Abstract
This unit presents a recombinant protein purification method that employs an Elastin-like polypeptide (ELP) as a purification tag. ELPs undergo a sharp and reversible phase transition when heated above their lower critical solution temperature (LCST). ELPs retain this behavior when they are fused to a protein, and thereby provide a simple method to isolate a recombinant ELP fusion protein from cell contaminants by cycling the solution through the insoluble and soluble phase of the ELP fusion protein by a method that we have termed Inverse Transition Cycling (ITC). This method does not require the use of chromatography, so that it is cost-effective, easy to scale up and to multiplex.
Keywords: Elastin-like polypeptides (ELP), inverse transition cycling (ITC), purification tag, recombinant protein, non-chromatographic purification, protein fusion, transition temperature
INTRODUCTION
We describe herein a simple method –Inverse Transition Cycling (ITC)– to purify recombinant proteins and peptides that was developed in our laboratory (Meyer and Chilkoti 1999). This technique exploits the unique phase transition behavior of elastin-like polypeptides (ELPs). In ITC, a target protein or peptide is fused to the ELP at the gene level, expressed in E. coli or another expression system, and purified using the ELP as a purification tag (Meyer and Chilkoti 1999). ITC is both cost- and time-efficient because this purification method eliminates chromatography. In addition, scale-up of this purification method is easy because it is not limited by resin capacity (Ge et al. 2006).
ELPs are peptide polymers that are composed of repeats of the pentapeptide (Val-Pro-Gly-Xaa-Gly)n where Xaa is the guest residue; this residue can be any amino acid except Pro; n describes the number of repeats and is typically between 20-330 (Meyer and Chilkoti 2004). ELP genes of different lengths can be synthesized by many methods (McPherson et al. 1996); (Chen et al. 2008) but are typically synthesized in our laboratory by a process that we developed, called Recursive Directional Ligation (Meyer and Chilkoti 2002).
ELPs undergo a sharp and reversible phase transition at a specific temperature known as the lower critical solution temperature (LCST) this temperature is also frequently referred to as the inverse transition temperature (Tt). Below its LCST, an ELP adopts a close to random coil conformation, is well solvated and is hence highly soluble in aqueous solution. When the solution is heated and the LCST is reached, ELPs become insoluble and form large micron-size aggregates that are visible to the naked eye (Luan et al. 1990; Meyer and Chilkoti 2002). This transition is completely reversible, so that the aggregated polypeptide completely dissolves when the temperature is lowered below the LCST of the ELP. The LCST decreases with increasing ionic strength, polymer concentration, and polymer length. The nature of the guest residue also affects the transition temperature, as more hydrophobic amino acid residues lower the transition temperature and vice versa. (Meyer and Chilkoti 2004)
ELPs are an attractive choice for a purification tag, as proteins or peptides that are fused to an ELP show similar stimulus responsive behavior (Meyer and Chilkoti, 1999). The physicochemical properties of the fused protein can affect the transition temperature of the ELP, though this effect is relatively minor for most soluble proteins. There are two exceptions to this statement: (1) proteins with a significant fraction of solvent accessible hydrophobic surface area will decrease the transition temperature; (2) proteins with a large fraction of charged solvent accessible area will increase the LCST of the ELP fusion protein compared to the free ELP (Trabbic-Carlson et al. 2004b).
To express and purify a target protein by ITC, the gene of the protein needs to be simply joined to a gene encoding an ELP. Once the plasmid encoding the fusion protein is constructed, and transformed into a suitable host cell (typically E. coli in our laboratory) the host cells are cultured and the ELP fusion protein is expressed in the host cells.
A parenthetical note on proteins that form inclusion bodies
We note that for ITC to be useful, a large enough fraction of the fusion protein must be expressed as soluble protein. For proteins that are known to form inclusion bodies, we have some preliminary evidence that fusion to an ELP can tilt the balance toward soluble protein expression, so that it may be worthwhile to explore the fusion of this class of proteins with an ELP.
Figure 1 shows the process of heterologous expression of a recombinant ELP fusion protein and its purification by ITC. The ELP tag provides a convenient stimulus responsive “handle” to separate the target protein from other contaminants by cycling the ELP fusion protein through its phase transition in cell lysate, as follows: after expression, the cells are lysed, and the cell debris is removed by centrifugation. The ELP fusion protein is then separated from soluble contaminants by triggering the phase transition of the ELP fusion protein (Figure 2). Increasing the solution temperature above the inverse transition temperature of the ELP fusion protein or increasing the salt concentration to depress the transition temperature below solution temperature are two orthogonal and convenient methods to trigger the phase transition of an ELP fusion protein. As increasing temperature might denature some proteins, it is often preferable to isothermally trigger the phase transition by increasing the salt concentration. Triggering the phase transition causes the ELP fusion protein to form micron size aggregates. These aggregates are typically highly enriched in the ELP fusion protein and are separated from the soluble fraction of the cell lysate by centrifugation at the same temperature or filtration. The supernatant is decanted and discarded. We call this centrifugation step a “hot spin” –a term that a bit of a misnomer– because it is carried out at around ambient temperature in the centrifuge (simply by turning off the refrigeration) and to distinguish it from the next step of the process that also involves centrifugation. In this subsequent step, termed the “cold spin”, the pellet of the aggregated ELP fusion protein is dissolved in cold buffer, and then centrifuged at 4 °C to remove any insoluble contaminants that may have been trapped in the mass of the aggregated ELP fusion protein. The soluble protein is decanted and retained, and this completes one cycle of ITC.
Figure 1. Flowchart of ITC purification process.
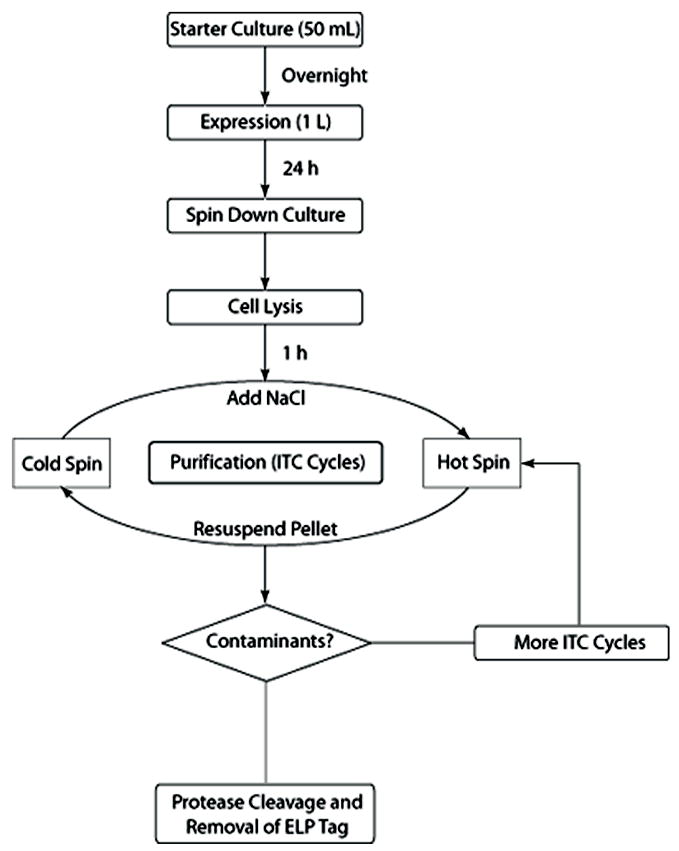
The initial steps of ITC are similar to most recombinant protein purification methods. The separation of target protein from contaminants occurs in each cycle of ITC.
Figure 2. Schematic of purification of recombinant ELP fusion proteins.
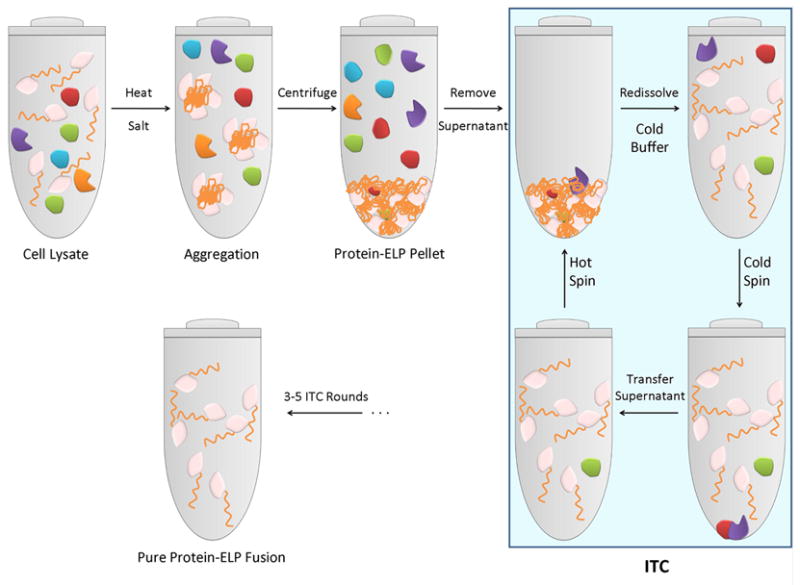
Addition of salt (typically NaCl) to cell lysate with a recombinantly expressed ELP fusion protein at room temperature triggers the phase transition, causing the ELP fusion protein to aggregate. Centrifugation of the sample separates the aggregated ELP fusion from other contaminants (“hot spin”). The pellet of aggregated ELP fusion protein contains a low, though variable amount of contaminants, which depends upon the target protein. The following steps, highlighted in the blue box, remove most contaminants: the aggregated ELP fusion protein in the pellet from the hot spin is dissolved in cold PBS (4 °C) followed by centrifugation at 4°C (“cold spin”). The cold spin removes insoluble contaminants that were trapped with the aggregated ELP fusion protein. This step is followed by a hot spin, which separates the ELP fusion from soluble contaminants. Typically, three to five rounds of ITC are sufficient to purify an ELP fusion sample from cell lysate to homogeneity.
This process can be repeated multiple times if needed, to increase the purity of the ELP fusion protein. The decision on the number of cycles to use for a particular protein is dependent on the protein, as some proteins are effectively purified by a few cycles while others may require 3-5 cycles to attain the desired purity. Increasing purity, obtained by increasing the number of cycles, has to be balanced by the increased time and expense for purification and the potential loss of protein in every cycle; for many proteins these losses are insignificant, though for some –typically hydrophobic– proteins these losses may represent a considerable fraction of the total ELP fusion protein in the cell lysate.
STRATEGIC PLANNING
Although ELPs can be used as purification tags for a wide range of proteins and peptides, each target protein or peptide will have unique properties that may require adjustment of the purification protocol. We discuss below the list of variables that can be altered to optimize the ITC purification protocol for an ELP fusion of interest.
Nomenclature
We use the following nomenclature to identify ELPs: ELP[XiYjZk-n] where X, Y, Z indicate the one letter code of each amino acid in the guest residue position, i, j, k specify the number of the particular amino acid in the monomer gene, and n describes the number of pentapeptide repeats.
Choosing the correct ELP
To date, the most widely used ELP in fusion constructs is ELP[Val5Ala2Gly3-90] (Meyer and Chilkoti 1999; Trabbic-Carlson et al. 2004a; Trabbic-Carlson et al. 2004b). We note that its ubiquitous use is largely because of the following reasons: first, it was the first, neutral, somewhat hydrophilic ELP that we designed that provided high yields of many fusion proteins in a T7 expression system in E. coli. Second, it has a convenient transition temperature of ~40 °C in PBS at the micromolar concentrations that are typical of the expression levels of many soluble proteins, so that the addition of 1-3 M NaCl depresses the transition temperature below room temperature, and hence provides a convenient isothermal method for purification by ITC, especially for thermally labile proteins. (Meyer et al. 2001). Shorter more apolar ELPs that solely contain valine (Val) as the guest residue (ELP [Val5]) have also been fused to proteins to compare the efficiency of purification by these two different ELP tags. The potential advantage of this ELP is that because it is more apolar, it has a lower transition temperature than ELP[Val5Ala2Gly3] for the same chain length, so that one can use a shorter ELP [Val5] and still maintain a transition temperature of ~40 °C at micromolar concentrations. Nevertheless, this ELP is not our first choice for protein purification, as we have found that some ELP [Val5] fusion proteins are more difficult to purify than ELP[Val5Ala2Gly3] fusions because more apolar ELPs tend to carry more contaminants into the pellet during the hot spin. Furthermore, the ELP[Val5] fusions exhibit a more complex phase transition behavior for some proteins (Meyer et al. 2001). We hence recommend using the more hydrophobic ELP[V5-40] as a backup choice when expression levels of ELP[Val5Ala2Gly3-90] fusions are low, or if the longer ELP chain hinders correct folding of the protein.
Cloning the fusion construct
We have observed that the fusion order of the protein and ELP can affect expression levels. Proteins fused to an ELP at their C-terminus (protein-ELP) often have higher protein yields than the reverse order (Christensen et al. 2009). However, as an exception to this rule we have observed greater yields of short peptides (less than 5 kDa), that were fused at their N-terminus (ELP-peptide) as compared to their C-terminal ELP fusion (unpublished results). Given the intrinsic variation in protein expression and folding, we recommend testing ELP fusions in both orientations.
Size of the target protein
We have reported the purification of target proteins of sizes from 4 to 30 kDa (Trabbic-Carlson et al. 2004b). The levels of the expression and the ease of purification for the various constructs do not show a pattern with increasing size. Therefore, when expressing a new construct, the size of the protein will not necessarily dictate the yield of the fusion protein.
Buffer considerations
In our basic protocol, we use PBS; however, other buffers can be substituted. If the target protein is very hydrophobic, the pellet after the hot spin can be difficult to resuspend. In this case, distilled water can be used to resuspend the pellets, though it runs the risk of denaturing some proteins that are unstable in distilled water. The purification steps should be carried in the buffer of choice –typically PBS or HEPES– supplemented with a salt to trigger the phase transition of the ELP; these salts are typically NaCl; (1 M - 3 M), ammonium sulfate (0.4-1 M) or sodium citrate (0.15-0.5 M mM). During the last round of ITC, the pellets can be simply resuspended in the final desired buffer.
Hydrophobic/hydrophilic surface area
We have found that the transition temperature of some ELP fusion proteins differ from that of the free ELP (Trabbic-Carlson et al. 2004b). Proteins with a significant charged surface area increase the transition temperature while hydrophobic proteins depress the transition temperature. To maintain the transition temperature in the desired range of 35-55 °C in PBS, proteins with a large fraction of charged residues on the surface should be fused to a longer ELP or an ELP with a higher fraction of apolar guest residues, while hydrophobic proteins should be fused to a shorter or more polar ELP. Additionally, lowering the pH of the solution can lead to neutralizing the surface charges and lead to lower transition temperatures, though care must be exercised to keep the pH away from the pI of the protein, so as not to denature the protein (likely at extremes of pH) or aggregate the protein by isoelectric precipitation if the solution pH approaches the pI of the protein.
Cleavage site
For applications in which the target protein is required as the final product, the ELP tag can be cleaved using a protease of choice, by engineering a peptide cleavage site for that protease between the target protein and the ELP (we typically use thrombin or tobacco etch virus (TEV) protease). Cleavage of the ELP fusion protein liberates the target protein from the ELP, and another round of ITC is sufficient to recover the protein in the supernatant after the hot spin.
BASIC PROTOCOL
PURIFICATION OF WELL EXPRESSED ELP FUSION PROTEINS BY ITC
This protocol is useful for soluble and over-expressed ELP fusion proteins. It uses the phase transition behavior of an ELP tag to separate the fusion protein from E. coli contaminants. By increasing the salt concentration of the solution, the transition temperature of the protein ELP fusion is lowered below the ambient temperature, causing aggregation of the ELP fusion protein and thus the formation of a pellet upon centrifugation of the sample (“hot spin”). Upon resuspending the sample in a low ionic strength buffer, the ELP fusion redissolves, and any residual insoluble contaminants are removed by centrifugation at low temperature (typically 4 °C, and hence termed “cold spin”). The supernatant from this step contains the purified ELP fusion and this process constitutes one round of ITC. This procedure can be iteratively performed to obtain increasingly pure ELP fusion protein.
Materials
BL21(DE3) E. coli or BLR E. coli competent cells
200 mL flask containing 50 mL Terrific Broth (TB) media (autoclaved) for overnight culturing
4L flask containing 1L TB media (autoclaved)
Ampicillin (100 mg/L of media) or kanamycin (45 mg/L of media) depending on plasmid resistance.
2 mL of 10 % (v/v) polyethyleneimine (PEI) in water, for 1 L of culture
NaCl (or other stronger kosmotropes such as sodium citrate and ammonium sulfate)
Saturated NaCl solution in PBS or H2O
Protein resuspension buffer (e.g., PBS or HEPES)
Note:The resuspension buffer in this protocol is assumed to be PBS.
0.5 M CuCl2 in H2O (filtered)
Additional reagents and equipment for one dimensional SDS-gel electrophoresis of proteins (Unit 10.1)
Equipment
Floor model low speed centrifuge to spin down the bacterial culture after growth.
Temperature controlled high-speed centrifuge with a rotor capable of spinning down material in 50 mL tubes
Sonicator (or other method of lysing cells, e.g. French Press)
Two microcentrifuges: one at room temperature and one at 4 °C (place one in a refrigerator with an internal power outlet)
Growing protein ELP fusion
- Day 1: Add ampicillin (5 mg) or kanamycin (2.25 mg) to the 50 mL TB overnight culture. Inoculate the culture from either a DMSO (or glycerol) stock or from a single colony on an agar plate. Grow the starter culture overnight at 37 °C while shaking at 225 rpm.The antibiotics should not be added until the media has cooled down to ambient temperature. The plated colonies should be obtained by transforming the expression plasmid into BL21(DE3) or BLR(DE3) E. coli cells as explained in Appendix 4D. DMSO stocks are obtained from cultures of the plated cells.
- Day 2: Transfer 10 mL of the starter culture to a sterile centrifuge tube and centrifuge for 15 min at 6,000 xg (4 °C). Discard supernatant and resuspend the bacterial pellet in 1 mL fresh, autoclaved TB media. Use 1 mL of the resuspended bacteria to inoculate 1 L autoclaved TB media in a 4 L flask. Incubate the 1 L inoculated media at 37 °C for 24 h while shaking at 225 rpm.Incubation time and temperature may vary. The suggested incubation time of 24 h without induction (“no induction” expression protocol) is based on our observation that expression levels resulting from the leaky T7 promoter are higher than those of cultures induced with IPTG and incubated for 4 h post-induction. However, the more common IPTG induction protocol may be useful for some proteins, as we have observed that some fusion proteins express at a higher level after induction with IPTG than with the 24 h “no induction” protocol (see Critical parameters – Low expression levels).
Day 3: Transfer the 1 L of bacterial culture to 1 L bottles, and centrifuge at 6,000 xg for 15 min at 4 °C. Discard the supernatant and resuspend the cell pellets in 35 mL PBS buffer and transfer to 50 mL conical tube. The purification process can be initiated at this point or the resuspended cells can be stored at −80 °C.
Lysing the cells and the first round of ITC
-
4Thaw the bacterial suspension from step 3 and surround the tube with ice/ethanol in a beaker to prevent sample from heating up during lysis. Lyse by ultrasonic disruption (10 sec on, 20 sec off, total on time is 3 min, power output of 85 W. Repeat the process twice).Protease inhibitors should be added to the thawed cell lysate before sonication in cases where the fusion construct is degraded. The exact parameters used for ultrasonic disruption may vary, depending on the model used.
-
5Split the lysed bacteria into two 50 mL centrifuge tubes and add 1 mL PEI solution to each tube. Save 20 μl for SDS-PAGE (not all 20 μl will be loaded on the gel but can be saved in case the gel needs to be run again). Centrifuge the rest of the lysate at 18,000 g for 15 min at 4 °C to separate cell debris from soluble cell lysate. Retain the supernatant for the first round of ITC (save 20 μl for SDS-PAGE gel).PEI is a positively charged polymer that will precipitate negatively charged contaminant molecules such as DNA from the cell lysate. If the protein of interest is highly negatively charged, then proceed with this step without adding PEI.
-
6Measure the volume of the supernatant in each tube, then add the required amount of NaCl to a final concentration of 3 M. As the NaCl dissolves and the solution returns to room temperature, the solution should turn visibly cloudy.The increase in turbidity of the solution indicates that the protein ELP fusion has undergone its phase transition and formed aggregates. The choice of salt to trigger the phase transition will depend on the target protein and level of expression [see critical parameters].
-
7To perform the first hot spin, centrifuge the cloudy suspension at 18,000 xg for 15 min at room temperature. A pellet should form at the bottom of the tube (save 20 μl of the supernatant for SDS-PAGE). Decant and discard the supernatant.The aliquot of the supernatant from the first hot spin is visualized by SDS-PAGE to see if all the ELP fusion protein transitioned. This sample is high in salt and may not run well on a SDS-PAGE gel.
-
8Resuspend the pellet in 3-4 mL PBS (save 20 μl for SDS-PAGE). In this step, placing the pellet on ice or adding cold buffer (~4 °C) may aid in resuspending the pellet. Transfer the resuspended pellet to microcentrifuge tubes in aliquots of 1 mL.The pellet should change color to clear brown as the ELP fusion reverses from insoluble to soluble upon cooling. Extensive pipetting is required for complete resuspension. Another option is to place the tubes on a rotator that is placed in a refrigerator. However, resuspension will take longer by this method.
-
9
To perform the first cold spin, centrifuge the samples 18,000 xg for 10 min at 4 °C. A pellet, consisting of the insoluble contaminants, will be formed. Transfer the supernatant to clean microcentrifuge tubes in aliquots of 1 mL while exercising care to not disturb the contaminant pellet (save 20 μl for SDS-PAGE).
Subsequent rounds of ITC
In the following rounds of ITC, it is important to use as little NaCl as possible to trigger the phase transition. Although, NaCl is very poor at “salting proteins out” as seen by its position on the Hofmeister series, reducing the NaCl concentration is still desirable, as it limits the propensity of contaminating proteins to co-precipitate during the phase transition of the ELP fusion protein. After the first round of ITC, the volume of the protein is considerably smaller, so that the concentration of the ELP fusion protein is increased. As the transition temperature is inversely proportional to concentration, increasing the concentration of the ELP fusion protein will lower the transition temperature, so that less salt is required in this and subsequent rounds of ITC, as compared to the first round, to lower the transition temperature below room temperature. The exact concentration of NaCl depends on the specific ELP fusion protein, but 1-2 M is often sufficient to trigger the phase transition in subsequent rounds.
-
10The following step describes the hot and cold spin:
- Either weigh the correct amount of NaCl for the desired sample volume to obtain a final concentration of 1.5 M NaCl or alternatively add 100-200 μl of a saturated NaCl solution (drop wise until the phase transition occurs).
- After the solution turns visibly turbid, centrifuge the tubes at 18,000 xg for 10 min at room temperature.
- Carefully remove and discard the supernatant.
- Resuspend the pellet in 500 μl chilled PBS (~4°C)
- Save a 10 μl aliquot for SDS-PAGE.
- Centrifuge the resuspended pellets at 4 °C at 18,000 xg for 10 min.
- Transfer the supernatant from each tube and combine them into one or more fresh microcentrifuge tubes (save 10 μL for SDS-PAGE).
-
11To remove all contaminants completely, repeat step 10 another 3-5 times. Decrease the volumes used to resuspend the pellets stepwise so that the pellet after 5 rounds of ITC is resuspended in 100-200 μL (or 500 μL for large pellets. Save 10 μL of sample after each hot and cold spin for SDS-PAGE.The contaminant pellet precipitating in the cold spin should diminish in size after each hot spin. In the last round of ITC, a contaminant pellet should not be visible. Not all collected aliquots are visualized on SDS-PAGE. Most often the samples collected from the lysate, the product from the first round of ITC and the aliquots from the cold spins in the following rounds of ITC are sufficient to track the purification of the ELP fusion protein. However, if problems arise during purification all samples from both hot and cold spin after every round of ITC should be run on a gel.
-
12Run the saved aliquots from previous steps on an SDS-PAGE gel (refer to UNIT 10.1). Stain the gel with either 0.5 M CuCl2 solution or Coomassie Blue stain (the gel can be stained with either – always start with the copper stain). CuCl2 negatively stains the gel, turning it white except for the protein bands, which stay clear. Coomassie Blue stain can be used as well but it should be noted that free ELP will not stain by Coomassie Blue as it is an uncharged polypeptide.Two important conclusions can be derived from SDS-PAGE. First, the lane with the cell lysate should indicate the level of expression of soluble ELP fusion protein. Second, the lane of the purified sample should indicate the presence of any contaminants (if there is more than one band) and the molecular weight of the purified fusion protein. Running the aliquots from each stage of the purification will indicate any loss of the target protein if it occurs, thereby simplifying troubleshooting procedures.
Protease cleavage and removal of ELP tag
At this point in the purification, the protease used and the specific steps depend on the cleavage site that is genetically engineered between the ELP and the protein. We commonly use thrombin or TEV for cleavage of the ELP from the target protein. Important considerations in the choice of the protease are: (1) cost (thrombin is cheap, while TEV is not); (2) the commercial availability of a biotinylated conjugate of the protease, which allows easy removal after cleavage by the addition of streptavidin-agarose beads to capture the protease, followed by centrifugation to isolate the beads (biotin conjugates of both thrombin and TEV are available); and (3) the extra amino acids that are left behind after cleavage with the protease: thrombin leaves behind a Leu-Val-Pro-Arg sequence if its substrate peptide (recognition sequence) is located on the N-terminus of the target protein and Gly-Ser if it is on the C-terminus of the target protein, while TEV leaves Glu-Asn-Leu-Tyr-Phe-Gln if it its recognition sequence is located on the N-terminus and Gly if it is on the C-terminal end of the protein. The residual amino acids on the cleaved peptide or protein are an important consideration in the selection of the protease only if they lower the activity of the protein or peptide, deleteriously alter a physico-chemical property that is important for their end-use (e.g., solubility or crystallization), lead to an immunogenic response, or create regulatory hurdles to their clinical use. If these issues are not important, criteria 1 and 2 should dictate the choice of the protease.
-
13Carry out the cleavage reaction on the purified ELP fusion protein as specified by the instructions provided with the protease and verify cleavage by running the product on an SDS-PAGE gel.If the reaction proceeds to completion, the SDS-PAGE gel should show two clear and sharp bands that correspond to the molecular weight of the target protein and free ELP and the absence of a band corresponding to the ELP fusion protein.
-
14
Remove cleaved ELP by adding salt to trigger its phase transition, then centrifuge at 18,000 g for 10 min at room temperature. Transfer supernatant to a clean microcentrifuge tube.
-
15
Add the suggested amount of streptavidin-agarose beads (see instructions provided by the manufacturer) to the supernatant. Incubate for the recommended time and then centrifuge to separate the beads.
-
16
To remove the excess salt added to trigger the phase transition of the ELP, one of two methods can be used. The first option is to dialyze the sample against the desired buffer. The second option is to use molecular weight cutoff filters. We recommend dialysis, as concentrating the protein during centrifugal ultrafiltration might result in significant sample loss due to adsorption on the filtration membrane.
-
17
Analyze the sample on an SDS-PAGE gel to verify that the free ELP and the protease have been completely removed.
ALTERNATIVE PROTOCOL
ELP fusion proteins that express poorly
Not all proteins are over-expressed in a heterologous expression system, and purification of proteins expressed at low levels can be a challenge. In this section, we show how to overcome this problem by co-aggregation: poorly expressing ELP fusion proteins can easily be purified by ITC by adding free ELP to aid the aggregation process in ITC (Christensen et al. 2007). After step 5 of the basic protocol, proceed with the following steps:
- Add 5 μM of free ELP (final concentration) composed of the same guest residues and molecular weight as the ELP tag in the ELP fusion protein.The protocol for the expression of free ELP is the basic protocol presented in this article; however we often use heat instead of salt to trigger the phase transition except for the first round of ITC. The plasmid construct is simply that of the desired ELP gene before fusion to the target protein.
Continue with step 6 in the Basic Protocol.
COMMENTARY
Background information
The first reports of protein purification efforts date back more than 200 years; in 1789 Antoine Fourcroy (1755-1809) identified three different animal proteins: albumin, fibrin and gelatin (Tanford and Reynolds 2003). In 1888 Hofmeister purified ovalbumin to obtain the first protein crystals (Hofmeister 1888). However, until the 1940s, protein purification was largely an academic exercise for research use. During World War II, the first therapeutic proteins were produced, and purified at large scale (Unit 1.1.1). At this time, highly pure proteins are critical for research, for use as reagents in biotechnology and medicine and as therapeutics. These scientific and commercial imperatives provide the driving force for the quest to develop methods to purify proteins that are: (1) efficient (result in low protein loss during purification); (2) scalable (from the milligram scale to the metric ton scale); (3) robust (can be applied to many proteins); (4) reliable; and (5) low-cost.
Over the past 50 years, various chromatography procedures have been developed, e.g. ion exchange, size exclusion, and hydrophobic interaction chromatography (Terpe 2003) and have matured to the point where they are now the dominant method for the purification of proteins, especially for large scale industrial production. In general, purification of untagged proteins is tedious, as it typically requires many sequential chromatographic steps that are typically orthogonal in their separation principle (e.g., charge versus hydrophobicity) to achieve the level of purity that is needed for most applications. This situation has seen dramatic improvement in the last two decades –especially for bench scale purification of proteins for research needs– with the development of affinity chromatography, as it can provide sufficiently pure protein after a single chromatography step for many applications (see Units 9.4 and 9.9). We parenthetically note that we distinguish the recent development of affinity chromatography for the purification recombinant proteins with peptide or protein affinity tags that are appended to the target protein at the gene level from the older incarnation of affinity chromatography, which was restricted to a few native proteins using their natural cofactors or dye ligands as immobilized capture agents for chromatography (Scopes 1993).
Immobilized metal ion affinity chromatography (IMAC) of recombinant proteins is the most widely used affinity chromatography method for bench-scale purification at the milligram-gram scale. IMAC is a powerful method for protein purification but it has limitations: (1) it requires expensive resins; (2) the resin leaches metal ions, and the preferred metal ions for IMAC, Ni2+ and Co2+ are highly toxic; and (3) some native proteins present in the cell lysate have sufficient affinity for metal ions to bind to the IMAC resin, which typically limits the level of purification achieved by IMAC. Despite the popularity of IMAC and other affinity chromatography methods for purification of recombinant proteins, these methods are still confined to the laboratory, and have not been applied to the large scale purification of proteins, with the exception of the purification of antibodies by immobilized protein A in a chromatographic setup.
In our laboratory, we developed a new protein and peptide purification method that uses ELPs as the purification tag (Meyer and Chilkoti 1999; Trabbic-Carlson et al. 2004a). The method exploits our observation that proteins or peptides that are fused to a stimulus responsive ELP retain this behavior in the complex milieu of contaminating cellular components. We showed that by adding salt to the fusion protein solution or by heating the solution –or by a combination of both stimuli– the phase transition of the ELP fusion can be triggered in cell lysate, resulting in the formation of micron–size aggregates that are highly enriched in the ELP fusion, and that can easily be pelleted by centrifugation or collected on a filter. After removal of the supernatant, the pellet containing the fusion protein is redissolved (or dissolved from the filter) in low ionic strength buffer. Following the resuspension step, insoluble E. coli contaminants trapped in the fusion protein pellet are removed during the “cold spin” where the solution is centrifuged under solution conditions (low temperature, low salt) in which the ELP fusion is soluble (see Figure 2). One cycle of ITC takes roughly 30 minutes, and pure protein is often obtained after three rounds of ITC, so that this is an extremely time efficient method. Using ELPs as the purification tag has other significant advantages compared to affinity purification. The purification process does not require any resins or specialized equipment such as a dedicated HPLC or FPLC (e.g., Akta); although centrifuges are required, these are standard in most laboratories that perform molecular biology. The yields are comparable to those obtained by IMAC. During ITC purification, the obtained pellets can be dissolved in a small volume after each cycle of ITC, which concentrates the fusion protein. Concentrating the protein during the purification process is limited in affinity chromatography, as the elution volume is dictated by the loading of the column, the slope of the gradient used to elute the protein, and the size of each fraction that is collected.
All protein purification methods using a recombinant tag have the disadvantage that the tag has to be fused to the target protein at the gene level. Some applications require removal of the purification tag after purification of the fusion protein; for affinity tags, the purification process will include running the cleaved sample through the affinity column. Removing the ELP tag is simple. After proteolytic cleavage, salt is added to trigger the ELP phase transition and the ELP tag is simply centrifuged out of solution leaving only the target protein and the protease in the supernatant. The use of a protease-ELP fusion would allow the removal of the protease during this step as well.
We have reported the purification of several protein ELP fusions. They encompass a range of molecular weights, hydrophobicities, guest residue compositions and expression levels. The table 1 and 2 list the protein ELP fusion reported in literature.
Table 1.
List of protein ELP fusions purified in the Chilkoti lab
| Fused protein | ELP guest composition | Number of pentapeptides | MW of protein | Tt* | Citation | Comments |
|---|---|---|---|---|---|---|
| Thioredoxin | V | 20 | 11.8 | NA | (Meyer et al. 2001) | Hydrophilic. High levels of expression. |
| V5:A2:G3 | 90 | 54.0 | (Trabbic-Carlson et al. 2004b) | |||
| Chloramphenicol Acetyl Transferase | V5:A2:G3 | 90 | 25.7 | 38.3 | (Trabbic-Carlson et al. 2004b) | |
| Blue Fluorescent Protein | V5:A2:G3 | 90 | 26.9 | 51.1 | (Trabbic-Carlson et al. 2004b) | |
| 120 | 48.2 | unpublished | ||||
| 180 | 44 | unpublished | ||||
| Green Fluorescent Protein | V5:A2:G3 | 90 | 26.9 | 52 | (Trabbic-Carlson et al. 2004b) | |
| WW domain | V5:A2:G3 | 90 | 4.7 | 45.6 | unpublished | Unfolded mutant must be fused to the C-terminus. |
| Tendamistat | V5:A2:G3 | 90 | 8 | 35.6 | (Trabbic-Carlson et al. 2004b) | Hydrophobic. Fused to Thioredoxin to solubilize |
| Calmodulin | V5:A2:G3 | 60 | 16.9 | NA | (Kim and Chilkoti 2008) | Final resuspension in HEPES to avoid Ca+2 precipitation. |
| 180 | 41** | |||||
| IL1-Ra | V | 30 | 17 | 32 | (Shamji et al. 2007) | |
| V5:A2:G3 | 90 | 34 |
These Tt are for protein ELP fusion concentrations of 25μM in PBS
Measured in HEPES buffer in presence of Ca+2
Table 2.
Examples of protein ELP fusions purified using isothermal cycling in literature
| Protein ELP fusion | ELP guest composition | Citation |
|---|---|---|
| Synthetic helix-ELP-Enhanced cyan fluorescent protein | V | (Fujita et al. 2007) |
| Synthetic helix-ELP-Enhanced yellow fluorescent protein | ||
| Tat-ELP | V5:A2:G3 | (Massodi and Raucher 2007) |
| Organophosphorus hydrolase-ELP | V | (Shimazu et al. 2002) |
| ELP-Protein G | V | (Kim et al. 2005) |
| ELP-Protein L | ||
| ELP-Protein GL | ||
| ELP-Calmodulin | V | (Jenikova et al. 2007) |
| ELP-mercury-responsive transcriptional activator | V | (Lao et al. 2007) |
| ELP-intein- Green Fluorescent Protein | V | (Wu et al. 2006) |
| ELP-intein- Chloramphenicol Acetyl Transferase | V | (Banki et al. 2005) |
| ELP-intein-NusA | ||
| ELP-intein-Maltose binding protein | ||
| ELP-intein-Glutathione S-tranferase | ||
| ELP-intein-Catalase | ||
| ELP-intein-β-galactosidase | ||
| ELP-intein-β-lactamase | ||
| ELP-intein-A-hemoglobin stabilizing protein | ||
Critical parameters
DMSO stocks
For repeated protein expression and purification, it is easier to prepare a DMSO stock of BL21(DE3) or BLR(DE3) cells that are transformed with the expression plasmid. A stock of 1:1 7.5% DMSO to cell culture can be stored in the -80 °C and used to inoculate the expression cultures.
Salt choice
Salt is an important parameter when purifying ELPs, as transition temperatures are affected by the type and concentration of salt present in solution. The effect of these salts corresponds to their position in the Hofmeister series (Cho et al. 2008). Anions higher on the series than Cl- have a salting out effect on proteins and are known as kosmotropes. Strong kosmotropes, such as citrate and sulfate ions, can be used to trigger ELP transition when NaCl is not sufficient. However, strong kosmotropes also decrease the solubility of the other contaminating proteins (Zhang and Cremer 2006) resulting in the presence of more contaminants in the pellet. In this regard, NaCl is a good choice for the purification of ELP fusions by ITC as it is midway in the Hofmeister series, so that it is not very effective at salting proteins out in the 1-2 M concentration that is typically used for ITC. We have found that sodium citrate at concentrations of 0.15-0.5 M is a good choice for proteins with higher transition temperatures or lower yields. Ammonium sulfate is a strong kosmotrope and can be used for proteins that are difficult to purify, either due to their hydrophilic nature or low expression levels, in the initial rounds of ITC, though care must be exercised in the concentrations that are used –we typically use it at a concentration of < 1 M– as it can salt contaminating proteins out, especially at higher concentrations. Note that the concentration of ammonium sulfate that is needed for ITC is typically 0.4-1 M, which is significantly lower than is used for salting proteins out of solution.
E. coli growth temperature
In some cases, it will be advantageous to lower the growth temperature (step 2) to 25 °C if the fusion protein tends to form inclusion bodies or low yields are observed. For certain proteins, incubation at higher temperatures can also cause incorrect folding of the protein, which will decrease the yield of soluble, active fusion protein. Lower temperatures are also speculated to lower degradation rates (Farewell and Neidhardt 1998) allowing the protein to express at higher yields.
Additional chromatography purification steps
A final purification step using size exclusion chromatography (SEC) is an option to ensure complete homogeneity of the purified protein sample. However, we have found it unnecessary for most in vitro applications of ELPs. Although ITC is fairly effective in removing endotoxins, for in vivo applications of ELPs, especially where the amount of injected or implanted protein may be large such as in tissue engineering or local drug delivery, it is useful to remove all traces of endotoxin by a final chromatography step. We suggest that a final chromatography step using an endotoxin removal kit or using ion exchange chromatography (IEC) may be useful to remove any residual endotoxin.
Troubleshooting
Contaminants
Cold and hot denaturation can assist in removing any soluble contaminants. Storing the resuspended samples at -20 °C or incubating at temperatures above 42 °C can denature contaminating proteins rendering them insoluble and causing them to precipitate in the cold spin. It is important to note that these steps should only be used if the target protein will not denature under these conditions.
Additionally, less kosmotropic salts can be used to reduce the amount of contaminants that co-precipitate (if stronger kosmotropes than NaCl were used). Protease inhibitors (Roche cOmplete protease inhibitor cocktail) added to the resuspended cell lysate from step 3 and a shorter expression time in step 2, such as 12 h for the no induction protocol, might also be useful in reducing contaminants.
Low expression levels
If the cell lysate visualized on an SDS-PAGE gel does not contain a band corresponding to the expected target protein molecular weight, the target protein expression levels are low. In the case where expression levels are low due to the protein’s toxicity or where the protein is expressed unfolded, one method of resolving this issue is by inducing expression of the ELP fusion protein. In the pET expression system (Novagen), isopropylβ-D-1-thiogalactopyranoside (IPTG) is typically used to induce protein expression. We note that this is the accepted method for protein expression using this T7 expression system, though we have found that the T7 promoter is sufficiently leaky that for many proteins the low baseline expression allows greater overall accumulation (on a volumetric basis in shaker flask culture) by culturing the cells for 24 h in the absence of IPTG as opposed to the standard IPTG induction protocol. However, IPTG induction remains a viable option and indeed should be explored with any new protein. To induce expression, add a final concentration of 1 mM IPTG to the culture when the cell density reaches the recommended range (OD600 nm of 0.6 - 1) and grow for an additional 4 h. Additional control over baseline expression can be obtained using BL21(DE3)pLys cells, which express T7 lysozyme to more tightly control baseline expression (Stano and Patel 2004).
As mentioned previously, growth temperature can affect expression levels of proteins that are unfolded or degraded during expression. Lowering growth temperature to 25 °C works to decrease the rate of degradation and favors protein folding, though it will also decrease bacterial growth rate, so that the total amount for recovered cells and hence mass of the expressed protein on a volumetric basis will also be lower. The other two parameters that may affect yield of the fusion protein are the order of protein-ELP fusion, and ELP composition. Reversing the order of the fusion so that the protein is at the C-terminus of the ELP and fusing the protein to a more hydrophobic ELP have been shown to increase yields in several cases (Christensen et al. 2009).
Premature cleavage of ELP tag
For certain fusion proteins, we have observed that the ELP and protein are cleaved during expression (unpublished). This most likely occurs due to a non-specific protease site in the protein sequence. There are two solutions to the problem. The first approach is to add protease inhibitors in the culture (Roche EDTA free protease inhibitor cocktail) and to the resuspended cell pellet from step 3 (Roche cOmplete protease inhibitor cocktail). The protease inhibitors’ concentration in culture should be a fifth of the concentration in the cell lysate. The second approach is to shorten the culture time. Instead of expression for 24 h with no induction, addition of a final concentration of 1 mM IPTG to the culture when the cell density reaches the recommended range (OD600 nm of 0.6 - 1) followed by growth for an additional 4 h may resolve this problem.
Anticipated results
Yields of target proteins range between 50-500 mg/L for many proteins, depending on the protein and the ELP length and composition. The amount of target protein lost increases with each subsequent ITC round. Therefore, the minimum number of rounds of ITC that provide the desired purity is recommended. In contrast, complete removal of cleaved ELP should be expected after cleavage and another round of ITC. The activity of the ELP fusion protein or cleaved target protein should be verified before further experiments are conducted. If the ELP tag is not cleaved, it can serve as a “handle” for capture and separation in downstream experiments.
Time considerations
The time required to express and purify a target protein ranges between 2 to 5 days depending on the number of stop points. Growing the starter culture (step 1) requires overnight incubation. If no DMSO stocks are available, then an extra step to transform the plasmid into BL21(DE3) cells is needed and the plated cells must be grown overnight. Setting up the expression cultures (step 2) requires < 1 h while the incubation is 24 h for the no induction protocol or 8 h for the IPTG induction protocol. Spinning down the cell culture the next day and resuspension requires < 1 h. At this point, the sample can be stored at -80 °C. The next step is to sonicate the cells, which requires < 1 h. One round of ITC (hot spin and cold spin) takes 2 h to complete. After the hot spin or cold spin in any round of ITC, the sample can be stored at -20 °C. Analysis by SDS-PAGE requires 1 h. Protease digestion can take up to 16 h, depending on the protease. Protease cleavage reactions and sample analysis are completed in two days. It is possible to process up to 12 L of cell culture of the same protein in one round of ITC using standard floor model laboratory centrifuges. If smaller culture volumes (1 L) of different protein are expressed, up to 4 samples can be processed in one round of ITC.
Figure 3. SDS-PAGE gels following the ITC purification process of BFP-ELP[Val5Ala2Gly3-90].
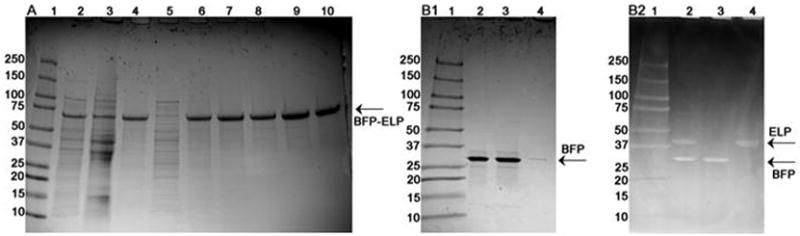
Gel A: Coomassie stained gel showing the molecular marker (lane 1), the insoluble fraction containing cell debris (lane 2), the soluble cell lysate (lane 3), the resuspended pellet from the first hot spin of ITC (lane 4), the supernatant from the first hot spin (lane 5), the supernatant from the first cold spin (lane 6), the supernatant from the second cold spin (lane 7), the supernatant from the third cold spin (lane 8), the supernatant from the fourth cold spin (lane 9), and the supernatant from the fifth cold spin (lane 10). Gel B1 (Coomassie stained) and B2 (Cu-stained) show the molecular marker (lane 1) BFP-ELP after thrombin cleavage (lane 2), pure BFP after thrombin cleavage and separation by ITC from the free ELP (lane 3), and the free ELP obtained after thrombin cleavage and ITC purification (lane 4). The purification process followed on gel A show that some BFP-ELP is trapped in the cell debris fraction (gel A, lane 2); the cell debris fraction can be washed with buffer in order to recover the trapped fusion protein. The supernatant from the first hot spin (lane 5) show no BFP-ELP; hence all the fusion protein aggregated and was collected in the pellet. 0.5 μl of each sample was added to the wells, the intensity of bands corresponding to BFP-ELP is increasing as the purification process progress due to concentrating the fusion protein. Thrombin cleavage is shown on gels B. Since ELP do not stain with Coomassie (B1) a copper stained gel is also shown (B2). Lane 2 shows the cleaved fusion protein before an additional ITC purification step. Lane 3 and 4 show BFP and ELP respectively after they have been separated by ITC.
Table 3.
Purification timeline
| Step | Time for completion |
Stop point | |
|---|---|---|---|
| Preparation | Incubation | ||
| Growing protein ELP fusions | |||
| Day 1:Starter culture *an extra day for transformation may be required prior to this step |
< 1 h | O/N | - |
| Day 2: Expression culture | < 1 h | 24 h (8h for induction protocol) | - |
| Day 3: Cell cultivation | 1 h | - | Store at -80° C |
| Lysing cells & ITC rounds | |||
| Day 4: Sonication | 1 h | - | Must continue to purification |
| Day 4 or 5: ITC rounds (each) | 2 h | - | After hot spin resuspension or cold spin store at -20° C |
| Day 4 or 5: Sample analysis | 1 h | - | - |
| Protease cleavage and ELP tag removal (Day 5 or 6) | 2 h | 16 h | Cleaved product can be stored at -20°C |
Abbreviations
- ITC
Inverse transition cycling
- ELP
elastin-like polypeptide
- TB
Terrific Broth
References
- Banki MR, Feng LA, Wood DW. Simple bioseparations using self-cleaving elastin-like polypeptide tags. Nat Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- Chen THH, Bae Y, Furgeson DY. Intelligent biosynthetic nanobiomaterials (IBNs) for hyperthermic gene delivery. Pharmaceutical Research. 2008;25:683–691. doi: 10.1007/s11095-007-9382-5. [DOI] [PubMed] [Google Scholar]
- Cho YH, Zhang YJ, Christensen T, Sagle LB, Chilkoti A, Cremer PS. Effects of Hofmeister Anions on the Phase Transition Temperature of Elastin-like Polypeptides. Journal of Physical Chemistry B. 2008;112:13765–13771. doi: 10.1021/jp8062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T, Amiram M, Dagher S, Trabbic-Carlson K, Shamji M, Setton L, Chilkoti A. Fusion order controls expression level and activity of elastin-like polypeptide fusion proteins. Protein Science. 2009;18:1377–1387. doi: 10.1002/pro.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T, Trabbic-Carlson K, Liu WG, Chilkoti A. Purification of recombinant proteins from Escherichia coli at low expression levels by inverse transition cycling. Analytical Biochemistry. 2007;360:166–168. doi: 10.1016/j.ab.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. Journal of Bacteriology. 1998;180:4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Funabashi H, Mie M, Kobatake E. Design of a thermocontrollable protein complex. Bioconjugate Chemistry. 2007;18:1619–1624. doi: 10.1021/bc070120x. [DOI] [PubMed] [Google Scholar]
- Ge X, Trabbic-Carlson K, Chilkoti A, Filipe CDM. Purification of an elastin-like fusion protein by microfiltration. Biotechnol Bioeng. 2006;95:424–432. doi: 10.1002/bit.21046. [DOI] [PubMed] [Google Scholar]
- Hofmeister F. Zur Lehre von der Wirkung der Salze. Naunyn-Schmiedeberg’s Archives of Pharmacology. 1888;24:247–260. [Google Scholar]
- Jenikova G, Lao UL, Gao D, Mulchandani A, Chen W. Elastin-calmodulin scaffold for protein microarray fabrication. Langmuir. 2007;23:2277–2279. doi: 10.1021/la0626151. [DOI] [PubMed] [Google Scholar]
- Kim B, Chilkoti A. Allosteric Actuation of Inverse Phase Transition of a Stimulus-Responsive Fusion Polypeptide by Ligand Binding. Journal of the American Chemical Society. 2008;130:17867–17873. doi: 10.1021/ja8059057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Mulchandani A, Chen W. Temperature-triggered purification of antibodies. Biotechnol Bioeng. 2005;90:373–379. doi: 10.1002/bit.20451. [DOI] [PubMed] [Google Scholar]
- Lao UL, Kostal J, Mulchandani A, Chen W. Affinity purification of plasmid DNA by temperature-triggered precipitation. Nat Protoc. 2007;2:1263–1268. doi: 10.1038/nprot.2007.171. [DOI] [PubMed] [Google Scholar]
- Luan CH, Harris RD, Prasad KU, Urry DW. DIFFERENTIAL SCANNING CALORIMETRY STUDIES OF THE INVERSE TEMPERATURE TRANSITION OF THE POLYPENTAPEPTIDE OF ELASTIN AND ITS ANALOGS. Biopolymers. 1990;29:1699–1706. doi: 10.1002/bip.360291403. [DOI] [PubMed] [Google Scholar]
- Massodi I, Raucher D. A thermally responsive Tat-elastin-like polypeptide fusion protein induces membrane leakage, apoptosis, and cell death in human breast cancer cells. Journal of Drug Targeting. 2007;15:611–622. doi: 10.1080/10611860701502780. [DOI] [PubMed] [Google Scholar]
- McPherson DT, Xu J, Urry DW. Product purification by reversible phase transition following Escherichia coli expression of genes encoding up to 251 repeats of the elastomeric pentapeptide GVGVP. Protein Expression and Purification. 1996;7:51–57. doi: 10.1006/prep.1996.0008. [DOI] [PubMed] [Google Scholar]
- Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nature Biotechnology. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: Examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- Meyer DE, Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- Meyer DE, Trabbic-Carlson K, Chilkoti A. Protein purification by fusion with an environmentally responsive elastin-like polypeptide: Effect of polypeptide length on the purification of thioredoxin. Biotechnology Progress. 2001;17:720–728. doi: 10.1021/bp010049o. [DOI] [PubMed] [Google Scholar]
- Scopes R. Protein Purification: principles and practice. 3. Springer-Verlag; New York and Heidelberg: 1993. [Google Scholar]
- Shamji MF, Betre H, Kraus VB, Chen J, Chilkoti A, Pichika R, Masuda K, Setton LA. Development and characterization of a fusion protein between thermally responsive elastin-like polypeptide and interleukin-1 receptor antagonist - Sustained release of a local antiinflammatory therapeutic. Arthritis and Rheumatism. 2007;56:3650–3661. doi: 10.1002/art.22952. [DOI] [PubMed] [Google Scholar]
- Shimazu M, Mulchandani A, Chen W. Environmentally triggered purification and immobilization of Elastin-OPH fusions. Abstracts of Papers of the American Chemical Society. 2002;224:U228–U228. [Google Scholar]
- Stano NM, Patel SS. T7 lysozyme represses T7 RNA polymerase transcription by destabilizing the open complex during initiation. Journal of Biological Chemistry. 2004;279:16136–16143. doi: 10.1074/jbc.M400139200. [DOI] [PubMed] [Google Scholar]
- Tanford C, Reynolds J. Nature’s robots: A history of proteins. Oxford Press; 2003. [Google Scholar]
- Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Applied Microbiology and Biotechnology. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- Trabbic-Carlson K, Liu L, Kim B, Chilkoti A. Expression and purification of recombinant proteins from Escherichia coli: Comparison of an elastin-like polypeptide fusion with an oligohistidine fusion. Protein Science. 2004a;13:3274–3284. doi: 10.1110/ps.04931604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabbic-Carlson K, Meyer DE, Liu L, Piervincenzi R, Nath N, LaBean T, Chilkoti A. Effect of protein fusion on the transition temperature of an environmentally responsive elastin-like polypeptide: a role for surface hydrophobicity? Protein Engineering Design & Selection. 2004b;17:57–66. doi: 10.1093/protein/gzh006. [DOI] [PubMed] [Google Scholar]
- Wu WY, Mee C, Califano F, Banki R, Wood DW. Recombinant protein purification by self-cleaving aggregation tag. Nat Protoc. 2006;1:2257–2262. doi: 10.1038/nprot.2006.314. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Cremer PS. Interactions between macromolecules and ions: the Hofmeister series. Current Opinion in Chemical Biology. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]


