Abstract
Because mutations in RAS and BRAF represent the most common mutations found in human tumors, identification of inhibitors has been a major goal. Surprisingly, new oncogenic BRAF specific inhibitors inhibit cells transformed with mutated BRAF but paradoxically stimulate the growth of cells transformed with RAS. Here, we show that the mechanism for activation is via drug-induced dimer formation between CRAF and kinase suppressor of Ras (KSR)1. To understand the function of KSR1, we generated a KSR1 mutant that cannot bind ATP but stabilizes the closed, active conformation of KSR1. Molecular modeling suggested that the mutant stabilizes the two hydrophobic spines critical for the closed active conformation. We, therefore, could use the mutant to discriminate between the scaffold versus kinase functions of KSR1. The KSR1 mutant bound constitutively to RAF and mitogen-activated protein kinase kinase (MEK) but could not reconstitute activity suggesting that the catalytic activity of KSR1 is required for its function. Analogous mutations in BRAF and CRAF allowed us to test the generality of the model. The mutation induced changes consistent with the active, closed conformation of both kinases and confirmed that BRAF functions distinctly from CRAF in the MAP kinase pathway. Not only does this work suggest that KSR1 may function as a kinase, we anticipate that the mutation that we generated may be broadly applicable to stabilize the closed conformation of other kinases many of which may also form dimers.
Keywords: cancer, protein kinase, signal transduction
Mutations in the small G protein RAS and the serine-threonine kinase BRAF (B-Raf) represent the majority of oncogenic mutations in most human cancers including malignant melanoma (1). Although BRAF specific inhibitors have shown promise in the clinic, some of them have a paradoxical effect, inhibiting cells with mutated BRAF but accelerating the growth of cells with mutated RAS (2–4). Recent studies suggest that, in RAS transformed cells, these drugs bind to and induce the closed, active conformation of wild-type BRAF and CRAF (also known as Raf-1, c-Raf or c-Raf-1) (2, 5). Drug binding allows dimers between BRAF and CRAF to form and through a mechanism that is unknown, dimerization results in the activation of CRAF and downstream signaling pathways.
Interestingly, one of the drugs tested, PLX4720, does not induce BRAF/CRAF dimers but can still activate mitogen-activated protein kinase kinase (MEK) and extracellular signal regulated kinase (ERK) in RAS transformed cells (2, 4, 5). This finding suggests that the mechanism of activation might not be related to BRAF/CRAF dimers but to other proteins that bind to the closed active conformation of BRAF and CRAF. Because the scaffold protein, kinase suppressor of Ras (KSR) can form dimers with both RAF isoforms (6, 7), we were interested to examine the role of KSR in BRAF inhibitor induced MEK activation.
KSR was first discovered in Drosophila and Caenorhabditis elegans as a positive effector of the RAS/MAP kinase signaling pathway (8–10). Genetic epistasis experiments place KSR in a position either upstream or parallel with RAF. Although KSR is closely related to RAF (see Supporting Information for an alignment), the absence of the critical catalytic lysine (in mammalian forms of KSR) and the lack of any convincing evidence for in vitro kinase activity (11) has led to the model that KSR functions mainly as a noncatalytic scaffold for the RAS/MAP kinase signaling pathway (11). Recently, it was shown that KSR1, BRAF, and MEK form a ternary complex (6). Based on the symmetric packing of RAF molecules in the crystal structures, Therrien and coworkers suggested that a side-to-side dimer interface, conserved in KSR and in all isoforms of RAF, mediates the ability of RAF to bind with itself or with KSR (7). Because BRAF activation of CRAF requires binding but not kinase activity (2–4), we were interested to explore the role of KSR in this system.
Because genetic and biochemical proof for KSR kinase activity is still lacking, KSR is considered to be a pseudokinase that scaffolds components of the MAP kinase pathway. Mutagenesis strategies that impair kinase catalytic activity, however, result in dynamic structures that also have impaired scaffold activity, making it difficult to distinguish between the scaffold and catalytic function of kinases using traditional mutagenesis approaches. Thus, a mutant that impaired the potential kinase activity of KSR without effecting the scaffolding function would allow the potential kinase activity of KSR to be directly tested.
Here, we found that two different BRAF inhibitors (PLX4720 and GDC0879) induced CRAF/KSR1 dimers. Importantly, we found that the ability of these inhibitors to activate MEK and ERK in RAS transformed cells also required KSR. To distinguish between the scaffold and catalytic activity of KSR, we generated a mutated form of KSR1 that dimerized constitutively with CRAF but could not bind ATP. The failure of this mutant to reconstitute KSR function suggests that the scaffolding function of KSR1 with CRAF is not sufficient for its function. We therefore tested whether KSR1 was a kinase. Although KSR1 exhibited no kinase activity when expressed alone, coexpression and binding of KSR with CRAF resulted in kinase activity for MEK. Our work suggests that KSR1 is a bona fide kinase whose activity is required to cooperate with RAF for the activation of MEK.
Results
BRAF Inhibitors Induce KSR1/RAF Complexes.
For our experiments, we used two different RAF inhibitors, GDC0879 and PLX4720. Although the drugs are structurally unrelated, both drugs were selected for their ability to inhibit a constitutively active form of BRAF (V600E) but also inhibit, at lower affinities, each of the wild-type RAF isoforms (2, 10). Crystallographic studies show that both drugs are type I inhibitors that induce formation of the closed, active conformation of RAF (2, 5). Previous reports show that type I RAF inhibitors induce the formation of BRAF/CRAF complexes supporting dimerization as a potential mechanism for RAF activation (2–4). This proposed mechanism, however, is not supported by the behavior of PLX4720, which does not induce binding of BRAF to CRAF and by data showing that ERK stimulation induced by GDC0879 and PLX4720 does not require BRAF (2–4).
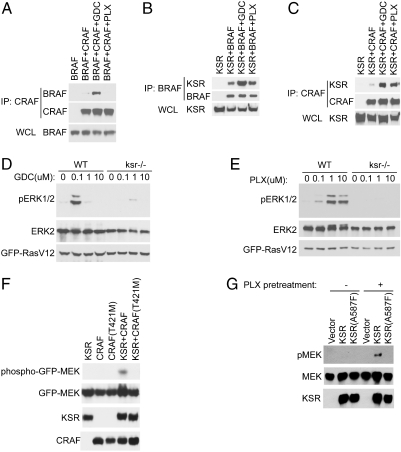
Because KSR1 can also form complexes with BRAF and with CRAF (6, 7), we tested whether RAF inhibitors could enhance formation of complexes between RAF and KSR1. Cells grown in serum, expressing combinations of KSR1, BRAF, and CRAF were treated with both drugs. Coimmunoprecipitations were performed to examine complex formation. As reported previously (2–4), GDC0879 but not PLX4720 induced BRAF/CRAF dimer formation (Fig. 1A). However, both drugs induced complexes between KSR1 and CRAF and enhanced interactions between KSR1 and BRAF (Fig. 1 B and C), which suggested that KSR1/RAF complexes induced by the drug might explain the effects of the type I BRAF specific inhibitors.
Fig. 1.
RAF inhibitors induce dimer formation between KSR and RAF, and activate KSR by CRAF. (A) GDC0879 but not PLX4720 induces BRAF/CRAF dimers. Cells overexpressing myc-CRAF and BRAF were treated with drug for 1 h and CRAF immunoprecipitates were immunoblotted for BRAF and CRAF (epitope tagged with myc). (B) GDC0879 but not PLX4720 enhances KSR/BRAF complexes. KSR immunoprecipitates were prepared from cells overexpressing FLAG-KSR and BRAF after treatment with the indicated drug for 1 h and immunoblotted using antibodies to BRAF. (C) Both GDC0879 and PLX4720 induce KSR/CRAF complexes. KSR immunoprecipitates were prepared from cells overexpressing FLAG-KSR and myc-CRAF after treatment with the indicated drug for 1 h and immunoblotted for CRAF using myc antibodies. (D and E) Requirement of KSR for drug-induced ERK activation. Lysates from wild-type and KSR deficient fibroblasts, transfected with RASV12, were treated with the indicated doses of either GDC-0879 (D) or PLX4720 (E) for 1 h. Lysates were immunoblotted for phospho-ERK1 and 2, ERK2, and RASV12. (F) KSR and CRAF cooperate to activate MEK. Cells expressing the indicated constructs were treated with a 50 uM PLX4720 for 2 h before cell lysates were prepared and analyzed for pMEK by immunoblotting. CRAF(TM) refers to the T421M gatekeeper mutant that cannot bind to the drug(4). (G) KSR in vitro kinase reactions. Cells were cotransfected with WT or ATP binding deficient KSR and CRAF and immunoprecipitates prepared after cells were treated with an activating dose of PLX (10 uM) for 1 h. KSR immunoprecipitates were prepared, pretreated with 50 uM PLX4720 to inhibit coprecipitating RAF activity, and then tested for kinase activity using purified MEK. MEK phosphorylation was detected using a pMEK specific antibody.
BRAF Inhibitor Induced ERK Activation Requires KSR1.
We used KSR1 deficient cells (12) to determine whether KSR was required for the ability of the drugs to induce ERK activation. Whereas mammals have two KSR genes, KSR2 is not expressed in fibroblasts, thus the cell line is deficient for both KSR isoforms (13). Cells transduced with constitutively active RAS(V12) or grown in serum were treated with various doses of each drug. Activation was assessed by immunoblotting cell lysates with an antibody that detects active ERK. As reported previously, treatment of wild-type cells with either drug strongly induced ERK activation at low to intermediate doses but inhibited ERK activation at higher doses (2–4) (Fig. 1 D and E). Similar results were obtained with cells expressing constitutively active RAS (Fig. 1 D and E) or after serum treatment. Strikingly, in KSR deficient cells, ERK activation was significantly attenuated after drug treatment (Fig. 1 D and E), which demonstrates that the ability of PLX4720 and GDC0879 to activate ERK requires the presence of KSR1. Given previous reports demonstrating that CRAF is required for the positive effect of the drugs on ERK activation (2, 4), our data suggest that drug-induced complexes of CRAF and KSR1 may be responsible for MEK/ERK activation.
KSR1 Is a MEK Kinase Activated by CRAF.
We tested the function of the CRAF/KSR1 dimer by coexpressing both proteins with and without treatment with PLX4720 (Fig. 1F) or GDC0879 to induce complex formation. Because drug treatment can induce MEK and ERK activation, we treated cells with an inhibitory dose (50 µM; see Supporting Information) that induces complex formation but should also inhibit CRAF activity. Under these conditions, we found that, when KSR1 was overexpressed with CRAF, MEK activation was induced by drug treatment (Fig. 1F). Because a mutated form of CRAF (CRAF TM) that is unable to bind to the drug (2, 4) did not result in phosphorylation of MEK, this result suggested that induction of the CRAF/KSR1 complex might be important in the activation of MEK.
We tested the possibility that KSR1 might have kinase activity by performing KSR1 in vitro kinase reactions. Consistent with previous reports (11), we failed to detect KSR1 kinase activity in vitro against purified RAF or MEK (Fig. 1G) when KSR was expressed alone. To test whether KSR1 might be activated by CRAF, we coexpressed KSR1 and CRAF and induced dimerization of CRAF with KSR1 by adding a low dose (10 µM) of PLX4720. KSR1 immunoprecipitates were then prepared and tested for kinase activity in vitro. To inhibit any contaminating RAF kinase activity coprecipitating with KSR1, we preincubated the immunoprecipitates with an inhibitory dose of PLX4720 (50 µM). Under these conditions, in vitro kinase activity toward MEK was detected but only after drug treatment (Fig. 1G), which suggests KSR1/CRAF complex formation kinase activity toward MEK. Importantly, no MEK kinase activity was seen using a mutated KSR1 (A587F, described below) that is unable to bind to ATP. We infer that KSR1 has kinase activity that is required for its ability to cooperate with CRAF to phosphorylate MEK.
ATP Binding to KSR1 Is Required for Its Function.
To confirm the ability of KSR1 to function as a kinase, we wanted to generate a kinase-deficient KSR1 mutant. Typically, substitution of the catalytic lysine with arginine or methionine can be used to ablate catalytic activity in most kinases (14). Mammalian KSR1 lacks this catalytic lysine, one reason that KSR1 is considered to be an inactive pseudokinase. In recent years, however, several kinases once considered to be pseudokinases because they lack critical catalytic residues have been shown to be bona fide kinases (15), suggesting that new mutagenesis strategies might be needed to generate true kinase-inactive mutants.
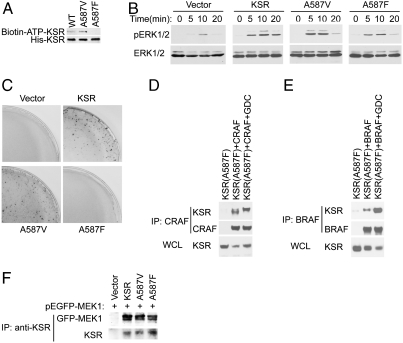
We therefore sought to generate a KSR1 mutant that could not bind ATP and thus could not possess any catalytic activity. Based on the conserved structure of protein kinases, we reasoned that placing a bulky hydrophobic residue in place of the highly conserved alanine (A587) residue, located in the back of the KSR1 ATP binding pocket, might block ATP binding. We tested substitutions of A587 with phenylalanine or valine for ATP binding using a photoactivatable biotin-ATP analog (Fig. 2A). These experiments showed that substitution of A587 with phenylalanine (A587F), but not valine (A587V), disrupted ATP binding.
Fig. 2.
The ability of KSR to bind ATP is required for the function of KSR. (A) Mutagenesis was used to substitute phenylalanine or valine for A587 of mouse 6xHis-KSR1. Each mutant was expressed in cells, purified using Ni2+-agarose and tested for ATP binding using a biotinylated-ATP analog after UV cross-linking and immunoblotting for the presence of biotin. (B) KSR deficient fibroblasts were reconstituted with YFP fused to wild-type or mutated KSR and sorted to generate cell lines with similar expression levels (see Supporting Information). Cells were stimulated with EGF for the indicated times and cell lysates were immunoblotted with an antibody to phosphorylated ERK (pERK). (C) Stably transfected KSR deficient cell lines, described in B, were transfected with an expression vector for RASV12 and assessed for transformed colony formation. (D) Constitutive KSR/CRAF complexes induced by A587F. FLAG-KSR immunoprecipitates were prepared from lysates from cells expressing A587F FLAG-KSR with myc-CRAF and immunoblotted for CRAF (myc). This experiment was performed with Fig. 1D and is directly comparable. The complete blot is shown in Supporting Information. (E) A587F KSR mutation does not affect basal binding to BRAF. Experiment was performed as described in E, except BRAF was used instead of CRAF. (F) A587F KSR mutation does not affect MEK binding. KSR immunoprecipitates from cell coexpressing GFP-MEK1 and WT or KSR mutants were immunoblotted for GFP-MEK and KSR (FLAG).
The function of the ATP binding deficient (A587F) KSR1 mutant was tested by reconstituting KSR1 deficient cells with either wild-type or one of the two KSR1 mutants, A587F or A587V. Because expression levels can affect the function of KSR, we monitored expression using a KSR1-YFP fusion protein and isolated stable cell lines with equivalent levels of KSR1-YFP expression using cell sorting (see Supporting Information). KSR1 function was tested by measuring ERK activation after EGF treatment. Whereas wild-type KSR1 and the ATP binding A587V mutant were both able to rescue ERK activation, the ATP binding deficient A587F mutant did not (Fig. 2B). Because cell transformation by constitutively active RAS requires KSR, we confirmed the function of the KSR1 mutants using a RAS transformation assay (Fig. 2C). Cell lines generated above were transduced with RASV12 and cell transformation assessed by focus formation (Fig. 2C). The wild-type and A587V mutants supported RASV12 transformation, but the A587F mutant could not. Lastly, we confirmed the function of the mutants using Drosophila KSR and RAF. Unlike mammalian KSR which has no effect when overexpressed, overexpression of Drosophila KSR (dKSR) with Drosophila RAF (dRAF) results in strong MEK activation (7). Valine and phenylalanine mutations were generated in the analogous alanine residue (A703) in dKSR and each mutant was coexpressed with dRAF in Drosophila S2 cells. Although the wild-type and the A703V mutant were able to strongly activate MEK, substitution with phenylalanine abrogated the effect (see Supporting Information).
As described above (Fig. 1G), the KSR1 A587F mutant was tested for its ability to cooperate with CRAF to phosphorylate MEK. Whereas wild-type KSR1 immunoprecipitates showed kinase activity toward MEK, no kinase activity was detected in KSR A587F immunoprecipitates. Thus, replacement of alanine 587 of KSR1 with phenylalanine disrupts ATP binding and abrogates KSR1 function potentially via the abrogation of KSR1 kinase activity.
KSR1 A587F Mutant Binds Constitutively to CRAF.
Kinases have catalytic and scaffold functions. Because the scaffold function of KSR is defined by its ability to bind RAF (6, 7) and to bind MEK, we tested the KSR1 A587F mutant for its ability to bind RAF (Fig. 2 D and E) and MEK (Fig. 2F). Complexes between KSR1 and RAF were measured by comparing the amount of RAF that coimmunoprecipitated with wild-type or mutated KSR1 (Fig. 2 E and F). Although little to no CRAF coprecipitated with wild-type KSR1, the A587F mutant formed constitutive complexes with CRAF. In contrast, the A587F mutation had little to no effect on the level of complex formation with BRAF presumably because of the high basal levels of KSR/BRAF complexes (Fig. 2). Similarly, the A703F mutation in dKSR strongly enhanced the stability of dKSR/dRAF complexes (see Supporting Information). The specific effect of the A587F mutant on CRAF and not BRAF supports the idea that the CRAF/KSR1 complex may have a function distinct from the BRAF/KSR1 complex. Lastly, the A587F mutation did not affect KSR1 binding to MEK in mammalian (Fig. 2F). Thus, the two known scaffold functions of KSR1 are preserved.
Molecular Modeling Suggests That the A587F Mutation Induces the Closed, Active Conformation of KSR.
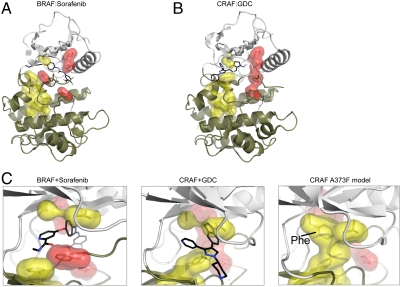
The ability of the A587F mutant of KSR1 to induce constitutive binding to CRAF suggested that the phenylalanine substitution might be affecting the conformation of the kinase domain of KSR1 by stabilizing the dimer interface. A recent study of features conserved in the structures of active kinases and not present in the structures of inactive kinases suggests that kinase activation involves the formation of two hydrophobic spines, the catalytic and regulatory hydrophobic spines (16) (Fig. 3A). The formation of these two hydrophobic spines during the process of kinase activation serves to generate a hydrophobic core that stabilizes the active conformation of the kinase. In the catalytic hydrophobic spine of protein kinase A, a conserved alanine (A70) from the upper lobe and a conserved leucine (L173) from the lower lobe interact with the top and bottom of the adenine ring from ATP to bring the two lobes of the kinase together. Alanine 587 of KSR1 corresponds to the conserved alanine (A70 of PKA) from the upper lobe and the leucine in the lower lobe (173 of PKA) corresponds to phenylalanine (690) of KSR1.
Fig. 3.
Modeling the structural effects of the alanine to phenylalanine change in CRAF and BRAF. The position of residues constituting the hydrophobic spines of CRAF crystallized with a type I inhibitor (stabilizes the closed and ATP bound form of the kinase) are shown in B, and the hydrophobic spine residues in BRAF bound to a type II inhibitor (binds to the open conformation preventing closing of the cleft) is shown in A. Components of the catalytic hydrophobic spine are shown in yellow, whereas components of the regulatory hydrophobic spine are shown in red. Note the contiguous residues of red and yellow induced by the type I inhibitor in B, whereas the pattern of these residues is interrupted in A. Note also how the drug molecule in B functions to connect components of the catalytic hydrophobic spine in the upper and lower lobes of the kinase. In C, a simulated structure of CRAF where A373 is replaced with Phe is shown (Right). Energy minimization was done using the program TINKER. The minimized structure is rotated 90° in D to show more clearly how the C spine is stabilized and completed by the Phe mutation. For each structure, the N lobe is shown in white and the C lobe in tan.
We first analyzed the published structure of CRAF bound to GDC0879(2), a type I inhibitor, and confirmed that drug binding resulted in the formation of both the catalytic and regulatory spines (Fig. 3). In contrast, analysis of a structure of BRAF complexed with Sorafenib (17), a type II inhibitor, was consistent with an inactive kinase without assembly of either of the hydrophobic spines (Fig. 3). Using energy minimization modeling, the structure of CRAF with alanine replaced by phenylalanine was modeled. The results showed that the phenylalanine residue in CRAF position 373 interacts with phenylalanine 475 in the lower lobe completing the catalytic hydrophobic spine and stabilizing the closed, active conformation of the kinase (Fig. 3). This model suggested that the A587F mutant of KSR1 results in a true pseudokinase that is in the active conformation but catalytically inert because it can no longer bind to ATP.
Analogous A to F Mutations in BRAF and CRAF Induce Dimer Formation.
To test the generality of this hypothesis, we generated analogous mutations in BRAF and CRAF. Coimmunoprecipitation assays showed that BRAF A481F bound constitutively to CRAF and that CRAF A373F bound constitutively to BRAF (Fig. 4A). The CRAF A373F mutant also bound constitutively with KSR1 but the BRAF A481F mutant did not appear to enhance the basal association with KSR1 (Fig. 4B).
Fig. 4.
A to F mutation in RAF induce dimmer formation and activate ERK signaling. (A) Constitutive CRAF/BRAF dimer formation induced by A to F mutation. The CRAF A373F and the BRAF A481F mutants were coexpressed with wild-type BRAF or wild-type CRAF(myc), respectively, and heterodimers assessed by coimmunoprecipitation. (B) The CRAF but not the BRAF A to F mutation induces constitutive KSR complexes. The CRAF A373F(myc) and the BRAF A481F mutants were coexpressed with wild-type KSR (FLAG) and heterodimers assessed by coimmunoprecipitation. (C) Expression of BRAF A481F stimulates RAS independent ERK activation in cells. Cells were transiently transfected with expression constructs for BRAF A481F, CRAF A373F(myc), or KSR A587F(FLAG) mutants. Lysates were immunoblotted with antibodies to pERK after 18 h. Coexpression of dominant negative RAS (N17) was to determine RAS dependence. (D) WT and ksr-/- mouse embryonic fibroblasts (MEFs) were transfected with BRAF(A481F). Total ERK and pERK were assessed by immunoblotting. (E) WT and ksr-/- MEFs were transfected with BRAF(V600E) and prepared as described in D.
Because the AF mutants appear to induce the closed, active conformation of all three kinases, we reasoned that we could use these mutants to distinguish between their functions as enzymes or as scaffolds. All three AF mutants (BRAF, CRAF, and KSR1) were overexpressed in cells and tested for their effects on endogenous ERK activation (Fig. 4C). Consistent with previous work showing that kinase-inactive forms of BRAF can stimulate the activation of MEK and ERK in the presence of active RAS (3, 17), overexpression of the BRAF A481F mutant resulted in constitutive activation of ERK. However, coexpression of dominant negative RAS (N17) showed that ability of the BRAF A481F mutant to activate ERK was RAS independent (Fig. 4C). The RAS independence of A481F BRAF thus resembles the V600E mutant of BRAF and suggests that the greatly increased kinase activity of BRAF V600E (17) may not be the only reason it is oncogenic. Rather, it suggests that the scaffold function of BRAF and not its kinase activity may be required for its transforming activity.
Finally, we tested whether ERK activation by BRAF A481F or BRAF V600E required KSR1 by expressing each construct in the KSR1 deficient cell line (Fig. 4 D and E). The ability of both proteins to activate ERK was significantly compromised in the absence of KSR1, which supports the idea that the mechanism of action of both A481F and V600E are similar and dependent on the presence of KSR1. In contrast, overexpression of CRAF A373F or KSR A587F had no effect on ERK activation (Fig. 4C). As both mutants form complexes with each other, these results suggest that ERK activation requires that both proteins be enzymatically active.
Discussion
Understanding how the MAP kinase signaling complex functions has been particularly challenging given that there are at least three kinases in the cascade and an even larger number of components identified by genetic epistasis whose function is still unknown (18, 19). Although the canonical pathway involving RAS, RAF, MEK, and ERK has been known for over a decade, important details about the mechanism of activation are still unknown, especially regarding the role of KSR and the function of the different RAF isoforms. Our data suggest that KSR plays an integral role as a kinase upstream of MEK activation.
Recent data suggest that the roles of the three RAF isoforms, ARAF (A-Raf), BRAF, and CRAF, are more complex than initially thought (20). BRAF and CRAF are widely expressed and expressed together in most cells, whereas ARAF expression is restricted mainly to germ cells (21). Originally, each RAF isoform was thought to phosphorylate MEK independently. Recent studies, however suggest that the RAF isoforms have a hierarchy, with BRAF able to activate CRAF but not the other way around (17, 22). By a mechanism that does not require kinase activity, association of BRAF with CRAF induces the activation of CRAF (3, 17, 22). This discovery is based on the finding that oncogenic forms of BRAF that lack kinase activity can still drive activation of the pathway via activation of CRAF (3). The function of these catalytically impaired mutants, however, requires RAS and CRAF. RAS functions presumably to induce the active conformation of BRAF whereas CRAF is required to convey the signal downstream. The mechanism of CRAF activation is not known but could be either through an allosteric mechanism or by the recruitment of accessory proteins associated with BRAF (or KSR) to modify and activate CRAF. In contrast, oncogenic forms of BRAF that have enhanced kinase activity like the V600E mutant are both CRAF and RAS independent (22) suggesting that they can directly phosphorylate and activate MEK.
We found that both kinase-active and kinase-inactive BRAF mutants required KSR1 for their function, suggesting that, regardless of whether BRAF bypasses CRAF to phosphorylate MEK (V600E) or whether it activates MEK through activation of CRAF (kinase-dead BRAF), it still requires KSR1. One possible explanation is that KSR1 functions to bring both MEK and BRAF to CRAF. Because MEK and BRAF binding to KSR1 are constitutive (6), activation of the pathway may involve the induced recruitment of CRAF. Complex and perhaps, dynamic, kinase-substrate interactions between BRAF, CRAF, KSR1, and MEK underlie a complicated activation mechanism that we do not yet fully understand. Understanding this stoichiometry is clearly an important future challenge.
Our study extends previous studies using BRAF specific inhibitors to reveal insights into the mechanism of RAF activation. Because RAF is downstream of RAS, it was originally expected that RAF specific inhibitors would inhibit the downstream activity of oncogenic forms of RAS. Because catalytically inactive BRAF can activate CRAF, it was plausible to suggest that drug inhibition of BRAF could explain the activity of the drugs (2–4). But the stimulatory activity of the drugs does not require BRAF, and not all of the drugs induce BRAF/CRAF dimers (2, 4). The drugs can induce and activate CRAF/CRAF homodimers explaining how they might activate ERK in the absence of BRAF (2, 4). Not tested in these previous studies, however, was the role of KSR. Our finding that (i) the positive activities of the drugs requires KSR1, that (ii) they specifically induce KSR1/CRAF complexes, and (iii) combined with previous studies showing that CRAF is essential (2, 4) suggest that the induced KSR1/CRAF dimer explains how the drugs activate ERK.
Our data suggest that MEK phosphorylation by CRAF requires KSR1 catalytic activity. Because the known inhibitory concentration of PLX4032 is in the nanomolar range (2, 4), the concentration of the drug that we used to inhibit contaminating RAF (50 µM) should be sufficient to block RAF activity, which suggests that KSR may function as a kinase downstream of CRAF. It is also possible, however, that binding of KSR to CRAF inhibits binding of the drug to CRAF leading to incomplete inhibition. Lastly, it is possible that KSR may phosphorylate MEK and this phosphorylation facilitates CRAF phosphorylation of the activation loop.
By mutating the conserved Ala in the catalytic spine to Phe in KSR, CRAF, and BRAF, we created an adenine mimetic that stabilizes the closed conformation of the kinase core that induces the dimer interface but renders the kinase inactive. The pseudokinases that we generated assume a conformation that resembles the active kinase, but because they cannot bind ATP, they are unambiguously catalytically dead. Previously known strategies to inactivate kinase activity result in dynamic structures with potentially impaired scaffolding function.
Because some of the scaffolding functions of kinases require the active conformation, our mutant is unique because it stabilized the scaffolding function, which allowed us to use the mutants to separate the scaffolding properties of BRAF, CRAF, and KSR from their catalytic activity. In the case of BRAF, the A481F mutant was able to constitutively activate MEK and ERK in a manner that was independent of kinase activity and RAS but KSR dependent. The RAS independence suggests that both the V600E and A481F mutations uncouple the inhibitory amino-terminal domain from the kinase domain. However, because the AF mutant lacks catalytic activity, one implication is that the scaffolding and not the kinase function of the BRAF V600E mutant might be sufficient to account for its transformation activity.
Since its identification 15 y ago, the exact function of KSR has been controversial. Although there have been claims that KSR functions as a kinase (23, 24), most studies have failed to show convincing evidence, resulting in the consensus that KSR most likely functions as a scaffold (25). Because KSR binds to RAF, MEK, and ERK, it can clearly function as a scaffold, but separating its potential role as a kinase from its role as a scaffold has been difficult to parse genetically. Complementation studies with “kinase-dead” mutants of KSR in C. elegans and Drosophila have given different results (26, 27). Our A587F mutant of KSR that still retains scaffolding function, because it can bind with BRAF, CRAF, and MEK, allowed us to separate these two functions of KSR and establish clearly that ATP binding and kinase activity are required for KSR function.
Our work supports the idea that eukaryotic protein kinases evolved to be highly sophisticated allosteric proteins that function as sensitive and dynamic molecular switches functioning both as catalysts and as scaffolds. Using a mutant that induces the closed, active conformation but is catalytically active allowed us to separate these two different functions of kinases. Whereas BRAF could function as a scaffold alone, the requirement for both CRAF and KSR to bind to ATP for downstream activation of MEK and ERK suggest that both are kinases and have distinct functions from BRAF. Because all of the RAF isoforms are capable of phosphorylating MEK, the significance of KSR’s ability to phosphorylate MEK is unclear. Our work does suggest that understanding the effect of Raf inhibitors will require a better knowledge of Raf and KSR interactions. Given the long coevolutionary history between KSR and RAF, we suspect that these complex interactions allowed for important regulatory controls for this highly conserved and ubiquitous signaling pathway.
Materials and Methods
Chemicals
PLX4720 and GDC0879 were purchased from Selleck Chemicals.
Antibodies
Phospho-ERK (T202-Y204) and phospho-MEK (S217/S221) antibodies were purchased from Cell Signaling.
Dimerization Experiments
For most experiments, constructs for wild-type and mutated BRAF, CRAF, and KSR1 were appended with epitope tags (FLAG, 6xHis, Myc, GFP) and were expressed by transient transfection into 293 T cells. Cells were lysed in a buffer containing 1% Nonidet P-40 (NP40) and 0.1% deoxycholate. Cells were pretreated with drugs for 1 h prior to lysis. Immunoprecipitates were analyzed by gel electrophoresis and immunoblotted after transfer to nitrocellulose membranes using standard methods.
ATP Binding Assay
WT and mutated KSR1 constructs epitope tagged with 6xHis were expressed in 293 T cells and purified using Ni-nitrilotriacetate agarose. ATP binding was assessed by incubating the samples with 100 µM biotin-azido-ATP (Affinity Probes) in a buffer containing 20 mM Na2HPO4/NaH2PO4 (pH 7.2) and 10 mM MgCl2. After incubation on ice for 5 min, samples were UV irradiated for 2 min. ATP-cross-linked KSR mutants were examined by SDS-PAGE and Western blotting with strepavidin-HRP.
Kinase Reactions
Cells transfected with various constructs were treated or not with PLX4720 for 1–2 h. Cells were lysed with 1% NP40 and immunoprecipitates prepared. In vitro kinase reactions were performed in a standard buffer with 10 mM MgCl2, with 1 µg of kinase-dead MEK, and 100 µM cold ATP. To inhibit contaminating Raf activity, 50 µM PLX4720 was preincubated with some reactions.
Supplementary Material
Acknowledgments.
We thank Kevan Shokat, Richard Marais, Olga Lubman, and Daved Fremont for discussions and suggestions about how to generate the ATP binding defective KSR1 mutant; Shiva Malek and Richard Marais for helpful discussions; and Marc Therrien for providing reagents. The work was supported by The Howard Hughes Medical Institute and initially by the Washington University/Pfizer Biomedical agreement.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102554108/-/DCSupplemental.
References
- 1.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 2.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 3.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci USA. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 9.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 10.Therrien M, et al. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 11.Michaud NR, et al. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen A, et al. Kinase suppressor of Ras (KSR) is a scaffold which facilitates mitogen-activated protein kinase activation in vivo. Mol Cell Biol. 2002;22:3035–3045. doi: 10.1128/MCB.22.9.3035-3045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo-Garvey DL, et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009;10:366–378. doi: 10.1016/j.cmet.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs CS, Knighton DR, Sowadski JM, Taylor SS, Zoller MJ. Systematic mutational analysis of cAMP-dependent protein kinase identifies unregulated catalytic subunits and defines regions important for the recognition of the regulatory subunit. J Biol Chem. 1992;267:4806–4814. [PubMed] [Google Scholar]
- 15.Taylor SS, Kornev AP. Yet another “active” pseudokinase, Erb3. Proc Natl Acad Sci USA. 2010;107:8047–8048. doi: 10.1073/pnas.1003436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 18.Rubin GM, et al. Signal transduction downstream from Ras in Drosophila. Cold Spring Harb Symp Quant Biol. 1997;62:347–352. [PubMed] [Google Scholar]
- 19.Therrien M, Morrison DK, Wong AM, Rubin GM. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics. 2000;156:1231–1242. doi: 10.1093/genetics/156.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr Opin Genet Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Niault TS, Baccarini M. Targets of Raf in tumorigenesis. Carcinogenesis. 2010;31:1165–1174. doi: 10.1093/carcin/bgp337. [DOI] [PubMed] [Google Scholar]
- 22.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Kolesnick R, Xing HR. Inflammatory bowel disease reveals the kinase activity of KSR1. J Clin Invest. 2004;114:1233–1237. doi: 10.1172/JCI23441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing HR, Kolesnick R. Kinase suppressor of Ras signals through Thr269 of c-Raf-1. J Biol Chem. 2001;276:9733–9741. doi: 10.1074/jbc.M008096200. [DOI] [PubMed] [Google Scholar]
- 25.Morrison DK. KSR: A MAPK scaffold of the Ras pathway? J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 26.Stewart S, et al. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.