure. Protein was purified by Ni-NTA chromatography and gel filtration with B7-H6 proteins eluting at a volume indicative of a monomer species.
Crystallization of Human NKp30.
Wild-type NKp30 was concentrated to 6 mg/mL and dialyzed into 10 mM Tris-HCl at pH 9 for crystallization trials. Initial crystallization screening was carried out using a Mosquito robot system with 0.1 μL mother liquor added to 0.1 μL protein solution using a hanging drop vapor diffusion system over commercially available crystallization screens at both 4 °C and 18 °C. Needle-shaped crystal clusters were observed to grow in the presence of low molecular weight PEG at 18 °C. Following optimization, larger crystals grew in a few days using 1.6 μL of mother liquor containing 40% PEG 200, 1.4% PEG400, 10 mM CaCl2, and 50 mM Hepes pH 7.4, 2 μL of protein followed by addition of 0.4 μL of 0.1 M EDTA.
Data Collection and Structure Determination.
Crystals were transferred into a solution containing mother liquor plus 20% glycerol for 10–20 s before rapid plunging into liquid nitrogen. Data were collected remotely using Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. All data were integrated and scaled using HKL2000 (32). Initial molecular replacement searches with Phaser (33), MolRep (34), and AmoRe (35) using all data did not yield any solution. Data were reprocessed in HKL2000 with a sigma cutoff of 2 during scaling to remove weak reflections that gave a complete dataset to 2.5 Å for use in molecular replacement searches. The structure was solved using Balbes (36) with an optimized search model based on the shark New Antigen Receptor V domain PDB#2COQ (37). Initial Rfree was 0.52 and did not decrease upon rigid body refinement with Refmac5 (38). However, following density modification, Non-Crystallographic Symmetry (NCS) averaging and phase extension to 1.85 Å resolution using DM (39) and Refmac5, the map was readily interpretable. Cycles of model building followed by refinement were carried out using COOT (40) and Refmac5. Final refinement was carried out using phenix.refine (41) and the structure was validated using Molprobity (42). The DALI server was used to identify the homologous structures to NKp30 (43). All figures were generated using PyMol (44).
Target Cells and Natural Cytotoxicity Assays.
Human NK cells were isolated from the peripheral blood of healthy donors using an NK cell isolation kit (Miltenyi Biotech). NK cell purity was assessed by FACS and the cell population was over 98% CD3−CD56+CD16+. IL-2 activated NK cells were cultured as previously described (45). The target cell lines K562 (myleocytic leukemia), H9 (T lymphoblast), and Raji (B lymphocyte; Burkitt's lymphoma) were cultured in RPMI medium supplemented with 10% FBS, 2 mM l-glutamine in a humidified atmosphere of 5% CO2 at 37 °C. Monoclonal anti-NKp30 antibodies used are P30-15 (BioLegend), Z25 (Beckman Coulter), clones 210845 and 210847 (R&D Systems). NK cytotoxicity was determined using a europium release assay (Perkin-Elmer) as previously reported (46). Labeled target cells were transferred into a 96-well V-bottom culture plate (Costar) at 5,000 cells per well density and IL-2 activated primary NK cells were added at a 5:1 or 2.5:1 effector-to-target ratio. In antibody-blocking experiments, NK cells were preincubated with anti-NKp30 antibodies at 1.5 μg/mL for 30 min at 37 °C before incubation with target cells. Clone 210845 but not 210847 from R&D Systems partially blocked the polyclonal NK cells lysis of target cells. After a 2-h incubation at 37 °C, europium release was measured using a time-resolved fluorimeter.
Peptides and Surface Plasmon Resonance Analysis.
NKp30 protein structure was examined and a group of seven biotinylated peptides of 14–17 amino acids in length were synthesized by Thermo Scientific. Peptides are designated as 1, Sheet_A ALWVSQPPEIRTLEGS; 2, BC_loop SFNASQGRLAIGSVT; 3, CDR2 VVPGKEVRNGTPEFRGR; 4, Sheet_D TPEFRGRLAPLASSRF; 5, Sheet_E&F AELHIRDVRGHDASIYV; 6, CDR3 RVEVLGLGVGTGNGT; and 7, Sheet_G AAAGNGTRLVVEKE. A 14-residue control peptide was also synthesized to act as a control during binding assays (SAGGSAAADAAAL). Peptides were immobilized on a streptavidin sensor chip and binding of NKp30 antibody was analyzed by surface plasmon resonance using a Biacore 3000 instrument (GE) at a flow rate of 20 μL/min. The goat antihuman NKp30 polyclonal antibody used to assess peptide binding was purchased from R&D Systems. Binding curves were adjusted by subtraction of the control peptide curve.
The binding of recombinant NKp30 and NKp30 mutants to polyclonal NKp30 antibody and B7-H6 was carried out by immobilizing polyclonal NKp30 antibody or the recombinant B7-H6 proteins corresponding to either the V domain or the V- and C-domain constructs onto a CM5 sensor chip, respectively, and using a constant concentration of either wild-type or mutant NKp30 proteins as analytes. Binding affinities were calculated using a 1:1 Langmuir fitting-based curve using BIAcore BIAevaluation software with an average value determined over the range of concentrations assessed.
Supplementary Material
Acknowledgments
We thank Christine Foster and Julius Chung for preliminary work at the beginning of this project. We also thank Greg Snyder and Tsan Xiao for instruction and the use of a Mosquito crystallization robot for initial high throughput crystallization screening. Use of the Advanced Photon Source was supported by the Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This research is supported by the intramural research funding from the National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors (code 3NOI) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100622108/-/DCSupplemental.
References
- 1.Fan QR, et al. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature. 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 2.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci USA. 1999;96:3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 4.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 5.Tormo J, Natarajan K, Margulies DH, Mariuzza RA. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature. 1999;402:623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 6.Li P, et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 7.Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity. 2001;15:1039–1049. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- 8.Moretta A, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 9.Pende D, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferlazzo G, et al. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnon TI, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 12.Mavoungou E, Held J, Mewono L, Kremsner PG. A Duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. J Infect Dis. 2007;195:1521–1531. doi: 10.1086/515579. [DOI] [PubMed] [Google Scholar]
- 13.Pogge von Strandmann E, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27:965–974. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Simhadri VR, et al. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS ONE. 2008;3:e3377. doi: 10.1371/journal.pone.0003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht ML, et al. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 2009;8:712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 16.Brandt CS, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casasnovas JM, Stehle T, Liu JH, Wang JH, Springer TA. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 19.Foster CE, Colonna M, Sun PD. Crystal structure of the human natural killer (NK) cell activating receptor NKp46 reveals structural relationship to other leukocyte receptor complex immunoreceptors. J Biol Chem. 2003;278:46081–46086. doi: 10.1074/jbc.M308491200. [DOI] [PubMed] [Google Scholar]
- 20.Ponassi M, et al. Structure of the human NK cell triggering receptor NKp46 ectodomain. Biochem Biophys Res Commun. 2003;309:317–323. doi: 10.1016/j.bbrc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Cantoni C, et al. The three-dimensional structure of the human NK cell receptor NKp44, a triggering partner in natural cytotoxicity. Structure. 2003;11:725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37(Database issue):D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geysen HM, Rodda SJ, Mason TJ, Tribbick G, Schoofs PG. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 26.Stamper CC, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001;410:604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- 28.Lin DY, et al. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci USA. 2008;105:3011–3016. doi: 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lázár-Molnár E, et al. Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc Natl Acad Sci USA. 2008;105:10483–10488. doi: 10.1073/pnas.0804453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May AP, Robinson RC, Vinson M, Crocker PR, Jones EY. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhuravleva MA, Trandem K, Sun PD. Structural implications of Siglec-5-mediated sialoglycan recognition. J Mol Biol. 2008;375:437–447. doi: 10.1016/j.jmb.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Processing x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagin AA, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 35.Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 36.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streltsov VA, Carmichael JA, Nuttall SD. Structure of a shark IgNAR antibody variable domain and modeling of an early-developmental isotype. Protein Sci. 2005;14:2901–2909. doi: 10.1110/ps.051709505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Cowtan K. “dm’: An automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter. 2005;42(8) [Google Scholar]
- 42.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLano W. Mannheim, Germany: Schrödinger, LLC; 2008. The PyMOL Molecular Graphics System, Version 1.3. [Google Scholar]
- 45.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- 46.Nagao F, et al. Application of non-radioactive europium (Eu3+) release assay to a measurement of human natural killer activity of healthy and patient populations. Immunol Invest. 1996;25:507–518. doi: 10.3109/08820139609055739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.