INTRODUCTION
The Aedes aegypti genome project revealed mosquito homologues of many genes known to regulate development in other arthropods. This protocol can be used for analysis of gene expression in Ae. aegypti embryos and larvae, which is a critical aspect of understanding developmental gene function in this vector mosquito.
MATERIALS
Equipment
Depression slide
Dissection microscope
Floating rack
Heat block or boiling bath
Ice bucket with ice
Microfuge tubes
Micropipetter
Timer
Water bath (60° C)
Reagents
Anti-DIG antibody (Roche Diagnostics)
AP buffer <R>
AP-NBT/BCIP <R>
Detergent solution <R>
DIG-labeled riboprobe (sense control and antisense)
Glycerol solution (50% and 70%) <R>
Hyb <R>
Hyb-DNA-SDS <R>
Methanol
PBS <R>
PT <R>
PTw <R>
Tissues (dissected embryos or sonicated larvae)
METHOD
Riboprobes should be used in conjunction with this protocol (see Patel, 1996 for a detailed discussion of riboprobe synthesis). This procedure can be completed in 2-3 days and can be used in conjunction with the accompanying immunohistochemistry protocol for combined analysis of protein expression.
Day One
Rehydration and detergent treatment
1. Fix and prepare tissues as described in the accompanying protocol. The tissue can remain in microfuge tubes for the duration of the experiment. Unless otherwise indicated, use 1 ml rinse/wash volumes for this step and throughout the protocol. If the tissue has been stored in methanol in the freezer, remove the methanol and rehydrate the tissue with 50% methanol / 50% PBS for 5 min. Proceed with a 5 min rinse in PBS and 2× 10 min rinses in PTw. Larvae will need to be sonicated as described in the fixation/tissue preparation protocol before proceeding to step 2.
2. Remove the PTw and add 1 ml of Detergent solution. Incubate tissues in the Detergent solution for 30 min with shaking.
3. Rinse 2× 10 min in PTw.
Blocking and hybridization
4. Remove the PTw and rinse 1× 5 min with 500 μl of 50% PTw/50% Hyb solution. Replace the 50% PTw/50% Hyb with 500 μl of Hyb solution for 10 min. While the tissues are incubating in Hyb, begin step 5.
5. Boil the Hyb-DNA-SDS solution for 10 min to denature the DNA. Store on ice until you are ready to use it.
6. Remove the Hyb solution from the tissues and add 100 μl of denatured Hyb-DNA-SDS solution. Place these tubes in a floating rack located in a 60° C water bath for 60 min. A shaking bath is preferred, though not required for this blocking step and all subsequent steps completed in the water bath.
7. Toward the end of the pre-hybridization step, heat-denature the riboprobe resuspended in Hyb-DNA-SDS solution by boiling it for 5 min. Boiling helps to remove secondary structure in the riboprobe.
8. Collect tubes from the water bath. Remove the Hyb-DNA-SDS used in the blocking step (#6) and add your denatured probe. Mix the tissues and the probe by gently stirring with your pipette tip.
9. Place tubes in the 60° C water bath, where they will remain overnight.
Day Two
Washes and Probe Detection
10. Perform the following washes at 60° C (use 1 ml volumes of prewarmed solutions):
1× 30 min Hyb solution
5× 30 min with PTw
Bring the tubes to room temperature and wash for an additional 30 min with PT.
11. Remove the PT and add 300 μl of anti-DIG antibody at a dilution of 1:2000 in PT. Mix gently with your pipette tip. Leave the antibody on overnight at 4° C. Alternatively, for embryos, it is possible to leave the antibody on at room temperature for 2 hrs.
12. Rinse the tissue 4× 30 min with PT at room temperature. It is possible to extend these washes overnight if preferred.
Color reaction (for AP-conjugated anti-DIG antibodies)
13. Rinse the tissue 3× 5 min in AP buffer (performing this step as described helps to ensure quicker color reaction times).
14. Add 300 μl of AP-NBT/BCIP solution. The majority of this step, the color reaction, should be performed in the dark. Occasionally transfer a few embryos/larvae (using a cut pipette tip) to a depression slide in order to observe the progress of the reaction; brief exposure to light will not disrupt the progress of the reaction, which can take anywhere from several minutes to several hours to complete. If the color reaction takes longer than 3 hrs, then it may be helpful to add fresh AP-NBT/BCIP in newly made AP buffer.
15. Stop the reaction by removing the AP-NBT/BCIP and rinsing 4× 15 min with PT.
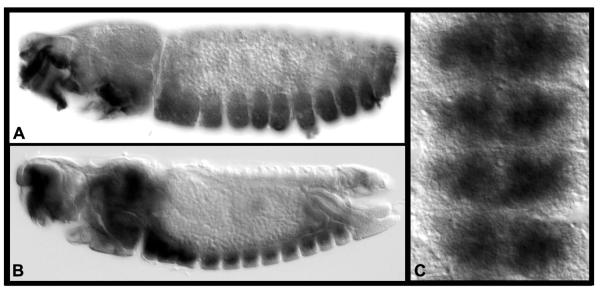
16. Remove the PT and rinse 1× 5 min with PBS. Remove the PBS and add 500 μl of 50% glycerol solution. After 60 min, replace the 50% glycerol with 70% glycerol. Leave the tissues at room temperature overnight for clearing. The tissues can be mounted and analyzed in 70% glycerol. Examples of tissues that we’ve processed in this manner are shown in Fig. 1.
Fig. 1. Gene expression in Ae. aegypti embryos.
(A) fra and casein kinase (B, C) are expressed ventrally in the developing nerve cord at 55 hrs. of development. Lateral views of whole-mount embryos stained with the accompanying protocol are shown in A and B (anterior is oriented left). A filleted nerve cord is shown in C (anterior is oriented up).
Combining mRNA and protein localization
If a combination of mRNA and protein expression analysis (double label) is desired, then proceed directly from step 15 to the blocking and primary antibody incubation steps of the accompanying immunohistochemistry protocol. If the anti-DIG AP antibody is used for the in situ, then a brown HRP reaction should be used for the protein localization.
TROUBLESHOOTING
Problem: Loss of embryos (throughout procedure); Solution: When exchanging solutions, it may be helpful to pipette the solution to be discarded into a petri dish and to look for/retrieve any embryos. This is particularly important when embryos are in Hyb solution, as they tend to float.
Problem: Poor staining of tissues (step 14); Solutions: If tissues are under- or over-stained, the concentration of the probe may need to be adjusted. Also, be certain that tissues have not become desiccated or stuck to the sides of the tube during the procedure, either of which can lead to poor staining.
DISCUSSION
This protocol is an adaptation of standard Drosophila whole-mount in situ hybridization protocols (Tautz and Pfeifle, 1989; Patel, 1996). In recent years (VanZomeren-Dohm et al., 2008), we have eliminated the time-consuming and sometimes technically challenging xylene, post-fix, and proteinase K treatment steps described in the original Drosophila protocols. Replacement of the proteinase K treatment step with detergent treatment (day one, step 2) yields excellent results (Patel et al., 2001; Duman-Scheel et al., 2002). Use of this methodology is providing new insight into the function of developmental genes in Ae. aegypti.
RECIPES.
AP BUFFER and AP-NBT/BCIP
| Reagent | Quantity (for 50 ml) | Final Concentration |
|---|---|---|
| MgCl2 | 250 μl 1M | 5 mM |
| NaCl | 5 ml 1M | 100 mM |
| Tris (pH 9.5) | 5 ml 1M | 100 mM |
| Tween-20 | 50 μl | 0.1% |
| Add dH20 to a total volume of 50 ml. Make just prior to use and store for no more than three hours. | ||
| For color reactions, add 20 μl of NBT/BCIP solution (Roche Cat. #11681451001) to 1 ml AP buffer and use immediately. | ||
DETERGENT SOLUTION
| Reagent | Quantity (for 50 ml) | Final concentration |
|---|---|---|
| SDS | 5 ml 10% SDS | 1.0% |
| Tween-20 | 250 μl | 0.5% |
| Tris-HCl (pH 7.5) | 5 ml 0.5 M Tris-HCl | 50 mM |
| EDTA (pH 8.0) | 100 μl 0.5 M EDTA | 1 mM |
| NaCl | 7.5 ml 1 M NaCl | 150 mM |
| Add dH20 to a total volume of 50 ml and store at room temperature. | ||
GLYCEROL SOLUTION
50% and 70% glycerol solutions can be prepared by mixing the appropriate volumes of ultrapure glycerol with 1× PBS. Place the solution on a rocker at room temperature for 30 min to ensure thorough mixing of PBS and glycerol. Check the final pH to be sure that it is near 7.4. Store at room temperature.
HYB and HYB-DNA-SDS
Prepare Hyb as follows:
| Reagent | Quantity (for 50 ml) | Final concentration |
|---|---|---|
| Deionized formamide | 25 mL deionized formamide | 50% |
| SSC | 12.5 mL of 20× SSC | 5× |
| Heparin | 2.5 mg heparin | 50 μg/mL |
| Tween-20 | 50 μl 100% Tween-20 | 0.1% |
| Adjust the pH to 5.0 using HCl and bring the final volume to 50 ml with sterile dH20. Hyb buffer is stored at −20° C. | ||
| For Hyb-DNA-SDS, combine 14.85 ml Hyb solution and 150 μl of 10 mg/ml sonicated salmon sperm DNA (Invitrogen Cat. No. 15632-011). Store at −20° C. Prior to use, warm the solution and add SDS to a final concentration of 0.3%. Use this solution in the blocking step prior to hybridization and during the actual hybridization. | ||
PBS (10× stock)
| Reagent | Quantity (for 1 L) | Final concentration |
|---|---|---|
| Na2HPO4 | 11.9 g | 84.1 mM |
| NaH2PO4 (anhydrous) | 2.23 g | 18.6 mM |
| NaCl | 102.2 g | 1.75 M |
| Bring the volume to 1 L with distilled water. Adjust the pH to 7.4 and autoclave before use. Prepare the working strength solution (1×, simply referred to as PBS in the protocol) by diluting 1:10 with sterile dH20. Both 1× and 10× PBS are stored at room temperature. | ||
PT
| Reagent | Quantity (for 1 L) | Final concentration |
|---|---|---|
| 10× PBS | 100 ml | 1× |
| 100% Triton X-100 | 1 ml | 0.1% |
| Bring final volume to 1 L with sterile dH20. Mix and store at room temperature. | ||
PTw
| Reagent | Quantity (for 1 L) | Final concentration |
|---|---|---|
| 10× PBS | 100 ml | 1× |
| 100% Tween-20 | 1 ml | 0.1% |
| Bring final volume to 1 L with sterile dH20. Mix and store at room temperature. | ||
ACKNOWLEDGEMENTS
Development of the protocol described was funded by the following awards to MDS: NIH/NIAID Award R01 AI 081795-01 and NIH/NINDS Award R15 NS 048904-0. Kristopher Kast and Caitlin Jacowski were supported by the University of Notre Dame College of Science Summer Undergraduate Research Fellowship program.
Footnotes
Conflicts of interest: none declared
REFERENCES
- Duman-Scheel M, Pirkl N, Patel NH. Analysis of the expression pattern of Mysidium columbiae wingless provides evidence for conserved mesodermal and retinal patterning processes among insects and crustaceans. Dev Genes Evol. 2002;212:114–123. doi: 10.1007/s00427-002-0215-6. [DOI] [PubMed] [Google Scholar]
- Patel N. In situ hybridization to whole mount Drosophila embryos. In: Krieg PA, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. Wiley-Liss; New York, NY: 1996. pp. 357–370. [Google Scholar]
- Patel NH, Hayward D, Lall S, Pirkl N, DiPietro D, Ball E. Grasshopper hunchback expression reveals conserved and novel aspects of axis formation and segmentation. Development. 2001;128:3459–3472. doi: 10.1242/dev.128.18.3459. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- VanZomeren-Dohm A, Flannery E, Duman-Scheel M. Whole mount in situ hybridization detection of mRNA in GFP-marked Drosophila imaginal disc mosaic clones. Fly. 2(6):323–325. doi: 10.4161/fly.7230. [DOI] [PubMed] [Google Scholar]