Abstract
Mutations in ALS2, carrying three putative guanine exchange factor (GEF) domains, are causative for a juvenile, autosomal recessive form of amyotrophic lateral sclerosis (ALS), primary lateral sclerosis, and infantile-ascending hereditary spastic paralysis. Endogenous ALS2 is shown here to be enriched in nervous tissue and to be peripherally bound to the cytoplasmic face of endosomal membranes, an association that requires the amino-terminal “RCC1 (regulator of chromatin condensation)-like” GEF domain. Disease-causing mutants and a naturally truncated isoform of ALS2 are shown to be rapidly degraded when expressed in cultured human cells, including lymphocytes derived from patients with ALS2 mutations. Thus, mutations in the ALS2 gene linked to early-onset motor neuron disease uniformly produce loss of activity through decreased protein stability of this endosomal GEF.
ALS2 was originally described as a chronic juvenile autosomal recessive form of amyotrophic lateral sclerosis (ALS) discovered in a large consanguineous Tunisian family. The clinical features were progressive spasticity of limb and facial muscles, bulbar or pseudobulbar symptoms, and distal muscle weakness and atrophy, suggesting the degeneration of upper motor neurons (1). In some cases, involvement of lower motor neurons was reported. Recently, mutations in the ALS2 gene were identified as causative for this disease (2, 3), and a growing number of genetically heterogeneous progressive neurodegenerative disorders, including a recessively inherited juvenile form of primary lateral sclerosis in Kuwaiti and Saudi Arabian families (3) and infantile-ascending hereditary spastic paralysis in French, Italian, Algerian, Pakistani, and Israeli families (4–6). To date, nine homozygous ALS2 mutations from nine independent families have been identified. All of these mutations result in predicted premature translation termination caused by the recessive frameshift or nonsense mutation.
The ALS2 gene is encoded by 34 exons and has been reported to produce at least two splice variants, a long form (6.5-kb transcript) and a short form (2.6-kb transcript), in a variety of human tissues including the brain and spinal cord. The ALS2 polypeptide (also referred to as Alsin) is predicted to be 184 kDa and encode three putative guanine exchange factor (GEF) domains. These include an amino-terminal region that shares homology with RCC1 (regulator of chromatin condensation), a known GEF for the small GTPase Ran (7); a centrally located tandem organization of Dbl homology and pleckstrin homology (PH) domains characteristic of GEFs for the Rho GTPase family (8); and a carboxyl-terminal vacuolar protein sorting 9 (VPS9) domain that is also found in GEFs for the GTPase Rab5 (9, 10). There are also seven membrane occupation and recognition nexus (MORN) repeats (11). Furthermore, the presence of both the PH domain and the MORN repeats suggest that ALS2 is associated with intracellular membranes. In vitro the VPS9 domain can interact with the endosome-associated small G protein Rab5a, and ALS2 with or without an intact amino-terminal RCC1 domain can act as a GEF for Rab5a (12). When expressed at high levels, immunolocalization of full-length ALS2 has further suggested an association with endosomal membranes mediated by the carboxyl-terminal VPS9 domain (12).
We have now determined that although enriched in neural tissues endogenous ALS2 is a low abundance protein that reversibly associates with the cytoplasmic face of endosomal membranes, an interaction requiring an intact amino-terminal RCC1 domain. Moreover, the predicted protein derived from the short 2.5-kb transcript of ALS2 and the disease-associated mutant forms are inherently unstable and are targeted for rapid proteasome-mediated degradation in cultured human cells, including transformed lymphocytes derived from patients with ALS2 mutations.
Materials and Methods
Plasmids. Full-length ALS2 cDNA was assembled by using the cDNA clone KIAA1563, containing nucleotides 1034–6271 of the 6.5-kb ALS2 transcript (obtained from T. Nagase, Kazusa DNA Research Institute, Kisarazu, Japan). The remaining 5′ portion of the long form and ALS2 short-form cDNA were cloned by RT-PCR of mRNA extracted from HeLa cells.
Mammalian expression constructs used for this study were pCIneoFL, which was constructed by insertion of the FLAG tag into pCIneo vector (Promega), effectively generating cDNAs fused in-frame with an amino-terminal FLAG tag, pCIneoFL-ALS2 long form (WT) and ALS2 short form, disease-associated mutants of ALS2 [pCIneoFL-T475fs (frameshift), V491fs, L623fs, N846fs, M1207st (stop codon), and V1574fs], and amino-terminal truncated mutants [pCIneoFL-ΔRCC1–1 (amino acids 264-1657), ΔRCC1–2 (amino acids 459-1657), and ΔRCC1–3 (amino acids 608-1657)]. Details are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. For baculoviral expression, FLAG-tagged ALS2 was cloned into pFastBacHT, and recombinant baculovirus was generated according to the protocol provided by Invitrogen. His-FLAG double-tagged ALS2 was produced as described (13).
Antibodies. The polyclonal antibody pAb-ALS21082 was generated against a synthetic peptide (DGYGEYRIPNKAMNKEDHYC), of which the 18 amino-terminal amino acids correspond to residues 1082–1100 of human ALS2. This antibody was conjugated to keyhole limpet hemocyanin and affinity-purified by binding to the peptide coupled to Sulfolink (Pierce). For pAb-ALS2RCC1, a portion of ALS2 (amino acids 1–617) was expressed in bacteria as an amino-terminal His-tag fusion, purified, and used as an immunogen in rabbits. Antibody was affinity-purified by binding to the antigen coupled to CNBr-activated Sepharose 4B (Amersham Pharmacia Biosciences). These antibodies were used at 1:600 dilution for immunoblot and 1:250 for immunofluorescence study. Other antibodies used in this study are listed in Supporting Materials and Methods.
Cell Culture, Transfection, Preparation of Cell Lysates and Tissue Extract, Immunoblot, and Immunofluorescence Microscopy. Details of these methods are available in Supporting Materials and Methods.
Subcellular Fractionation, Sucrose Gradient, Protease Protection Assay, and Alkali Extraction. Details of these methods are available in Supporting Materials and Methods. For the sucrose gradient, total membranes (P100) purified from rat cerebral cortex were adjusted to 40% sucrose and loaded on the bottom of a one-step sucrose gradient as described (14). Both the protease protection assay and alkali extraction were performed as described (15) with minor modifications.
Assessment of ALS2 Protein Stability. For assessment of protein stability, HEK293 cells were transiently transfected with ALS2 constructs. Twenty-four hours after transfection, 100-mm dishes of transfected HEK293 cells were split into two 60-mm dishes, then incubated an additional 24 h. These dishes were treated with or without 20 μM MG132 (Calbiochem) for 10 h before analysis. For pulse–chase experiments, 24 h posttransfection, 100-mm dishes of transiently transfected HEK293 cells were subdivided into four wells of a six-well dish. After an additional 24-h incubation, cultures were metabolically radiolabeled for 45 min with 0.3 mCi of35S methionine/cysteine per ml of medium by using the Easy Tag cell labeling kit (Perkin–Elmer). Cells were harvested immediately or at 3, 6, and 24 h after the addition of radiolabel. Lysates from labeled cells were prepared in the same condition as described above, then an equal amount of radiolabel from each extract was incubated with anti-FLAG M2 beads (Sigma) and resolved by means of SDS/PAGE. Each radiolabeled ALS2 band was quantified by Phosphor-Imaging.
Results
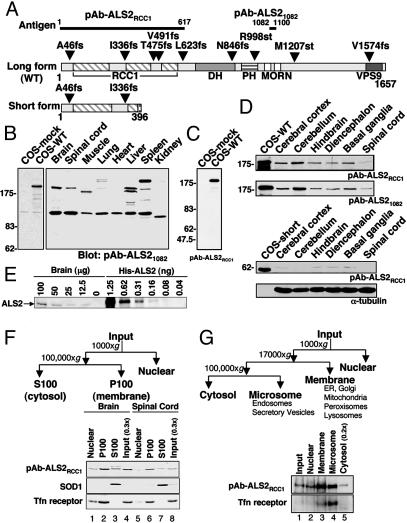
The 184-kDa ALS2 Is Rare Even in the Neuronal Tissues in Which It Is Enriched. To identify the accumulated ALS2 isoforms and the tissue types where they are expressed, two rabbit polyclonal antibodies (pAb-ALS2RCC1, pAb-ALS21082) were raised (Fig. 1A). After affinity purification, pAb-ALS21082 and pAb-ALS2RCC1 identified a polypeptide of ≈180 kDa in cells transiently transfected to express the predicted long form of ALS2 (to be referred to as WT ALS2) (Fig. 1 B and C). Using this antibody (Fig. 1B) or pAb-ALS2RCC1 (data not shown), immunoblotting of various mouse tissues identified this 180-kDa species to be most abundant in brain, with lower levels in spinal cord, liver, and spleen. A smaller ≈160-kDa species was also observed in liver and muscle (Fig. 1B), whereas more slowly migrating species were also present in lung and spleen, strongly suggesting the presence of tissue-specific variants of ALS2. [A prominent 120-kDa band was also seen in all tissues: this must represent a non-ALS2-encoded polypeptide that shares epitopes in common with WT ALS2, because it is identified with both antibodies, but its abundance is not reduced in samples from human patients with homozygous premature translation termination mutations in the ALS2 gene (see Fig. 5).] Immunoblotting of various regions of mouse brain also demonstrated that ALS2 was most abundant in cerebellum (Fig. 1D Upper), consistent with previous reports of ALS2 mRNA abundance (2). Even in these tissues, however, comparison by quantitative immunoblotting to known amounts of purified recombinant ALS2 revealed that endogenous ALS2 was relatively rare, representing only ≈0.0003% of the total detergent-soluble fraction of mouse brain lysate (Fig. 1E). Further, no species corresponding to the predicted shorter ALS2 isoform could be detected in any tissue (Fig. 1D Lower).
Fig. 1.
Endogenous WT ALS2 is enriched in neuronal tissues and membrane-containing fractions. (A) Predicted domain structures of ALS2 long (WT) and short form with reported sites for disease-associated mutations. The two antigens (ALS2RCC1, ALS21082) used for antibody production are also indicated. DH, Dbl homology; PH, pleckstrin homology; MORN, membrane occupation and recognition nexus. (B and C) Immunoblots of mouse tissue extracts and COS cell extracts. Aliquots of 70 μg of various mouse tissue extracts together with 5 μgof untransfected (COS-mock) and WT ALS2-transfected COS cell (COS-WT) lysates were immunoblotted as indicated. (D) Immunoblots of mouse nervous tissue extracts. Aliquots of 100 μg of mouse neuronal tissue extract and 5 μg of ALS2-transfected COS cell lysate were immunoblotted as indicated. (E) Relative abundance of endogenous ALS2 in mouse brain. Indicated amounts of mouse brain lysate and recombinant ALS2 (His-ALS2) were immunoblotted with pAb-ALS2RCC1. (F and G) Subcellular fractionation of mouse and human neuronal tissues as illustrated. Equal proportions of fractionated mouse brain and spinal cord (F) or human cerebral cortex and 0.2 times of the cytosol fraction (G) were immunoblotted as indicated. Tfn receptor, transferrin receptor; ER, endoplasmic reticulum.
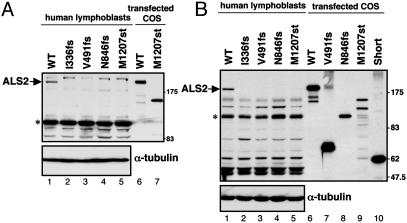
Fig. 5.
ALS2 protein is not accumulated in lymphoblasts of ALS2 patients. Immunoblot analysis of lymphoblasts from ALS2 patients and a nondiseased patient control. Aliquots of 40μg of lymphoblast lysates and 0.5μg of transfected COS cell lysates were resolved with SDS/PAGE and blotted with pAb-ALS21082 (A) and pAb-ALS2RCC1 (B), respectively. WT ALS2 is indicated. The asterisk (*) denotes a nonspecific ≈120-kDa band. Both blots were reprobed with anti-α-tubulin antibody to demonstrate equal loading.
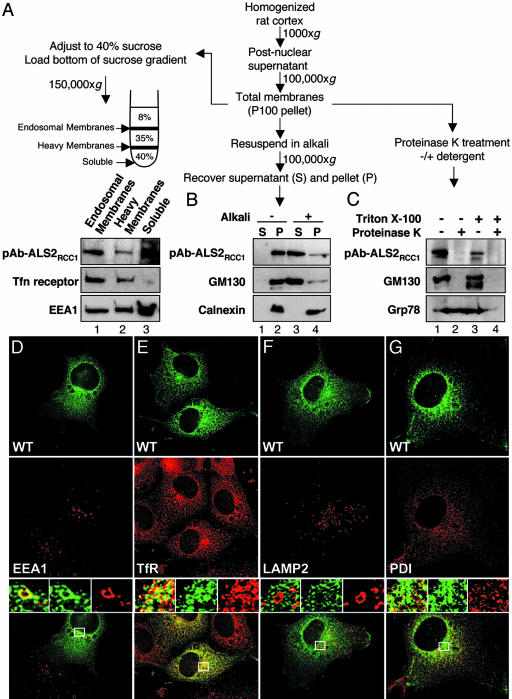
Endogenous ALS2 Is Peripherally Associated with the Cytoplasmic Face of Endosomal Membranes. To determine the localization of endogenous ALS2, cytosolic, nuclear, and membrane fractions from mouse brain and spinal cord were isolated by differential centrifugation. Although a cytosolic pool of ALS2 was also detected (Fig. 1F, lanes 3 and 7), WT endogenous ALS2 was enriched in the membrane (P100) fractions. Similarly, fractionation of human (Fig. 1G, lanes 3 and 4) and rat (data not shown) cerebral cortex revealed that ALS2 was enriched ≈3.5-fold in both a rapidly sedimenting membrane fraction that includes the endoplasmic reticulum, Golgi, and lysosomes and a more slowly sedimenting microsomal/vesicular fraction enriched in endosomes (including transferrin receptor-enriched recycling endosomes) and secretory vesicles. Further fractionation by flotation of total membranes derived from rat cerebral cortex showed that endogenous ALS2 was enriched in fractions containing transferrin receptor, a recycling endosome marker, and early endosome antigen 1 (EEA1) (Fig. 2A).
Fig. 2.
Endogenous WT ALS2 is a peripherally associated endosomal membrane protein oriented toward the cytoplasm. (A–C) Total membranes were processed for flotation centrifugation, alkali extraction, and proteinase sensitivity assays as illustrated. (A) Endogenous ALS2 is detected primarily in endosomal membrane fractions. After centrifugation, the membrane-containing interfaces and an aliquot of the layer containing soluble proteins (40%) were collected and immunoblotted as indicated. (B) Endogenous ALS2 is peripherally associated with intracellular membranes. After alkali extraction, aliquots of supernatants (S) and membrane-containing pellets (P) were immunoblotted as indicated. (C) Endogenous ALS2 resides on the cytoplasmic face of intracellular membranes. Aliquots of total membranes were treated with proteinase K in the presence or absence of detergent and immunoblotted as indicated. (D–G) Transfected ALS2 colocalizes with the endosomal markers EEA1 and transferrin receptor (TfR). Representative deconvolved images of COS cells transiently transfected with WT ALS2 (FITC, green, Top) and stained for endogenous membrane markers (Cy5, red, Middle): EEA1 (D), TfR (E), lysosome-associated membrane protein-2 (LAMP-2) (F), and protein disulfide isomerase (PDI) (G). Merged images together with enlarged boxed area that demonstrates merged (Left), ALS2 (green, Center), and endogenous marker (red, Right) images are shown (Bottom). (Magnifications: ×600.)
To directly distinguish whether the membrane-associated endogenous ALS2 is an integral membrane protein or is peripherally associated, the total membrane fraction (P100) from rat cortex was extracted with alkali. Similar to GM130 (a peripherally associated Golgi protein), alkali released the majority of ALS2 (Fig. 2B), while as expected the integral membrane protein calnexin remained membrane associated. Protease protection assays demonstrated that the ALS2 epitopes were accessible to digestion in the absence of detergent (as was also the case for GM130), indicating that membrane-associated ALS2 is oriented toward the cytoplasm (Fig. 2C). Confirming this notion, a significant portion of both ALS2 and EEA1 that initially copelleted in the total membrane fraction was apparently released from membranes during flotation and was recovered in the soluble fraction, demonstrating a labile association of ALS2 with one or more membranes (Fig. 2A, lane 3).
Exogenous ALS2 Partially Colocalizes with Endosomal Compartments. Although we were unsuccessful in using immunocytochemistry to locate endogenous ALS2 (data not shown), intracellular distribution of WT ALS2 was determined after transient expression of WT FLAG-tagged ALS2. pAb-ALS2RCC1, which detects transfected ALS2, easily distinguished transfected and untransfected cells (Fig. 2 D and E) and revealed that ALS2 was enriched in perinuclear, vesicular-like structures in monkey cells (Fig. 2 D–G), cultured human kidney cells (HEK293), and mouse neuroblastoma cells (data not shown). ALS2 was partially colocalized with both EEA1 (Fig. 2D) and transferrin receptor (Fig. 2E), markers for early and recycling endosomes, respectively. In contrast, there was no overlap with the late endosomal/lysosomal membrane protein LAMP-2 (Fig. 2F) or with protein disulfide isomerase (Fig. 2G), a marker of the endoplasmic reticulum.
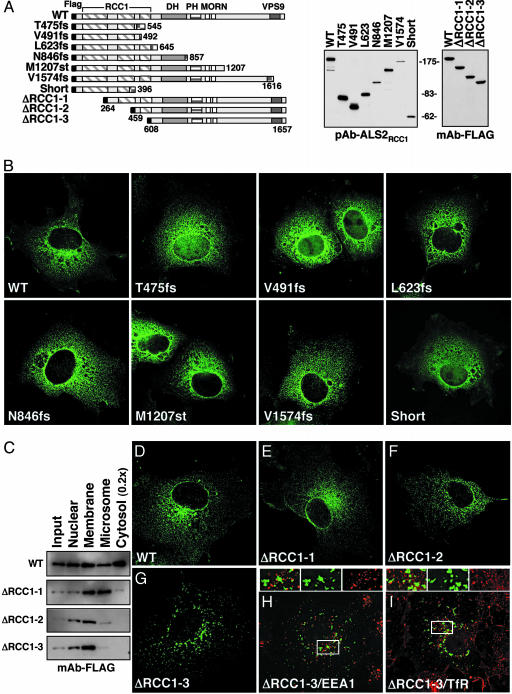
An Intact RCC1-Like Domain Is Required for Association with Endosomes. To test whether disease-causing ALS2 mutations altered the intracellular positioning of the corresponding mutants, six disease-associated mutants, representing the range of carboxyl-terminal truncations (Fig. 3A), were transiently expressed by using a replication-competent DNA construct to drive high levels of accumulation. All mutants (including L623fs, N846fs, M1207fs, and V1574fs) with an intact RCC1 domain, but missing most or all of the VPS9 region, were found in a vesiculated, perinuclear pattern indistinguishable from that observed for WT ALS2 (Fig. 3B). Despite continued association with the heavy membrane fraction (Fig. 3C), truncation or absence of the RCC1 domain (Fig. 3 E–G) eliminated colocalization with EEA1 or transferrin receptor (Fig. 3 H and I). Like WT ALS2, the predicted ALS2 short isoform, T475fs, and V491fs (all of which contain most of the RCC1 domain) were also found in a similar perinuclear vesicular pattern, in addition to some nuclear accumulation, presumably arising from passive diffusion of these small proteins through nuclear pores (Fig. 3B).
Fig. 3.
An intact amino-terminal RCC1-like domain is required for correct subcellular localization. (A) Schematic representation of the disease-associated ALS2 mutant and amino-terminal deletion expression constructs. Expression was verified by immunoblotting transiently transfected COS cell lysates as indicated. DH, Dbl homology; PH, pleckstrin homology; MORN, membrane occupation and recognition nexus. (B) Representative deconvolved micrographs of COS cells transiently transfected with indicated constructs and stained with pAb-ALS2RCC1 and FITC-conjugated goat anti-rabbit IgG. (C–I) Subcellular localization of ALS2 depends on an intact amino-terminal RCC1 domain. (C) HEK293 cells transiently transfected with amino-terminal-deleted ALS2 mutants were fractionated (see scheme in Fig. 1F) and immunoblotted with anti-FLAG M2 antibody. (D–I) Representative deconvolved images of COS cells transiently transfected with indicated constructs and stained with FITC-conjugated anti-FLAG M2 antibody (green, D–I) and endogenous (red) EEA1 (H) or transferrin receptor (TfR) (I). (Magnifications: ×600.)
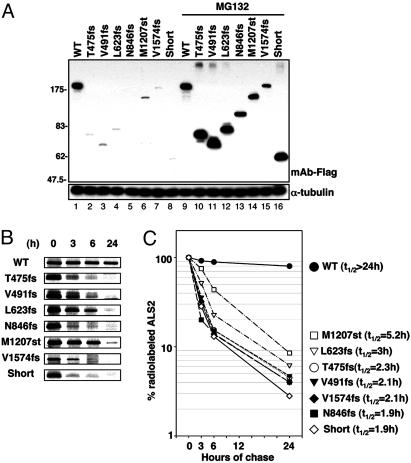
Disease-Causing Mutant ALS2 Proteins and the Predicted Short Form of ALS2 Are Rapidly Degraded by the Proteasome. Intracellular stabilities of WT ALS2, disease-causing ALS2 mutants, and the predicted short form of ALS2, were initially estimated by their accumulation levels relative to WT ALS2 when expressed by transfection in cultured human cells. All six truncation mutants (including V1574fs in which only the last 83 aa of ALS2 are missing) and the predicted short form accumulated to levels >20-fold lower than WT ALS2 (Fig. 4A). After proteasome inhibition by addition of MG132 for the last 10 h of culture, however, all of the mutants and the short form accumulated to at least 4-fold higher levels, indicating that each has a <5-h half-life in the absence of proteasome inhibition. In contrast, WT ALS2 was already completely stable (t1/2 > 24 h), with no increase in its level after 10 h of proteasome inhibition (Fig. 4A, compare lanes 1 and 9).
Fig. 4.
Disease-causing ALS2 mutants and the short form of ALS2 are unstable proteins. (A) ALS2 mutant proteins are unstable compared with the WT ALS2 protein. Mutant ALS2 was transiently transfected in HEK293 cells in the presence or absence of 20 μM MG132 for 10 h. Lysates were analyzed by immunoblot with anti-FLAG M2 antibody (Upper). The same membrane was blotted with anti-α-tubulin antibody to demonstrate equal loading (Lower). (B and C) Half-lives of ALS2 and mutant proteins. Transfected HEK293 cells were metabolically radiolabeled, immunoprecipitated, and analyzed by SDS/PAGE (B). The radiolabeled ALS2 bands were quantified by densitometry and plotted. Calculated half-lives of the ALS2 proteins are as indicated (C).
To test instability of the ALS2 mutants more directly, newly synthesized proteins in cultured human cells transfected to express each mutant were briefly radiolabeled for 45 min with [35S]methionine and cysteine, and the persistence of radiolabeled WT or mutant ALS2 was followed by immunoprecipitation for up to 24 h. As before, WT ALS2 was very stable with an extrapolated half-life of >24 h (Fig. 4B). All of the disease-associated mutants of ALS2, and the short form, had t1/2s that were at least 4.5–13 times shorter than the WT (Fig. 4 B and C).
ALS2 Is Not Detectable in Lymphoblasts from Patients with Confirmed ALS2 Mutations. To measure ALS2 mutant polypeptide stability most directly, accumulation of ALS2 mutants I336fs, V491fs, N846fs, and M1207st was analyzed in Epstein–Barr virus-immortalized lymphoblasts from patients with confirmed homozygous mutations in ALS2 (4, 16). None of the expected truncated protein products could be detected by using either of our ALS2 antibodies (Fig. 5), although endogenous ALS2 was found in lymphoblasts from a normal individual. In addition, despite the presence in all of the lymphoblast samples of a polypeptide of mobility similar to the proposed 62-kDa ALS2 short form, this cannot be the predicted ALS2 short form because in the I336fs cells it is present at the same levels as in normal lymphoblasts, despite the fact that this mutation would truncate this isoform to a predicted 53-kDa fragment.
Discussion
WT ALS2 contains RCC1-, Rho-, and VPS9-like GEF domains, each of which has been implicated in intracellular membrane trafficking or microtubule dynamics, the latter itself an integral component of such trafficking. Proportions of ALS2 have been shown here to be present in the cytoplasm and peripherally associated with the cytoplasmic face of intracellular membranes including early and recycling endosomes. Analysis of amino- and carboxyl-terminal truncation mutations and disease-associated mutants lacking both Rho-like and VPS9-like GEF domains (e.g., L623fs and N846fs) has further shown that an intact RCC1 domain is both necessary and sufficient for labile association with endosomes and accumulation of a significant cytosolic pool. Thus, instead of suppressing its endosome association as predicted (12), the RCC1-like domain can direct ALS2 localization and may be essential for its recycling from membranes into a cytoplasmic pool.
All known disease-causing ALS2 mutations are carboxyl-terminal truncations for which there is no apparent correlation between the degree of truncation and severity of disease. We have shown here that a common feature of the mutants, however, is that all have very significantly reduced stability, leading to undetectable levels in patient lymphoblasts, despite the ability of each to localize appropriately when synthesized at high levels. Because the longest mutant polypeptide (V1574fs, deleted in only the 83 carboxyl-terminal amino acids of the VPS9 domain) is as short-lived as the other mutants tested, the simplest view is that the similar severity of disease (onset at 1.5 years old) between this apparently mildest mutant and the much larger truncations (4, 5) probably arises from loss of overall ALS2 function, rather than from a specific requirement for the VPS9 domain, as has been proposed (12).
Our data also call into question the existence of an endogenous short form of ALS2, because we have shown that this predicted isoform has a short half-life, comparable to the mutant proteins that could not be detected in patient samples. Consistent with this view, an initial correlation (3, 17) between upper and lower motor neuron degeneration arising from mutations that affect both the long and predicted short form of ALS2, whereas mutations affecting the long form alone yield only upper motor neuron degeneration, has not been confirmed by latter mutations (4). Altogether, we propose that disease arises from a true loss of function caused, at least in part, by protein instability and therefore implies that all of the putative domains in ALS2 may potentially contribute to its endogenous function.
Our localization here of endogenous ALS2 to an endosomal membrane fraction, combined with its reported in vitro GEF activity for the Rab5 GTPase (12), already known to be required for endosomal membrane fusion, internalization, and motility (18), implicate defects in intracellular trafficking as one of the functions whose loss leads to clinical disease. This follows from identification of several causative mutations for the upper motor neuron disorder hereditary spastic paraplegia (HSP). These include atlastin, a GTPase mutated in early-onset autosomal dominant HSP (AD-HSP; SPG3A) (19); spastin, a microtubule binding AAA (ATPase associated with diverse cellular activities) family protein mutated in the most common form of AD-HSP (SPG4) (20); KIF5A, a microtubule-dependent motor implicated in intracellular trafficking mutated in pure AD-HSP (SPG10) (21); and spartin, mutation in which is causative for an autosomal recessive HSP known as Troyer syndrome (SPG20) and whose function is implicated in endosomal trafficking and microtubule binding (22). Further functional studies focused on each GEF domain in ALS2 will now be necessary to clarify the function of ALS2 in specific aspects of intracellular trafficking and how/why ALS2 is most essential to upper motor neurons, such that its loss leads to their killing beginning early in postnatal life.
Supplementary Material
Acknowledgments
We thank Drs. M. Cookson (National Institute of Aging, Bethesda), Y. Shen (Sun Health Research Institute, Sun City, AZ), K. Okumura, and Y. Mao (Ludwig Institute for Cancer Research, La Jolla, CA) for pAb-ALS21082, human samples, the pCIneoFL vector, and cloning of ALS2 cDNA, respectively. This work was supported by grants from the National Institutes of Health (NS 27036) and the Center for ALS Research at Johns Hopkins (to D.W.C.). C.V.V. is supported by a postdoctoral fellowship from the Paralyzed Veterans of America Spinal Cord Research Foundation. Salary support for E.E.-P. is provided by the Jean Pierre and Nancy Boespflug Myopathic Research Foundation. This work was supported by grants from Groupement d'Intérêt Scientifique-maladies rares (SPATAX network: European Network for Hereditary Spinocerebellar Degenerations), Ricerca Finalizzata Strategica on genetic leukodystrophies and spastic paraplegia from the Italian Ministry of Health, and the Association pour la Recherche sur la Sclerose Latérale Amyotrophique. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ALS, amyotrophic lateral sclerosis; GEF, guanine exchange factor; VPS9, vacuolar protein sorting 9; EEA1, early endosome antigen 1; HSP, hereditary spastic paraplegia; Rcc1, regulator of chromatin condensation.
References
- 1.Ben Hamida, M., Hentati, F. & Ben Hamida, C. (1990) Brain 113, 347-363. [DOI] [PubMed] [Google Scholar]
- 2.Hadano, S., Hand, C. K., Osuga, H., Yanagisawa, Y., Otomo, A., Devon, R. S., Miyamoto, N., Showguchi-Miyata, J., Okada, Y., Singaraja, R., et al. (2001) Nat. Genet. 29, 166-173. [DOI] [PubMed] [Google Scholar]
- 3.Yang, Y., Hentati, A., Deng, H. X., Dabbagh, O., Sasaki, T., Hirano, M., Hung, W. Y., Ouahchi, K., Yan, J., Azim, A. C., et al. (2001) Nat. Genet. 29, 160-165. [DOI] [PubMed] [Google Scholar]
- 4.Eymard-Pierre, E., Lesca, G., Dollet, S., Santorelli, F. M., Di Capua, M., Bertini, E. & Boespflug-Tanguy, O. (2002) Am. J. Hum. Genet. 71, 518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gros-Louis, F., Meijer, I. A., Hand, C. K., Dube, M. P., MacGregor, D. L., Seni, M. H., Devon, R. S., Hayden, M. R., Andermann, F., Andermann, E., et al. (2003) Ann. Neurol. 53, 144-145. [DOI] [PubMed] [Google Scholar]
- 6.Devon, R. S., Helm, J. R., Rouleau, G. A., Leitner, Y., Lerman-Sagie, T., Lev, D. & Hayden, M. R. (2003) Clin. Genet. 64, 210-215. [DOI] [PubMed] [Google Scholar]
- 7.Ohtsubo, M., Kai, R., Furuno, N., Sekiguchi, T., Sekiguchi, M., Hayashida, H., Kuma, K., Miyata, T., Fukushige, S., Murotsu, T., et al. (1987) Genes Dev. 1, 585-593. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt, A. & Hall, A. (2002) Genes Dev. 16, 1587-1609. [DOI] [PubMed] [Google Scholar]
- 9.Burd, C. G., Mustol, P. A., Schu, P. V. & Emr, S. D. (1996) Mol. Cell. Biol. 16, 2369-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiuchi, H., Lippe, R., McBride, H. M., Rubino, M., Woodman, P., Stenmark, H., Rybin, V., Wilm, M., Ashman, K., Mann, M., et al. (1997) Cell 90, 1149-1159. [DOI] [PubMed] [Google Scholar]
- 11.Takeshima, H., Komazaki, S., Nishi, M., Iino, M. & Kangawa, K. (2000) Mol. Cell 6, 11-22. [DOI] [PubMed] [Google Scholar]
- 12.Otomo, A., Hadano, S., Okada, T., Mizumura, H., Kunita, R., Nishijima, H., Showguchi-Miyata, J., Yanagisawa, Y., Kohiki, E., Suga, E., et al. (2003) Hum. Mol. Genet. 12, 1671-1687. [DOI] [PubMed] [Google Scholar]
- 13.Iwai, K., Yamanaka, K., Kamura, T., Minato, N., Conaway, R. C., Conaway, J. W., Klausner, R. D. & Pause, A. (1999) Proc. Natl. Acad. Sci. USA 96, 12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, L. A., Fialka, I., Paiha, K., Hunziker, W., Sacks, D. B., Bahler, M., Way, M., Gagescu, R. & Gruenberg, J. (2000) Traffic 1, 494-503. [DOI] [PubMed] [Google Scholar]
- 15.Lin, P., Le Niculescu, H., Hofmeister, R., McCaffery, J. M., Jin, M., Hennemann, H., McQuistan, T., De Vries, L. & Farquhar, M. G. (1998) J. Cell Biol. 141, 1515-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesca, G., Eymard-Pierre, E., Santorelli, F. M., Cusmai, R., Di Capua, M., Valente, E. M., Attia-Sobol, J., Plauchu, H., Leuzzi, V., Ponzone, A., et al. (2003) Neurology 60, 674-682. [DOI] [PubMed] [Google Scholar]
- 17.Shaw, P. J. (2001) Nat. Genet. 29, 103-104. [DOI] [PubMed] [Google Scholar]
- 18.Zerial, M. & McBride, H. (2001) Nat. Rev. Mol. Cell Biol. 2, 107-117. [DOI] [PubMed] [Google Scholar]
- 19.Zhao, X., Alvarado, D., Rainier, S., Lemons, R., Hedera, P., Weber, C. H., Tukel, T., Apak, M., Heiman-Patterson, T., Ming, L., et al. (2001) Nat. Genet. 29, 326-331. [DOI] [PubMed] [Google Scholar]
- 20.Hazan, J., Fonknechten, N., Mavel, D., Paternotte, C., Samson, D., Artiguenave, F., Davoine, C. S., Cruaud, C., Durr, A., Wincker, P., et al. (1999) Nat. Genet. 23, 296-303. [DOI] [PubMed] [Google Scholar]
- 21.Reid, E., Kloos, M., Ashley-Koch, A., Hughes, L., Bevan, S., Svenson, I. K., Graham, F. L., Gaskell, P. C., Dearlove, A., Pericak-Vance, M. A., et al. (2002) Am. J. Hum. Genet. 71, 1189-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, H., Cross, H., Proukakis, C., Hershberger, R., Bork, P., Ciccarelli, F. D., Patton, M. A., McKusick, V. A. & Crosby, A. H. (2002) Nat. Genet. 31, 347-348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.