Abstract
Plants adapted to environments where light is abundant are especially sensitive to competition for light from neighboring vegetation. As a result, these plants initiate a series of changes known as the shade avoidance syndrome, during which plants elongate their stems and petioles at the expense of leaf development. While the developmental outcomes of exposure to prolonged shade are known, the signaling dynamics during the initial exposure of seedlings to shade is less well studied. Here, we report the development of a new software-based tool, called HyDE (Hypocotyl Determining Engine) to measure hypocotyl lengths of time-resolved image stacks of Arabidopsis wild-type and mutant seedlings. We show that Arabidopsis grows rapidly in response to the shade stimulus, with measurable growth after just 45 minutes of shade exposure. Similar to other mustard species, this growth response occurs in multiple distinct phases, including two phases of rapid growth and one phase with slower growth. Using mutants affected in shade avoidance, we demonstrate that most of this early growth requires new auxin biosynthesis via the indole-3-pyruvate pathway. When activity of this pathway is reduced, the first phase of elongation growth is missing and this is correlated with reduced activity of auxin-regulated genes. Finally, we show that varying shade intensity and duration can affect the shape and magnitude of the growth response, indicating a dynamic spectrum of elongation response to shade.
Introduction
Plants need light to survive and are often in competition with other plants for photosynthetically active wavelengths of light (Franklin and Quail, 2010). As such, plants have evolved sophisticated photoreceptors capable of sensing their neighbors by monitoring the ratio of red light (R) (which chlorophyll absorbs as part of the photosynthetically active light spectrum) to far-red light (FR) (which chlorophyll does not absorb, and is thus reflected and transmitted through leaves) (Kasperbauer, 1987, Ballaré et al., 1990). A high R:FR ratio indicates a sparsely populated environment and abundance of photosynthetically active radiation. Conversely, a low R:FR ratio (<1) signals the presence of nearby vegetative neighbors that may soon compete for available light. This low R:FR ratio initiates a suite of responses, termed the Shade Avoidance Syndrome (SAS) that are of physiological and agricultural importance. The SAS has been extensively studied in the reference plant, Arabidopsis thaliana (Smith and Whitelam, 1997; Franklin, 2008). Plants grown under shade have decreased germination rate, increased hypocotyl and petiole elongation, inhibition of both leaf expansion and root elongation, reduced chlorophyll content, a tendency to flower early, reduced fecundity, and an increased susceptibility to herbivory (Izaguirre et al., 2006, Smith and Whitelam, 1997).
The Arabidopsis hypocotyl is an excellent model for studying shade phenotypes due to its simple structure, sensitivity to light, small cell number (~20 cells in each cell file) and reliance on cell expansion versus cell division for elongation growth (Gendreau et al., 1997; Chen et al. 2004). Seedlings foraging for light have long hypocotyls, while those in bright light (R:FR >1) have shorter hypocotyls. Hypocotyl length is inversely proportional to the fluence rate of white light. End-point assays quantifying shade avoidance phenotypes are typically performed by measuring hypocotyl length after several days of growth in light supplemented with far-red radiation (e.g. Lorrain et al, 2008; Sessa et al., 2005; Salter, et al., 2003). Through various studies under these conditions, a tentative model has emerged whereby the photoreceptors, phytochromes B, D, and E, perceive the shift in the ratio of R:FR light (Child and Smith, 1987; Smith, 2000). This allows the accumulation of at least two phytochrome-interacting basic Helix-Loop-Helix (bHLH) transcription factors, PIF4 (PHYTOCHROME INTERACTING FACTOR 4) and PIF5 (Lorrain et al., 2008; Khanna, et al., 2004; Huq and Quail, 2002), which promote growth through gene transcription (Lorrain et al., 2008). Out of the three phytochromes in the pathway, phyB, plays the strongest role, while phyD and phyE play relatively minor roles in the SAS (Franklin et al., 2003). A third bHLH protein, HFR1 (LONG HYPOCOTYL IN FAR-RED LIGHT), whose transcript is rapidly induced upon exposure to shade, constitutes a negative feedback mechanism by binding to PIF4 and PIF5, and inhibiting their DNA binding (and thus growth-promoting) activity (Fairchild, et al., 2000; Duek and Fankhauser, 2003; Sessa, et al., 2005; Hornitschek et al., 2009). Seedlings with mutations in the photoreceptor, phyA exhibit exaggerated hypocotyl elongation upon transfer to shade after 2 days of growth in continuous white light (Johnson et al., 1994), consistent with phyA signaling through HFR1 (Fairchild, et al., 2000). However, the role of phyA in shade avoidance is unclear as older seedlings do not display this phenotype (Zheng and Chory, unpublished data).
Gene expression studies have implicated other transcription factors with mild hypocotyl length phenotypes, including the bHLH-containing negative regulators PIL1 (PHYTOCHROME INTERACTING FACTOR LIKE 1), PAR1, and PAR2 (PHY RAPIDLY REGULATED) proteins, as well as a positive regulator, ATHB-2 (A. THALIANA HOMEODOMAIN BOX 2), an HD-ZIP class protein (Salter, et al. 2003; Roig-Villanova, et al., 2006; Devlin, et al., 2003; Sessa et al., 2005; Carabelli et al., 1996; Steindler et al., 1999; Schena, et al., 1993), yet their immediate targets have not been characterized.
New auxin biosynthesis is necessary for shade-induced hypocotyl elongation, evidenced by the discovery of an auxin biosynthesis gene, SHADE AVOIDANCE 3 (SAV3), which codes for the tryptophan aminotransferase, TAA1 (TRYPTOPHAN AMONITRANSFERASE OF ARABIDOPSIS 1, that converts tryptophan to indole-3-pyruvic acid). Exposure to shade induces auxin biosynthesis in wild-type plants (producing up to 50% more auxin within 1 hour), but not in plants lacking functional SAV3, in which shade avoidance phenotypes are severely reduced (Tao et al., 2008; Stepanova, et al., 2008). Treatment with NPA (1-naphthylphylamic acid), a polar auxin transport inhibitor, also attenuates shade-induced hypocotyl elongation (Steindler et al., 1999). Polar auxin transport has thus been shown to mobilize auxin from the leaves to sink tissues, such as the hypocotyl and petiole, consistent with the expression of SAV3 preferentially in leaf tissue (Tao et al., 2008). Finally, gibberellic acid (GA) promotes the shade response by inducing degradation of DELLA-containing proteins (a 5-member family of growth repressors) that can bind to and inhibit the PIF transcription factors (Djakovic-Petrovic et al., 2007; de Lucas et al., 2007; Feng, et al., 2008). Thus, the shade avoidance response provides an informative platform to study the intersection of light and various hormone signaling pathways as they relate to plant physiology, growth regulation, and environmental adaptation (Jaillais and Chory, 2010; Vandenbussche, et al., 2005).
While identification of some of the components that drive the shade avoidance response has advanced, there remains a gap between the time at which physiological phenotypes (several days after exposure) and molecular events (usually within 1–4 hours) are studied. Physical methods have been used to quantify growth in Vigna sinensis epicotyls (Garcia-Martinez, et al., 1987) and Sinapis alba stems (Morgan, et al. 1980, Child and Smith, 1987, Casal and Smith, 1989), and demonstrated that a rapid multiphasic growth response can occur upon exposure to shade. Similar physical techniques have been applied to study etiolated growth in Arabidopsis seedlings undergoing photomorphogenesis in response to monochromatic light (Parks and Spalding, 1999), and revealed subtleties of growth patterns among different photoreceptor mutants. While informative, these studies relied on invasive strategies for recording elongation rates, difficult (though not impossible) to execute on delicate and morphometrically dynamic Arabidopsis seedlings (Folta and Spalding, 2001; Miller et al., 2007), for which a wealth of experimental tools exist. Recently, more non-invasive imaging and feature-detection technologies have enabled the study of real-time growth dynamics in etiolated seedlings (Miller et al., 2007; Wang et al., 2008, Wang, et al., 2009) representing a great improvement over previous methods. However, automated image analysis of dark-grown seedlings is different than that of light-grown seedlings, as light-grown petioles can grow independently of the hypocotyl, and existing software exploits the petiole/cotyledon junction for analyzing dark-grown seedlings (Wang, et al., 2009). Thus, for imaging light-grown seedlings, new imaging approaches are needed.
Our goal was to assay hypocotyl elongation growth on short timescales by developing an image-based phenotyping platform. Here, we present HyDE (Hypocotyl Determining Engine), a new software tool for quantifying hypocotyl length on time-resolved image stacks of single light-grown seedlings. We employed this tool to reveal multiphasic growth patterns of Arabidopsis seedlings in response to shade, and to study the short-term effect of mutations in the shade avoidance response pathway. We defined distinct periods of hypocotyl elongation after shade and correlated these periods to mRNA accumulation for known shade marker genes. Finally, we showed that the magnitude and shape of the shade response was dynamically altered by adjusting the light regime. As such, our results lay a kinetic framework upon which to further characterize early events and identify novel components of the shade avoidance perception and response pathways.
Results and Discussion
HyDE software can image light-grown hypocotyl dynamics in an automated assay
To establish correlations between shade-regulated molecular events and the phenotypes they are thought to control, we developed an assay capable of measuring physiological phenotypes along the same short time scales used to study molecular ones. Our new assay adapts the Phytomorph CCD-camera based imaging system developed by Miller et al. (2007) to image light-grown Arabidopsis hypocotyls in a three-channel LED chamber, simulating high- and low-R:FR light conditions (Fig. S1a). To avoid interference from far-red light used to simulate shade, we utilized an IR filter with a cut-off at 790 nm, slightly longer than in Miller et al. (2007; 720nm), but still able to detect the 880 nm backlight source. To accompany the imaging setup, we devised a software tool, HyDE (for Hypocotyl Determining Engine) for automatically measuring the length of hypocotyls in a time-resolved image series (the HyDE user interface is detailed in the user guide provided with the software). The software first converts a cropped raw seedling image (Fig. S1b) to a binary (foreground/background) format (Fig S1c), and computes a Euclidean Distance Transform (EDT; Fig S1d). It identifies local maxima within the EDT, indicating centroid points of major organs (e.g. cotyledons and the shoot apical meristem). HyDE then constructs a digital hypocotyl based on the center line of the imaged hypocotyl using the bottom horizontal pixel slice as a reference to determine the horizontal position of the hypocotyl (the hypocotyl should be the only structure present on the lowest portion of the image). HyDE terminates the digital hypocotyl where it intersects the first local maximum corresponding to the shoot apical meristem, which forms a bulge where the petioles initiate (Fig. S1e). This hypocotyl center line is then spline-smoothened and its length recorded for all images in the stack (Fig. S1f). Our method works best on young (4–7 day old) seedlings, as they lack fully emerged true leaves, which can obscure morphological information of the hypocotyl-petiole junction. While we have not tested our software on older seedlings, we expect this 4–7 day window would represent the most opportune time to capture dynamic information, as older Arabidopsis hypocotyls are less responsive to growth stimuli. Similarly, we had to select seedlings prior to imaging that grew such that the cotyledons extended perpendicular (±15°) to the plane of imaging, so they would not interfere with hypocotyl length measurements. This software is available for download from: http://cactus.salk.edu/hyde.html.
We tested this image-processing algorithm on 5-day old long-day entrained (16 hours fluorescent white light/8 hours dark) wild-type Arabidopsis accession Columbia (Col-0) seedlings to assay for growth when released (at ZT12) into circadian conditions (LED simulated daylight). Under these release conditions, wild-type seedlings did not elongate their hypocotyls during the majority of the subjective dark period, rather exhibiting the major growth peak during the middle of the day, beginning at ZT2 and ending at ZT12 (Fig. S2). This is consistent with published circadian and diurnal growth data (Nozue, et al., 2007). We conclude from these experiments that the HyDE software is capable of measuring hypocotyl length on very short time-scales, being able to detect significant length differences in as little as 10 minutes, a resolution useful for understanding growth kinetics of shade avoidance responses.
Arabidopsis thaliana seedlings respond to shade within 1 hour in a multi-phasic growth pattern that varies with the R:FR ratio
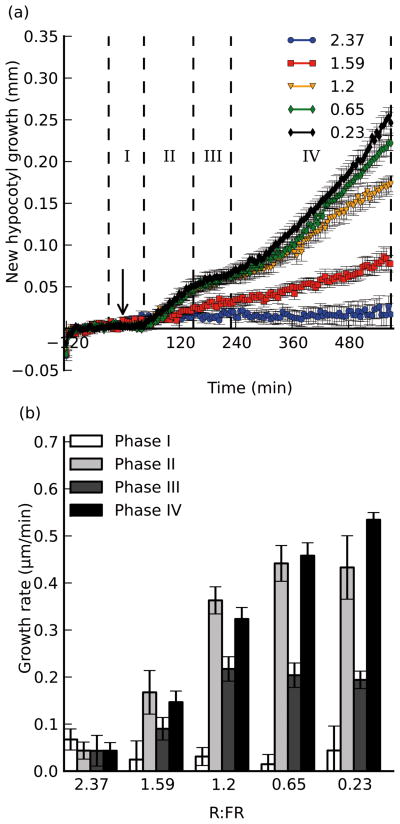
To assay for hypocotyl-specific growth during the shade avoidance response, we manipulated the far-red light intensity to mimic the very earliest “shade” conditions that plants encounter: the threat of shade (reflected FR light) from neighboring vegetation. Based on field studies of soybean plantings, which indicated that reductions in R:FR can be more prevalent at dusk (Kasperbauer, et al., 1987), we decided to treat the seedlings towards the end of the subjective day under light cycling conditions. Five-day old Arabidopsis seedlings (grown in fluorescent lights under long day conditions) were transferred to the imaging platform housed in a 3-channel LED chamber at ZT12, under high R:FR conditions (R:FR = 2.37). The seedlings were imaged for two hours to establish a baseline growth pattern, before being challenged with a marked increase in supplementary FR light, lowering the R:FR ratio to 0.23. After the start of treatment, we imaged seedlings for a further 9.5 hours, and hypocotyl lengths were measured using HyDE (Fig. 1a). As shown in Figure 1a, wild-type seedlings exhibited a lag period of about 45 minutes after exposure, growing at a basal rate of about 0.1 μm/min, before rapidly elongating at a rate of 0.45 μm/min until approximately 150 minutes post-exposure. The hypocotyl growth then slowed to a rate of 0.2 μm/min, until about 230 minutes after exposure, when it accelerated to ~0.55 μm/min for the duration of the experiment.
Figure 1. The magnitude of the growth response is inversely correlated with the R:FR ratio.
Seedlings were exposed to varying intensities of supplemental FR light, and imaged for 2 hours before and 9.5 hours after exposure. a), new hypocotyl growth observed for seedlings exposed to supplemental FR light such that the R:FR ratio became 0.23 (black), 0.65 (green), 1.2 (orange), 1.59 (red) or 2.37 (blue; control). Arrow indicates time of treatment (t=0). Dotted lines and roman numerals indicate phases of growth with which growth rates are calculated in b). b), growth rates calculated over selected time periods to reflect the different stages of growth. Error bars indicate ±s.e.m for at least 8 replicates in each assay.
Based on the shape of the curve, we defined four phases of growth under shade: a lag phase (measured from ~30 minutes pre-exposure to 45 minutes post-exposure), an initial growth phase (45–150 minutes post-exposure), a slowdown phase (150–230 minutes post-exposure), and a second growth phase (230 minutes post-exposure to the end of the assay). These phases are similar to those observed in Sinapis alba primary stems and first internodes (Morgan et al., 1980; Child and Smith, 1987; Casal and Smith, 1988); however, the observed lag times in Arabidopsis are significantly longer than those reported previously for Sinapis. Additionally, while it has been reported that Sinapis alba grows quicker in the first phase than the second, we observed that Arabidopsis hypocotyls grew at a slightly faster rate during the second growth phase than the first. Even more distinct are results from cowpea, which (while rapidly initiating growth within 60 minutes after exposure to supplementary FR), did not begin slowing until 10 hours into the experiment (Garcia-Martinez, et al., 1987). These disparities in kinetics may reflect the differences in species, tissues or experimental setup.
To further develop our assay and explore whether the severity of shade has a strong effect on growth kinetics, we treated wild-type seedlings in a similar assay as above, but decreased the R:FR to 0.65 (mild shade), 1.2 and 1.6 (non-shade), comparing growth curves from these seedlings to those that were not treated, or had the R:FR lowered to 0.23 (full shade). We found the elongation response to be very sensitive, showing a measurable growth rate increase even during the two least intense treatments (R:FR=1.6 and R:FR=1.2), previously assumed to be at or above the threshold of eliciting a shade avoidance response (Fig. 1; Smith, 2000). Mild shade treatment results in slower growth in both elongation phases (phase IV being slower than phase II) when compared to full-shade, in which seedlings grew faster in phase IV than in phase II (Fig. 1). Overall, there was an inverse correlation between the severity of the shade treatment and the magnitude of the elongation responses. All treatments exhibited a slowdown period, during which seedlings treated with 0.23, 0.65 and 1.2 R:FR light grew at approximately the same rate (0.2 μm/min), but those treated with 1.6 R:FR grew at a slower rate (~0.09 μm/min), by virtue of the reduced initial elongation growth rate, suggesting that this slowdown period does not need a threshold of growth to occur in order to be observed. Therefore our sensitive hypocotyl measurement method suggests that either the threshold R:FR ratio that elicits a shade response in Arabidopsis hypocotyls might be higher than previously thought, or that plants are sensitive to the change in the ratio, and not the absolute ratio itself. For further experiments, we decided to use the most severe treatment (R:FR 0.23), as it yielded the most dramatic results.
The relative time of treatment has a minimal effect on the quality and magnitude of the response within an 8-hour window
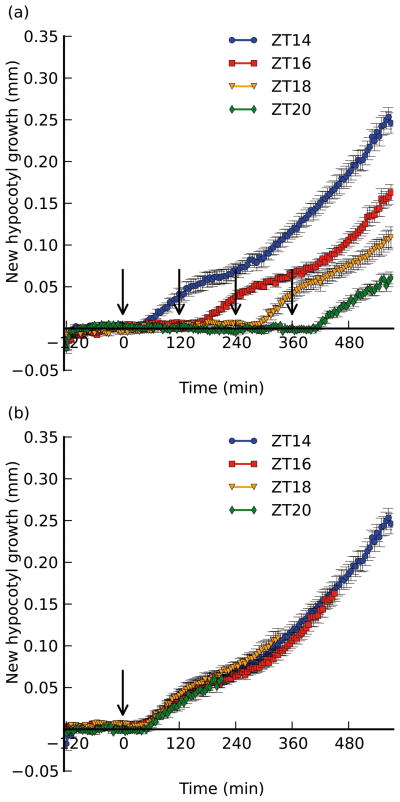
While growth in the absence of shade treatment appears to be negligible (Figs. S2 and S3), we decided to assess whether treating seedlings at different times would impact the magnitude or quality of the resulting growth curve. We subjected wild-type seedlings to the same pre-treatment conditions as before, but applied supplemental FR light 2, 4, or 6 hours later than our other assays (at ZT16, ZT18, or ZT20, respectively; Fig. 2). We observed the that quality of the growth response was nearly identical to that for seedlings treated at ZT14: seedlings had a sharp initial growth rate increase ~45 minutes after treatment, slowing after ~150 minutes, and then growing at a faster rate after ~230 minutes (Fig. 2a). The similarity of these responses is particularly apparent when the data are superimposed such that the subjective time of treatment is the same (0 minutes; Fig. 2b). Because we did not keep the time length constant for each assay, we cannot say that growth during phase IV of those seedlings treated at ZT20 is identical to that exhibited by seedlings treated at ZT14, but we observed that the lag period (Phase I) is identically timed, the slope and duration of the first growth phase (Phase II) is similar, and that these seedlings have a similar slowdown at the proper time (Fig. 2). Thus, it appears that if the time of treatment has a strong gating role in regulating hypocotyl growth in the shade, it is not immediately apparent during the 10 hour period we treat seedlings (at the end of the day), though we cannot rule out long-term temporal effects manifested after 12 hours of treatment. Our data suggest that factors that affect the qualitative parameters of the shade growth response (e.g. the slowing observed in Phase III) are likely to be initiated by the shade treatment itself. Thus, we saw little role for the circadian clock in promoting growth of hypocotyls within our time frame, in contrast to previous studies in which a more prominent role for clock regulation was apparent. However, we should emphasize that we did not perform a full-scale circadian clock assay that would directly test this hypothesis, and so we do not attempt to contradict previous reports suggesting a more prominent role for the clock (e.g. Salter, et al., 2003).
Figure 2. Shifting the time of shade treatment has no effect on the shape or magnitude of the growth response.
Col-0 seedlings were maintained in the same conditions as before (see Fig. 1), but were kept in high R:FR for 2, 4, or 6 hours longer than the original assay (treated with shade at ZT16, 18, or 20, respectively). Shown for reference is the ZT14 assay as described in Fig. 1. a) Growth responses normalized to time of transfer to the imaging platform. Treatment times are indicated by arrows (drawn at 0 min for ZT14, 120 min for ZT16, 240 min for ZT18, and 360 min for ZT20). b) curves normalized to time of treatment (indicated by the arrow). Data was truncated for length measured prior to 120 min. before treatment. Error bars indicate ±s.e.m. for at least 6 replicates.
Auxin is necessary for proper initiation of the elongation growth response, and together with PIF4 and PIF5 regulates its magnitude
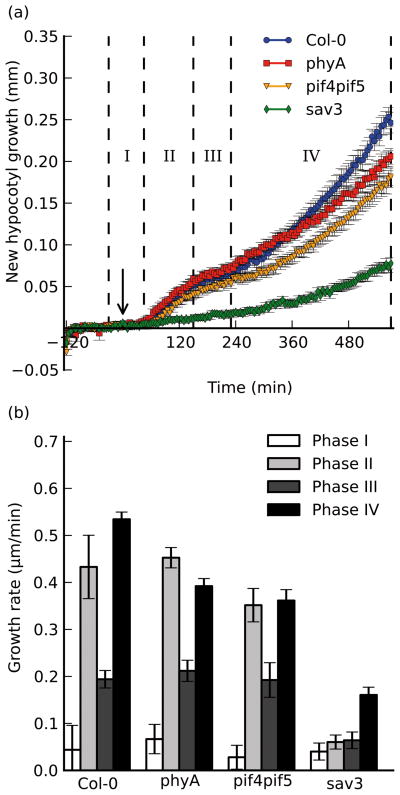
We next tested whether mutations within some of the shade avoidance pathway components would alter the quality or magnitude of the shade growth response. Previous studies, which quantified hypocotyl responses to shade in plants deficient in phytochrome A, suggested a negative role for this photoreceptor on shade-induced growth (Johnson, et al., 1994). Thinking that at least one, (if not all), growth phases in a phyA mutant would show higher growth rates, we imaged phyA-211 seedlings (a null allele; Nagatani, et al., 1993) in our shade assay. As shown in Fig. 3, phyA-211 seedlings had normal timing and magnitude of the initial elongation and slowdown phases; yet although they initiate phase IV at the same time as wild type, they fail to reach the same final growth rate as wild-type seedlings, resulting in slightly shorter hypocotyls at the end of the experiment. This is inconsistent with studies suggesting a negative effect of phyA action on the shade elongation response (Johnson, et al., 1994). Rather, our conditions (imaging seedlings older than those used in Johnson, et al., 1994) may highlight the proposed role of phyA in enhancing the response to low R/FR manifested by reduced phyB Pfr levels (Casal, 1996), as opposed to the role phyA plays in mediating the high irradiance response during de-etiolation.
Figure 3. Shade-induced growth occurs in multiple distinct phases at the end of the day.
a), Shade-induced growth of wild-type (Col-0; blue circles), phyA-211 (phyA; red squares), sav3-2 (sav3; green diamonds), and pif4-101pif5-1 (pif4pif5; orange triangles). Supplemental FR treatment was performed after 2 hours of acclimation (indicated by the arrow) at t=0 minutes. Seedlings were monitored for an additional 9.5 hours after exposure to shade. Phases of growth are indicated by roman numerals and dotted lines. b), Growth rates were calculated for defined periods after shade exposure. Error bars represent ±s.e.m. for at least 8 replicates in each assay.
We observed a more dramatic effect in sav3-2 seedlings, which did not respond at all during the first ~4 hours of the shade response, lacking growth in both phase II and phase III. They did, however, show a sharp increase in elongation rate towards the end of the assay at approximately the same time that wild-type seedlings began their second elongation phase, however this growth rate was only ~30% of that seen in wild-type seedlings in the same time period. These results suggest that new auxin biosynthesis made by the SAV3/TAA1 pathway plays an important qualitative role in initiating the elongation response, and a quantitative role in maintaining high rates of elongation growth after several hours of shade exposure. We also speculate that the lag time observed directly after treatment (phase I) could be due to biosynthesis and transport rates, as the site of synthesis (the cotyledon margins) is distinct from the hypocotyl (Tao, et al., 2008). Regulation of TAA1 expression alone cannot account for the phenotypes we observe, as its transcript abundance tends to decrease upon exposure to shade (Tao, et al., 2008). Recent results suggest that auxin transport in etiolated hypocotyls is linked to phytochrome activity (Wu, et al., 2010, Nagashima, et al., 2008), however the mechanism by which light regulates auxin biosynthesis or transport remains unclear.
pif4pif5 double mutant seedlings (Lorrain, et al., 2008; Fujimori, et al., 2004) exhibited all growth phases at the proper time, yet the magnitude of both growth phases is slightly less when compared to wild-type growth rates, and this defect is more pronounced for phase IV over phase II growth (Fig. 3). This mild phenotype suggests that while the PIF4 and PIF5 transcription factors have some quantitative role in modulating growth, they are either redundant with other growth regulators, or that transcription has only a minor role in mediating the early elongation response to shade.
Gene expression patterns of shade marker genes correlate with phases of growth
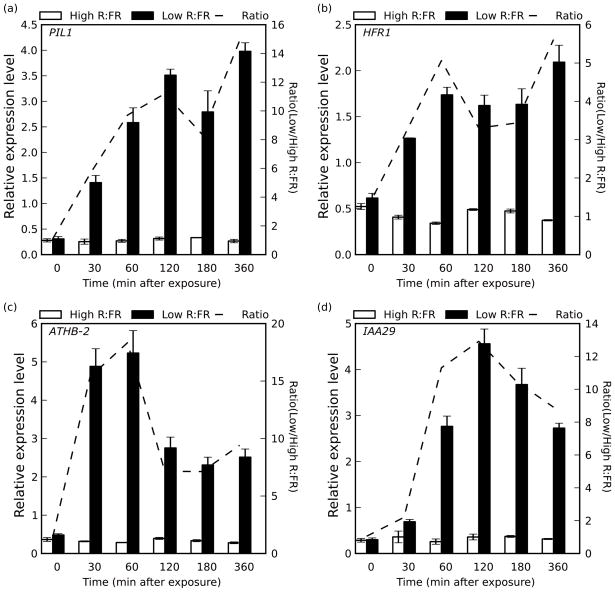
To assess whether gene transcription follows a similar multiphasic pattern, we prepared cDNA from wild-type plants at time points along the imaging assay reflecting each growth phase (0 min. and 30 min. for pre- and post-induction lag phase, 60 min. and 120 min. for the first elongation phase, 180 minutes for the slowdown phase, and 360 min. for the second elongation growth phase). We performed quantitative RT-PCR assays probing for PIL1 (Fig. 4a), HFR1 (Fig. 4b), ATHB-2 (Fig. 4c) and IAA29 (Fig. 4d) transcripts, previously shown to be strongly induced following one hour of shade exposure (Steindler, et al., 1999; Salter et al., 2003; Tao, et al., 2008). As expected, PIL1 expression was highly induced by the 30 minute time point, and continued to increase until at least 120 minutes post-exposure to shde light. We observed a marked decrease in expression level at the 180 minute time point, and a strong increase of expression again after 360 minutes, consistent with phase III and phase IV of our imaging assays (Figs. 1 and 4a). The same general pattern was observed for HFR1, although expression was merely maintained at a constant level through phase III, before increasing again towards the end of the assay (Fig. 4b). While PIL1 and HFR1 have both been shown to be negative regulators of the shade growth phenotype (Roig-Villanova, et al., 2006; Hornitschek, et al., 2009; Sessa et al., 2005), their expression patterns appeared to track growth. It is also important to note here that while expression levels fluctuated along the time course consistent with growth rates, shade treatment increased RNA levels at all time points assayed.
Figure 4. Gene expression patterns of shade marker genes correlate to hypocotyl growth phenotype.
Col-0 seedlings were transferred into two separate chambers 2 hours prior to treatment under identical light conditions. One chamber was treated with supplemental FR at time 0, bringing the R:FR down to 0.23 (Low R:FR; black bars), while the other left under control conditions (High R:FR; white bars). Tissue was harvested for RNA extraction at each time point indicated. Quantitative Real-Time PCR was performed for a) PIL1, b) HFR1, c) ATHB-2 and d) IAA29. Error bars indicate ±s.e.m. for 3 biological replicates. The dotted line indicates the ratio (High/Low R:FR) for each time point, and is plotted against a secondary y axis.
We also measured the expression pattern of two genes related to auxin signaling, ATHB-2 and IAA29 (Steindler, et al., 1999, Carabelli et al., 1996). mRNA levels of each were also induced quickly (ATHB-2 after 30 minutes, IAA29 after 60 minutes), and both continue to be induced into the 120 minute time point (phase II). However, both genes had reduced expression during phase III, and did not become re-induced during phase IV. This is consistent with auxin signaling playing a stronger role towards the beginning of the assay, indicated by the stronger sav3 phenotype during phase II than phase IV (Fig. 3). We speculate that the delayed induction response of IAA29 occurs as a consequence, and not a cause of the auxin-induced growth that occurs during the first elongation phase.
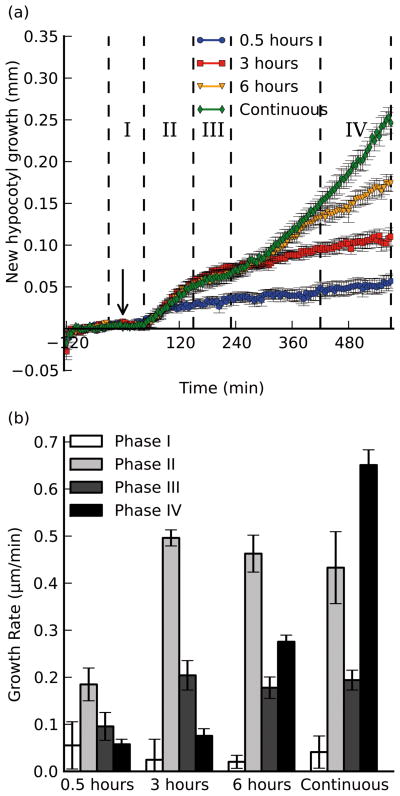
Elongation growth is reversible for at least 3 hours of shade treatment
To understand the extent of the elongation response that can be elicited by transient shade, we exposed plants to severe shade for 0.5, 3 or 6 hours before returning them to a high R:FR light condition (Fig. 5). Even though the shade treatment was reversed prior to the expected time of the initial response (phase II), seedlings still exhibited some elongation growth for at least 15 minutes before slowing their growth rate to phase I levels when exposed to just 30 minutes of low R:FR (Fig. 5). A similar reversal was seen for the 3 hour treatment, where seedlings never initiated the second phase of the growth response (phase IV), and instead slowed to phase I growth rates until the end of the experiment. Seedlings exposed to 6 hours of shade behaved identically to those exposed continuously until the 6 hour time mark (as expected); yet ~1 hour after reversion to high R:FR, the seedlings slowed to a level greater than phase I growth rates, indicating that some residual growth remains despite the light reversal. This does not appear to be a lag in reversal of growth, as the elevated rate persists for at least 3 hours, whereas both the 0.5-hour and 3 hour treated seedlings reverted to basal levels within 1 hour after the R:FR was raised (Fig. 5). This leads us to speculate that either some growth factor (perhaps auxin) remains at high levels after a longer duration of treatment, or that the cells have committed to an elongation program where low R:FR enhances, but is not necessary for continued elevated growth rates.
Figure 5. Elongation growth during shade is reversible by high R:FR light for at least 3 hours.
Seedlings were grown as described before, and treated with varying durations of low R:FR (0.23) light before the light was reverted to the high R:FR (2.37) control level. a), new hypocotyl growth (mm) plotted as a function of time for 3 treatments (continuous shade from Fig. 1 included for comparison). Arrow indicates time of shade treatment (t=0). Dotted lines indicate phases of growth (note last phase has different definition than Figs. 1 and 3 due to the close juxtaposition between the start of phase IV and the change of light treatment). b), growth rates measured over phases defined in a): the baseline rate (Phase I, 30 min. before treatment to 45 minutes after treatment; white bars), the first elongation phase (Phase II, 45 minutes to 150 minutes after treatment; light gray bars), the slowdown phase (Phase III, 150 minutes to 230 minutes after treatment; dark gray bars), and a section of the second elongation phase (Phase IV, 420 minutes to 570 minutes after treatment; black bars) chosen to highlight differences between the 6 hour and continuous treatments. Error bars indicate ±s.e.m. for at least 8 replicates in each assay.
In summary, using temporally resolved imaging assays, we have shown that shade avoiding hypocotyls exhibit multiple phases of elongation growth, controlled by separate, yet possibly integrated mechanisms, one of which being new auxin biosynthesis mediated by SAV3. This response can be mounted along a dynamic range of R:FR ratios, and the memory of the response can be maintained if longer durations of shade are applied. The time of treatment did not have a prominent role in promoting or restricting growth under our shade conditions within a 10 hour time frame. Rather, it is likely that the mechanism controlling the dynamic shape of the growth curve is internal to the treatment itself. Investigation into whether the clock has a strong role in controlling the shade avoidance response at different times of the day (e.g., at subjective dawn) would be an interesting avenue of future work. Initiating multiple layers of control of elongation by the shade stimulus may provide the plant with many decision points before commitment to a shade-avoiding lifestyle. An initial burst of growth after perception of a transient shade signal may give a plant just enough growth in the right direction to continue to intercept an optimal amount of light for photosynthesis, in which case it need not resort to the detrimental suite of responses characterized by plants growing in continuous shade. If, however, the signal is more persistent, such shade avoidance growth may be the best strategy to provide a competitive advantage under a less ideal environment. Further explorations of the mechanisms that control each phase of growth, and how a decision is made to commit to a shade-avoiding lifestyle will be informative.
Experimental Procedures
Plant growth and light conditions
Arabidopsis thaliana seeds were ethanol-sterilized and suspended in 0.1% agar for stratification prior to plating onto ½ Linsmaier and Skoog (LS) agar (0.8%) plates buffered with MES at pH 5.7. Plates were poured using a plastic mold occluding half of a circular petri dish, and the stratified seeds were plated on the ledge formed at the interface between the agar and mold (such that the apical portions of the seedling grow unobstructed in air, while the basal tissues grow into the agar plate). Plates were grown under 16hr/8hr day/night cycles in growth chambers (Percival, Perry, IA) supplemented with fluorescent and incandescent light (~37 μmol•m−2•s−1 blue light (400–600nm), ~30 μmol•m−2•s−1 red light (600–700nm), and ~11 μmol•m−2•s−1 far-red light) for 5 days before being transferred to the imaging platform in LED light (~37 μmol•m−2•s−1 blue light (400–600nm), ~30 μmol•m−2•s−1 red light (600–700nm), and ~11 μmol•m−2•s−1 far-red light) at ZT12 on the fifth day. Plants were then exposed to supplemental FR light by increasing the FR light intensity to lower the R:FR ratio as indicated in each experiment. For RNA experiments, seedlings were grown on Whattman filter paper atop ½ LS agar (0.8%) plates under the same light conditions as those for the imaging experiments.
Imaging Platform
Imaging machinery consisted of two CCD cameras, one Marlin and one Guppy (Allied Vision Technologies), coupled to macro video zoom lenses (Edmund Optics, NT54-363 for the Guppy, Qioptics MVZL for the Marlin) mounted to a rail on a thick aluminum surface (custom built). Plates are held in Plexiglas sample mounts for circular or square petri dishes, tightened by a thumb screw. The lenses were fit with IR long-pass cut filters (Edmund Optics NT54-755, Barrington, NJ), which allow only wavelengths longer than 790 nm through. Seedling samples were back-lit through an IR LED back-light emitting at 880 nm (Edmund Optics, Barrington, NJ). Image acquisition was attained via a FireWire connection to a laptop computer, using AVT SmartView software (Allied Vision Technologies, Stradtroda, Germany). This software allows imaging up to 15 frames per second for any duration of time, however in the interest of saving disk space and maximizing the number of genotypes and conditions to be sampled, we saved images every 5 minutes (every 240th frame with a frame rate of 0.8 fps). Image stacks were cropped with ImageJ (NIH) after image acquisition, but prior to processing with HyDE software such that only one seedling hypocotyl was visible (and the very base of the cropped image cut the hypocotyl off to remove interference with the seed coat and agar surface). The imaging platform and analysis software is diagramed in figure S1.
HyDE algorithm
HyDE input consisted of the cropped image stacks as described (see Imaging Platform in Materials and Methods), and were converted to binary image format using the MATLAB image processing toolbox, which automatically determined the threshold grayscale intensity value to assign foreground and background pixels. A coarse midline of the hypocotyl was determined by calculating the midpoint between two boundary pixels in successive horizontal pixel slices, ordered by the vertical position in the image. Each slice must be within a defined distance than that of the previous slice (this will break down within the petiole/cotyledon junction). A global Euclidean Distance Transform (EDT) using the city block method was calculated with the MATLAB image processing toolbox. The coarse hypocotyl midline was then refined by finding the maximum EDT value on the existing midline points, and terminating the hypocotyl at that point (which should correspond to the hypocotyl/petiole junction). The hypocotyl midline was then smoothened with a 4th-order spline (utilizing 3 segments), and the distance was measured by summation of 0.1 pixel size increments along the length of the spline. This process was repeated for each image in the stack, and automatically threw out data from images that deviated by more than a defined number of pixels from the previous image. Whole image stacks were removed from analysis if more than 10 consecutive images failed to give values.
RNA extraction and Quantitative RT-PCR
Seedlings grown in the indicated conditions were frozen in liquid nitrogen. Three biological replicates were collected per plate, ~10 seedlings/replicate. RNA was extracted from each seedling pool using the RNEasy Plant Mini Kit from Qiagen (Hilden,Germany) according to the manufacturer's instructions. cDNA was synthesized from 2 ug RNA using the Maxima cDNA synthesis kit from Fermentas (Burlington, Ontario) according to the manufacturer’s instructions. 10 μL of each 100X diluted cDNA sample was used for Quantitative RT-PCR, carried out with SYBR Green/Fluorescin dye on a BioRad iCycler (Hercules, CA) machine according to the manufacturer's instructions. Gene expression was quantified using the method presented in Pfaffl, et al., (2001). Primer pairs are listed in Supplementary Table 1.
Supplementary Material
Overview of imaging equipment and HyDE image analysis software
Circadian-regulated growth of long-day entrained wild-type seedlings.
Growth curves and growth rate of non-treated wild-type and mutant seedlings
Acknowledgments
We thank Dr. Edgar Spalding (U. of Wisconsin) for advice in developing the software, and adapting the imaging platform, Dr. Zuyu Zheng (Salk) for information concerning phyA shade phenotypes, and Dr. Christian Fankhauser (Lausanne) for the pif4pif5 double mutant line. Drs. Yvon Jaillais, Ullas Pedmale, Dmitri Nusinow, and Colleen Doherty provided critical feedback on this manuscript. This work was supported by grants from NIH (R01GM52413 to J.C. and R01GM56006 to S.A.K) and NSF (IOS-0649389 to J.C.). B.C. was supported by an NSF IGERT fellowship (0504645). J.C. is an investigator of the Howard Hughes Medical Institute.
References
- Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science. 1990;247:329–332. doi: 10.1126/science.247.4940.329. [DOI] [PubMed] [Google Scholar]
- Casal JJ. Phytochrome A enhances the promotion of hypocotyl growth caused by reductions in levels of phytochrome B in its far-red-light-absorbing form in light-grown Arabidopsis thaliana. Plant Physiol. 1996;112:965–973. doi: 10.1104/pp.112.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Smith H. Persistent effects of changes in phytochrome status on internode growth in light-grown mustard: occurrence, kinetics and locus of perception. Planta. 1988;175:214–220. doi: 10.1007/BF00392430. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Smith H. The 'end-of-day' phytochrome control of internode elongation in mustard: kinetics, interaction with the previous fluence rate, and ecological implications. Plant, Cell Environ. 1989;12:511–520. [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Child R, Smith H. Phytochrome action lin light-grown mustard: kinetics, fluence-rate compensation and ecological significance. Planta. 1987;172:219–229. doi: 10.1007/BF00394591. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R. DELLA protein function in growth responses to canopy signals. Plant J. 2007;51:117–126. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J. 2003;34:827–836. doi: 10.1046/j.1365-313x.2003.01770.x. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schäfer E, Fu X, Fan LM, Deng XW. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. Shade avoidance. New Phytol. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Bilingham OE, Halliday KJ, Whitelam GC. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Phys. 2003;131:1340–1346. doi: 10.1104/pp.102.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Spalding EP. Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J. 2001;28:333–40. doi: 10.1046/j.1365-313x.2001.01157.x. [DOI] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1078–1086. doi: 10.1093/pcp/pch124. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Keith B, Bonner BA, Stafford AE, Rappaport L. Phytochrome regulation of the response to exogenous gibberellins by epicotyls of Vigna sinensis. Plant Phys. 1987;85:212–216. doi: 10.1104/pp.85.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre MM, Mazza CA, Biondini M, Ballare CL. Remote sensing of future competitors: impacts on plant defenses. Proc Natl Acad Sci USA. 2006;103:7170–7174. doi: 10.1073/pnas.0509805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Bio. 2010;6:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions) Plant Phys. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperbauer MJ. Far-red light reflection from green leaves and effects on Phytochrome-mediated assimilate partitioning under field conditions. Plant Phys. 1987;85:350–354. doi: 10.1104/pp.85.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel recognition motif necessary for targeting photoactivated phytochrome to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Miller ND, Parks BM, Spalding EP. Computer-vision analysis of seedling responses to light and gravity. Plant J. 2007;52:374–381. doi: 10.1111/j.1365-313X.2007.03237.x. [DOI] [PubMed] [Google Scholar]
- Morgan DC, O'Brien T, Smith H. Rapid photomodulation of stem extension in light-grown Sinapis alba L. Planta. 1980;150:95–101. doi: 10.1007/BF00582351. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, Saji K, Furukawa T, Koshiba T, Sekimoto M, Fujioka S, Kuroha T, Kojima M, Sakakibara H, Fujisawa M, Okada K, Sakai T. Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. The Plant J. 2008;53:516–529. doi: 10.1111/j.1365-313X.2007.03358.x. [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and characterization of Arabidopsis mutants that are deficient in Phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloor JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Parks BM, Spalding EP. Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc Nat’l Acad Sci USA. 1999;96:14142–14146. doi: 10.1073/pnas.96.24.14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for real-time quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martinez-Garcia JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006;141:85–96. doi: 10.1104/pp.105.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Schena M, Llyod AM, Davis RW. The HAT4 gene of Arabidopsis encodes a developmental regulator. Genes Dev. 1993;7:367–379. doi: 10.1101/gad.7.3.367. [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Whitelam GC. The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant, Cell and Environ. 1997;20:840–844. [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants – an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer J, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, Zhao Y, Ballaré CL, Sandberg G, Noel JP, Chory J. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar F, Voesenek LA, Van Der Straeten D. Reaching out of the shade. Curr Op Plant Bio. 2005;8:462–468. doi: 10.1016/j.pbi.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Wang L, Assadi AH, Spalding EP. Tracing branched curvilinear structures with a novel adaptive local PCA algorithm. In: Arabnia HR, editor. Proceedings of the 2008 International Conference on Image Processing, Computer Vision, and Pattern Recognition. 2. CSREA Press; Athens, GA: 2008. pp. 557–563. [Google Scholar]
- Wang L, Uilecan IV, Assadi AH, Kozmik CA, Spalding EP. HypoTrace: Image analysis software for measuring hypocotyl growth and shape demonstrated in Arabidopsis seedlings undergoing photomorphogenesis. Plant Phys. 2009;149:1632–1637. doi: 10.1104/pp.108.134072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Cameron JN, Ljung K, Spalding EP. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. The Plant J. 2010;62:179–191. doi: 10.1111/j.1365-313X.2010.04137.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of imaging equipment and HyDE image analysis software
Circadian-regulated growth of long-day entrained wild-type seedlings.
Growth curves and growth rate of non-treated wild-type and mutant seedlings