Abstract
Persistent viral infections are often associated with serious diseases, primarily by altering functions of the host immune system. The hallmark of Kaposi’s sarcoma-associated herpesvirus (KSHV) infection is the establishment of a life-long persistent infection, which leads to several clinical, epidemiological and infectious diseases, such as Kaposi’s sarcoma, a plasmablastic variant of multicentric Castleman’s disease, and primary effusion lymphoma. To sustain an efficient life-long persistency, KSHV dedicates a large portion of its genome to encoding immunomodulatory proteins that antagonize the immune system of its host. In this article, we highlight the strategies KSHV uses to evade, escape and survive its battle against the host’s immune system.
Keywords: immune evasion, immune modulatory protein, Kaposi’s sarcoma, Kaposi’s sarcoma-associated herpesvirus, primary effusion lymphoma
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV), the most recent addition to the γ-herpesvirus family, was initially determined, from KS lesions of an AIDS patient, as an etiological agent in 1994 [1]. KSHV is also known as the causative agent of several B-cell malignancies, such as primary effusion lymphoma (PEL) and multicentric Castleman’s disease [1–3]. The large KSHV DNA genome consists of 145 kb of unique sequences bounded by 40 kb of terminal tandem repeats, and is most similar in structure and gene content to Herpesvirus saimiri (HVS) of New World primates [4–6]. As is typical of other herpesviruses, KSHV establishes a life-long persistent infection in host cells by adopting one of two lifestyles known as latency and lytic replication. During lytic replication, the full repertoire of the viral genome is expressed in a highly orchestrated temporal order from immediate-early (α), early (β) and, finally, late (γ) genes, thereby allowing viral replication and ultimately killing the infected cell. By contrast, viral gene expression is heavily restricted during latency, facilitating the maintenance of the viral genome and evading detection by the host.
The host immune system is constantly in a state of alert against pathogens, employing powerful antiviral mechanisms to deal with viruses on many levels as part of their innate and adaptive immune responses. The innate immune response is able to slow down viral replication and activate cytokines, which trigger the synthesis of antiviral proteins. The adaptive immune response neutralizes virus particles and destroys infected cells. Obligate intracellular parasites, such as KSHV, must cope with such immune responses in order to establish a persistent infection. Therefore, it is not surprising that viruses often resort to hijacking this cellular machinery to achieve a balance between activating the host defenses and escaping from these dangerous sentries. The viruses produce a wide range of immune modulator proteins and utilize a myriad of immune evasion mechanisms that exploit the host immune system to advance their own lifecycles. Due to the evolution of numerous immune-evasion mechanisms by KSHV, 25% of its proteins have been shown to regulate different aspects of the host immune system. In this article, we summarize the current state of research regarding KSHV-associated host innate and adaptive immune evasion strategies, and the implications these modulations have on the establishment and maintenance of a persistent infection.
KSHV: replication cycle & gene products
Kaposi’s sarcoma-associated herpesvirusis a member of the γ-herpesvirus subfamily, Rhadinovirus genus. Virus infection occurs when KSHV enters into the susceptible cell through membrane fusion by several enveloped glycoproteins [7–11]. Once the viral capsid is in the cellular cytoplasm, it is transported to the nucleus, upon which linear viral DNA rapidly circularizes to form a nuclear episome. Subsequently, it is decided which of the two known viral transcriptional phases of virus (latency or lytic phase) will arise. During latency, the viral genome is retained as a circular episome in the nucleus, expressing only a minimal number of the viral genes to facilitate the maintenance of viral genome as well as to escape host immune detection. To switch their lifecycle from latency to lytic replication, KSHV encodes ORF50 (replication and transcription activator [RTA]), a lytic master-switch protein that has been shown to be required and sufficient for the triggering of the lytic replication cycle, and hence the expression of the entire viral genome as well as the production of progeny virions [12–14].
As noted above, KSHV has a large dsDNA genome that consists of a 145-kb-long unique region encoding more than 80 genes, and a 40-kb terminal repeat region composed of highly GC-rich terminal repeat units in order to efficiently support such a highly regulated lifecycle [4,15,16]. Almost 50% of the KSHV genome is dedicated to modulating the environment within the host to promote its own survival, manipulating pathways such as cell cycle regulation, programmed cell-death, signal transduction and immune modulation. Moreover, several groups have collectively identified a total of 17 miRNAs that are transcribed from the latency-associated region of the KSHV genome, which includes several coding regions (latency-associated nuclear antigen [LANA], v-cyclin, v-FLIP and kaposin) and multiple overlapping transcripts [17–21]. Recent data and the fact that these KSHV miRNAs are involved in KSHV pathogenesis imply that they are essential for the control of KSHV replication and latency by regulating the expression of viral lytic genes and the host survival pathway [22–25]. In addition to miRNAs, KSHV encodes a noncoding RNA transcript, a 1077-nuclotide polyadenylated nuclear RNA [26]. It is highly expressed during the early lytic phase and confines intronless RNA within the nucleus to block the assembly of export-competent messenger ribonucleoprotein particles [26,27].
Innate immune evasion
The immune system is canonically divided into two major branches: innate and adaptive immunity. The innate response elicited by an invading pathogen involves the rapid recognition of general molecular patterns by non-immune cells or cells of the innate immune system, such as monocytes or macrophages, dendritic cells (DC), and natural killer (NK) cells. In the context of viral infections, the innate immune system acts as the first line of defense for sensing an infection, and responds by unleashing protective defense mechanisms. Hence, the first barrier to overcome for successful viral infection is the rapid innate immune response of the host, which are involved in several effector mechanisms, including complement cascade, type I IFNs, inflammatory cytokine, NK cell immunity, apoptosis, autophagy and Toll-like receptors (TLRs) pathway.
Complement system
The complement system has long been considered to be a key arm of the innate immune system response to pathogens, but there is also growing evidence that the complement system participates in the adaptive immune response [28]. The complement system is composed of several dozen proteins that are either circulating in serum or are attached to cell surfaces. They orchestrate three distinct cascades, termed the ‘classic’, ‘alternative’ and ‘lectin’ pathways, which result in the activation of antimicrobial effector activities that range from the opsonization of foreign particles and the recruitment of phagocytes, to the lysis of infected cells. All three cascades converge by assembling a C3 convertase that can either initiate the opsonization of the foreign body or continue to activate C5 and thus propagate the cascade [29]. The complement regulatory proteins share a common structural motif, known as the short consensus repeat. The complement cascade is under tight cellular control by host inhibitor proteins, and thus, it is perhaps not surprising that viruses have either hijacked or co-opted some of these as an anticomplement defense system [30].
The ORF4 of KSHV encodes a lytic cycle protein that not only eases viral entry by acting as an attachment factor for binding heparin sulfate, but also mediates evasion from the host complement system by inhibiting the complement-mediated lysis of infected cells [31–34]. The protein, appropriately named ‘KSHV complement control protein’ (KCP), contains four short consensus repeats or complement control protein motifs at its N-terminus, and a C-terminal transmembrane anchor [31]. KCPs regulate the complement system by accelerating the decay of the classical C3 convertase and act as cofactors for the inactivation of C3b and C4b, components of the C3 and C5 convertases. However, the ability of KCPs to promote the decay of activated C3 convertase in the alternative pathway is modest, indicating that this pathway may not have a significant role in the clearance of the virus in vivo. Other γ-herpesviruses, including HVS, rhesus monkey rhadinovirus (RRV) and murine γ-herpesvirus 68 (MHV68), can also interfere with the complement system by encoding host complement control protein homologs that disrupt the progression of the complement cascade [31–34]. Taken together, this suggests that the evasion of the complement system is essential for viral fitness.
Type I IFN-induced signaling
IFNs are signaling molecules that are essential for regulating the activation of immune cells during an antiviral response [35]. There are at least three distinct types of IFNs: type I (IFN-α/β), type II (IFN-γ), and type III (IFN-λ1, -λ2 and -λ3, also known as IL-29), which are classified based on their amino acid sequences [36]. Specifically, the type I IFNs, IFN-α/β, represent a powerful and universal intracellular defense system against viruses of all sorts, which is best demonstrated in knockout mice that are unresponsive to IFN-α/β due to targeted deletions in their type I IFN receptor [37]. These mice quickly succumb to viral infections despite retaining an intact adaptive immune system [38]. Likewise, humans with genetic defect in STAT-1, a protein involved in the IFN signaling cascade, succumb to viral diseases at an early age [39]. All nucleated cells in the mammalian body are able to synthesize and secrete type I IFNs in response to virus infections. Secreted IFNs are then recognized by neighboring cells, causing them to express potent antiviral proteins [38,40]. In order to complete their lifecycle, viruses must find a way to modulate the host IFN-mediated immune response to work in their favor. Notably, a portion of the KSHV genome spanning a cluster of loci is dedicated to encoding four viral IFN regulatory factors (vIRFs; vIRF1–4), which are homologous to cellular IRFs [4,5]. They are all expressed during lytic reactivation [41,42], but vIRF1 and vIRF3 have also been detected in latently infected cells of different KS lesions and PELs [43–45]. The significance of this differential expression of vIRFs is not yet known, but these observations suggest that vIRFs may function redundantly, but act independently (depending on the nature of the cell type and stage of viral infection) to thereby elicit disparate and distinct mechanisms of IFN evasion [46,47].
The N-termini of the vIRFs exhibit significant levels of homology to the N-terminal DNA-binding domain of cellular IRFs, but lack several tryptophan residues essential for DNA binding such that vIRFs do not directly bind DNA [48–51]. Although they do not prevent cellular IRFs from binding to specific target DNA sequence, they are able to interact with cellular IRFs, other transcriptional factors and co-factors to obstruct various downstream cellular IRF functions [49–51]. vIRF1, however, seems to be an exception, as demonstrated in a recent study in which vIRF1 was shown to bind directly to a specific DNA sequence in the promoter region of a polycistronic transcript encoding K3, viral dihydrofolate reductase (ORF2) and viral IL-6 (vIL6), which may function to regulate viral gene expression [52].
Viral IFN regulatory factors (K9) contains a DNA-binding domain but does not compete with IRF1 for DNA binding, nor does it bind to or suppress IRF1 [50,53]. Rather, vIRF1 directly targets and regulate p300 activity. The association of vIRF1 with p300 interferes with the formation of CBP/p300 complex, as well as p300 HAT activity to prevent the IRF3-mediated transcriptional activity of IFN-α [54,55]. In contrast to this functionally relevant binding to p300, vIRF1 interaction with IRF7 does not block the transactivation activity of IRF7 [56]. Despite this range of anti-IFN activity, the time windows in which vIRF1 is able to block IFN responses in KSHV-infected cells are limited [53], suggesting that vIRF1 may have other functions during the multifarious stages of the virus lifecycle or that other vIRFs and viral immunomodulators are required to completely inhibit IFN responses. Indeed, full-length vIRF2 (which is translated from exons K11.1 and K11) inhibits the expression of IFN-inducible genes driven by IRF1, IRF3 and IFN-stimulating gene factor-3 (ISGF3), but not IRF7 [57]. Unlike vIRF1, it has recently been shown that vIRF2 accelerates the decay of activated IRF3 through caspase-3, thus functioning as a catalyst in the IRF3 degradation process, and concomitantly inhibiting IRF3 transactivation and host antiviral response [58]. Moreover, the first exon of vIRF2 (K11.1) prevents the activation of PKR [59], reducing protein synthesis and blocking IFN-α/β signaling [53,60]. In contrast to vIRF1, the specific mechanisms of vIRF2 activity have yet to be defined. Unlike vIRF1 and vIRF2, which mostly target IRF3-mediated IFN signaling, vIRF3 (also known as LANA2), specifically interacts with either the DNA binding domain or the central IRF association domain of IRF7. This interaction consequently leads to the suppression of IRF7 DNA binding activity and, therefore, the suppression of IFN-mediated immunity through the inhibition of IFN-α production [61]. It was recently reported that vIRF3 also interacts with IRF5, abrogating IRF5 binding to IFN-responsive promoter elements to thereby block IRF5-mediated promoter activation. Furthermore, vIRF3 was shown to antagonize IRF5-mediated activation of the p21 promoter [62]. Consequently, vIRF3 is essential for the proliferation and survival of cultured PEL cells by releasing IRF5 from transcription complexes [62,63].
Taken together, the downregulation of the IFN regulatory pathway is a common characteristic of the three vIRFs whose functions have been well-studied because they target IRF3 and IRF7, key initial factors of host immune surveillance against viral infections. vIRF4, the most recently identified member of the vIRF family, has yet to be studied mechanistically, but it may also have the potential to affect IFN-mediated innate immunity. The apparent importance of the modulation of IRF3 and IRF7 activity by KSHV is made further evident by the fact that the latent protein LANA1 and the early protein K8 (K-bZIP) act to obstruct IRF3, while two immediate-early proteins, ORF45 and ORF50 (RTA), negatively regulate IRF7. First, LANA1 inhibits IFN-β transcription and synthesis by competing with IRF3 to bind to the IFN-β promoter, thus interfering with the formation of the IFN-β enhanceosome and subsequently preventing the expression of the CREB-binding protein [64]. Additionally, K-bZIP (known as K8) also directly interacts with the IFN-β promoter to impede its binding with IRF3, thus precluding efficient IFN-β gene expression [65]. Alternatively, after reactivation, ORF45 interacts with IRF7, preventing its phosphorylation and blocking its translocation to the nucleus, thus downregulating type I IFN production [66]. RTA, another immediate early protein, augments ORF45 activity by promoting IRF7 ubiquitination, resulting in the induction of the proteasomal degradation of IRF7 [67]. The manipulation of the stability, function and modification of host IRF3 and IRF7 by many KSHV gene products provides a number of novel regulatory strategies for circumventing the innate immune defense system whilst enabling them to act at different stages of the viral cycle, with some proteins playing a role during the lytic replication cycle and others during latency. Ultimately, the inhibition of the IFN pathway by KSHV may be the virus’ first line of protection against an innate immune reaction, enabling the successful establishment of viral persistence.
Modulation of inflammatory signaling
Virus infection provokes the production of cytokines and chemokines, which regulate leukocyte trafficking and effector functions [68], thereby playing a key role in directing immune and inflammatory responses as well as angiogenesis. Hence, many viruses would benefit from antagonizing normal cytokine activities [69]. Chemokines can be divided into four subfamilies defined by the pattern and number of conserved cysteine residues located near their N-terminus, which are involved in disulfide bond formation: CC-, CXC-, CX3C- and C-family [69]. The induction of particular chemokines in combination with the differential expression of specific seven-transmembrane-domain G-protein-coupled chemokine receptors (GPCR) by certain leukocyte subsets determines which immune cells migrate to the site of infection during inflammation [69,70]. Chemokines interact with both their specific receptors to transduce signaling, and with cell surface glycosaminoglycans to form immobilized chemotactic chemokine gradients used by immune cells to travel across endothelial cell monolayers and into tissues via distinct binding sites [69,71]. In addition, cytokines can be either positive or negative regulators of the immune responses, and for this reason, some viruses encode their own cytokines (virokines) or chemokine receptors (viroceptors), or secrete chemokine-binding proteins, that are presumed to interfere with normal antiviral chemokine functions [72,73].
vCCL-1, vCCL-2 & vCCL-3
Kaposi’s sarcoma-associated herpesvirus encodes three chemokine homologs produced during its lytic program: viral CC-chemokine ligand-1 (vCCL1; K6), vCCL2 (K4) and vCCL3 (K4.1), previously known as viral macrophage inflammatory protein (vMIP)-I, vMIP-II and vMIP-III, respectively. vCCL1 and vCCL3 are specific agonists of host CC-chemokine receptor-8 (CCR8) or CCR4, which are present on both Th2 and CD4+CD25+ regulatory T cells, and attract these cells to downregulate an immune response [74–77]. vCCL1, vCCL2 and vCCL3 appear to be involved in Th2 lymphocyte chemoattraction by specifically activating CCR3 and CCR8. Meanwhile, vCCL2 can also antagonize Th1 cells by inhibiting signaling mediated by many other classes of chemokine receptors with which it has a high affinity, including CCR1, CCR2, CCR5, CX3C-chemokine receptor-1 (CXCR1) and CXCR4 [78]. Consistent with these actions, vCCL2 inhibits signaling of these receptors for arrest of monocytes and Th1 cells in the presence of their authentic chemokine ligands, such as a RANTES or MIP1α I, as well as promoting the arrest of eosinophils and Th2 cells [79,80]. Thus, vCCL2 not only blocks Th2 responses due to its agonist actions, but also impairs Th1 responses due to its antagonist activities. Apart from their functions in immune modulation, vCCLs have all been shown to promote angiogenic responses through the induction of VEGF and other angiogenic factors, such as viral GPCR (vGPCR) and vIL-6 in cultured PEL cells [81,82].
vCD200
Another negative regulator of inflammatory signaling is the production of viral K14, also known as viral cluster of differentiation 200 (vCD200), previously designated as viral OX2. K14 is an early lytic gene and exhibits a significant level of homology to cellular CD200, a broadly distributed cell surface leukocyte glycoprotein [83]. CD200 acts as a tissue-specific negative regulator of target cells by interacting with a specific receptor, CD200R, which has restricted expression in myeloid and T cells [84,85]. It was initially reported that vCD200 targeted and stimulated monocytes, macrophages and DCs to produce inflammatory cytokines, potentially to promote the cytokine-mediated proliferation of KSHV-infected cells [83]. On the other hand, it was recently reported that vCD200 directly binds to CD200R, inhibiting myeloid cell activation and reducing Th1-associated cytokine production [86,87]. Furthermore, vCD200 suppressed acute inflammatory responses in mice in which neutrophil-mediated inflammation was induced by carrageenan [88], indicating that vCD200 may contribute to immune dysregulation. Taken together, it seems likely that a crucial function of vCD200 is its role as an anti-inflammatory agent, although its primary target cell types within the myeloid compartment during KSHV infection in vivo are still uncertain.
vIL-6
K2, an early KSHV lytic gene, encodes vIL-6 and exhibits 25% amino acid identity, as well as structural and functional homology, with cellular IL-6 [89,90]. Both the cellular and vIL-6 molecules transduce signals through the common receptor subunit, gp130, to orchestrate inflammatory responses, angiogenesis and oncogenesis. Cellular IL-6 binds to the CD126 component, which then forms a complex with the gp130 transducer component of the receptor, causing activated gp130 dimerization. By contrast, vIL-6 seems to circumvent the normal cellular checkpoint of CD126 coupling with gp130 by directly binding to gp130 to initiate downstream signaling [91]. Consequently, because most human B cells lack CD126, vIL-6 may act on a much wider spectrum of B cells than its human counterpart [92]. As noted earlier, vIL-6 expression can be detected at various levels in all lytically infected cells, as well as in some latently infected B cells [41,93–96]. Knockdown of vIL-6 expression in PEL cells leads to markedly reduced cell growth [97]. It is suggested that vIL-6 may support the growth and survival of latently infected cells in an autocrine manner via intracrine signaling, ultimately contributing to the maintenance of latently infected cells and to virus-induced neoplasis. vIL-6 also acts as an angiogenic factor inducing VEGF to promote the recruitment of permissive cells, such as lymphocytes, monocytes and endothelial cells, to primary infection sites, thereby enhancing viral production [98]. The vIL-6 promoter itself possesses an IFN-α stimulated response element sequence, such that vIL-6 expression is actually induced by IFN-α, essentially functioning as a feedback loop that ensures blocking of IFN-α antiviral infection [99].
vGPCR
The KSHV vGPCR (or ORF74) is a bona fide signaling molecule, as signaling by vGPCR elaborates many mitogenic and angiogenic cytokines that are vital to the biology of KS and other KSHV-driven malignancies. vGPCR is transcribed as an early gene during lytic infection and is a viral homolog of the cellular angiogenic IL-8 receptor. Unlike its cellular homolog, vGPCR signaling is ligand-independent and constitutively active [100]. It constitutively activates a series of transcription factors AP-1, NF-κB and HIF-1α, which, in turn, induce the expression of growth factors, pro-inflammatory cytokines and angiogenic factors such as VEGF [101]. Central to vGPCR-induced oncogenesis, it can also activate the MAPK, PI3k, Akt and mTOR cell survival pathways [102–104], and its oncogenic potential is demonstrated by the fact that it is sufficient to induce cellular growth transformation and VEGF-mediated angiogenesis in fibroblasts [101]. Interestingly, vGPCR is conserved in all γ2-hepresviruses members, such as Epstein–Barr virus, MHV68, HVS and rhesus monkey rhadinovirus [105–107]. EBV vGPCR inhibits antiviral PKR activity, and the vGPCR of MHV-68 is required for efficient and productive viral replication in culture and in vivo [107–110]. Thus, all this collectively indicates that vGPCRs activate gene expression and help establish an intracellular condition that promotes viral replication, be it during de novo or reactivated lytic replication.
vCKBP
Some viruses encode a vCKBP, which does not share any sequence similarity with any known chemokines or chemokine receptors, but instead binds to chemokines with a high affinity and efficiently neutralizes their ability to establish a gradient along which immune cells can migrate [111,112]. Similar to poxviruses, cowpox virus and vaccinia virus (A41L), MHV-68 (M3) and αherpesviruses (gG) encode different subfamilies of CKBP. They can all inhibit chemokine-mediated signal transduction by inhibiting chemokines from binding to their GPCR or glycosaminoglycan [113–115]. However, no vCKBP has yet been identified in KSHV.
Modulation of apoptosis & autophagy
A common cellular response to pathogenic invasion is to undergo cell death executed by two programs to limit the infection: apoptosis and autophagy. Apoptosis (Greek for ‘self-killing’) is the best-described form of programmed cell death, but autophagy (Greek for ‘self-eating’, a lysosomal degradation and recycling of cytoplasmic constituents pathway essential for homeostasis) has also been reported to contribute to cell death [116–118]. Even though they are characterized by distinctive morphological and biochemical changes, both are tightly regulated, are highly conserved processes that are essential for homeostasis and development, and have been implicated in diseases [118,119]. Apoptosis is a cellular suicide mechanism and it has been well-documented as a host defense mechanism against viral infections. There are at least two pathways that lead to apoptosis: an extrinsic death-receptor-mediated and intrinsic mitochondria-dependent pathway. Indeed, apoptosis represents the predominant demise of virally infected cells. By contrast, cellular autophagy has been historically recognized as a survival program vital for the maintenance of homeostasis by removing damaged or superfluous organelles, and the protection of cells during times of stress by providing a temporary source of energy and macromolecule in essential cellular processes until normal conditions return. Given this primordial role of autophagy in removing harmful components in the cytoplasm, it is not surprising that autophagy may be used as an intrinsic cellular defense against viruses. As intracellular parasites, viruses are bound to encounter and interact with autophagy proteins in the course of an infection. Hence, viruses generally employ some gene products to disrupt and overcome both these altruistic processes because they are part of the overall host surveillance mechanism for blocking viral replication and dissemination. While these two pathways are seemingly independent, apoptosis and autophagy are not mutually exclusive; the two are substantially interconnected and coordinately regulated [118]. For instance, several apoptosis signaling molecules (including TNF-α, TRAIL, FADD and p53) also mediate autophagy, while Atg5, an autophagy effector, triggers apoptosis through its interaction with FADD [120–127]. Furthermore, the inhibition of the class I PI3K, AKT and mTOR pathway suppresses both apoptosis and autophagy [123]. KSHV encodes proteins that target the crosstalk between apoptotic and autophagic signaling in the cell death pathway including: inhibition of death receptor signaling (as anti-apoptotic) or suppression of autophagy elongation by expressing viral FLICE/vFLIP; and negatively regulating both apoptosis and autophagy by binding specific effectors in these pathways with viral Bcl-2 (vBcl-2).
vFLIP (K13)
The KSHV-encoded anti-apoptotic protein, vFLIP (also known as K13), is a truncated homolog of the cellular FLIP (cFLIP) [42,128,129] and contains two death effector domains. Latent expression of vFLIP occurs via splicing of the LANA transcript from the tricistronic mRNA, and via the use of the IRES in vCyclin coding sequences [130–132]. The anti-apoptotic activity of vFLIP has been attributed to its inhibition of caspase-8 activation and more recently to its ability to induce the expression of anti-apoptotic proteins via activation of NF-κB, such as receptor-interacting protein, NF-κB-inducing kinase, IκB kinase (IKK)α, IKKβ and NF-κB essential modulator via the activation of NF-κB [133–136]. In support of this concept, vFLIP-expressing transgenic mice displayed constitutive NF-κB activation, ultimately leading to an increased incidence of lymphoma that was independent of its inhibition of Fas-induced apoptosis [137]. Furthermore, vFLIP likely contributes to the proinflammatory micro-environment because its expression was found to induce NF-κB-regulated cytokine expression and secretion [138,139]. The induction of NF-κB signaling has been linked to at least two aspects of KSHV-infected pathogenesis: viral latency and oncogenesis. On the one hand, NF-κB activation by vFLIP is critical for its inhibition of lytic replication via the AP-1 pathway [140,141]. On the other hand, enhanced NF-κB signaling may be an important component of the transforming and oncogenic potential of vFLIP, as demonstrated in Rat-1 fibroblast assays and tumors growth assays in nude mice [142]. Collectively, it may be stated that as a result of its strong activation of the NF-κB pathway, vFLIP plays a plethora of roles in viral infected cell survival, transformation, morphological change, inflammatory activation and latency control, thereby directly contributing to KSHV pathogenesis. More recently, Lee et al. revealed that vFLIP suppresses autophagy by preventing Atg3 from binding and processing LC3, a protein involved in the elongation step of autophagy [143]. Consequently, vFLIP not only possesses anti-apoptotic activities, but additionally serves as an anti-autophagy molecule through its inhibitory interaction with the E2-like enzyme, Atg3.
vBcl-2
Kaposi’s sarcoma-associated herpesvirus ORF16 encodes a homolog of cellular Bcl-2, a pro- and anti-apoptotic molecule, which forms homo- or heterodimers to regulate the release of mitochondria apoptotic components [144]. KSHV Bcl-2 possesses Bcl-2 homology (BH) 1 and BH2 domains characteristic of this family of proteins, allowing the viral protein to tightly bind proapoptotic Bak and Bax peptides, and thereby inhibit apoptosis [145–147]. Interestingly, other γ-herpesviruses, including EBV, HVS and MHV68, encode a Bcl-2 homolog [148–150]. The Bcl-2 of MHV68, also known as M11, has been implicated in preventing Bax toxicity in yeast and blocking apoptosis in culture cells when apoptosis is induced by various stimuli [151,152]. Recent advances demonstrate that the vBcl-2 of γ-herpesviruses (including KSHV and MHV68) can also inhibit autophagy via a direct interaction with Beclin1, adding autophagy inhibition as an additional function through which vBcl-2 advances viral lifecycle and pathology [153,154]. In fact, MHV68 vBcl-2 exhibits an even higher binding affinity to Beclin1 and thus a more robust and persistent inhibition of Beclin1-mediated autophagy upon environmental stress, compared with its cellular counterpart [155]. Very recently, Xiaofei et al. demonstrated that the inhibition of autophagy by vBcl-2 is important in maintaining latent infections, while the anti-apoptotic activity of vBcl-2 is largely involved in efficient viral reactivation from latency [156]. Overall, it can be postulated that vBcl-2 circumvents host immune responses to establish and/or maintain a persistent infection by concomitantly blocking both apoptosis and autophagy.
Modulation of TLRs signaling
Toll-like receptors are evolutionarily conserved innate receptors expressed by mammalian hosts in various immune and non-immune cells, and play a crucial role in sensing infectious agents through the induction of inflammatory cytokines and type I IFNs. Currently, 11 human TLRs and 13 mouse TLRs have been identified, and each TLR has been demonstrated to recognize a particular pathogen-associated molecular pattern derived from various microorganisms, such as bacteria, viruses and fungi [157]. All TLRs contain 21–25 leucine-rich repeats in their extracellular domains, which recognize and bind to pathogen-associated molecular patterns in their N-terminal regions. The intracellular signaling domain, known as Toll/IL-IR (TIR) resides in the C-terminal region and is required for interaction with many different kinds of adaptor molecules to subsequently activate downstream signaling pathways that culminate in the induction of cytokines and IFNs through NF-κB and IRFs. TLRs are expressed in distinct cellular compartments: TLR1, TLR2, TLR4, TLR5 and TLR6 are expressed on the plasma membrane, whereas TLR3, TLR4, TLR7, TLR8 and TLR9 are expressed in intracellular vesicles (e.g., endosome and endoplasmic reticulum) [157–159]. It has hitherto been reported that TLR3, TLR4, TLR7, TLR8 and TLR9 are involved in the recognition of viruses by way of binding to RNA, DNA or viral glycoproteins [160,161]. A recent growing body of information is beginning to shed light on how KSHV modulates the TLR pathway. The first report showed that KSHV primary infection of human monocytes upregulates cytokine and chemokine activation in a TLR3-dependent manner, especially CXCL10 and IFN-β [162]. In addition, infection of endothelial cells with KSHV suppresses TLR4 signaling by vGPCR-mediated ERK activation and vIRF1 [163]. Lastly, Gregory et al. screened several PEL cell lines with specific agonists of different TLRs. It was found that stimulation of TLR7 and TLR8 with a TLR8 ligand, single-stranded polyuridine (a synthetic single-stranded RNA homolog), strongly reactivated KSHV [164]. Taken together, there is evidence that KSHV has developed several different ways to escape TLR-mediated detection throughout its lifecycle. Moreover, a recent study demonstrates that another pathogen, vesicular stomatitis virus, induced KSHV reactivation in KSHV-infected PEL cells [164], thereby linking innate immune activation with viral reactivation from latency. Consequently, it has been suggested that either TLR agonists or oncolytic therapy may be a valuable treatment of KSHV-related neoplasms.
Adaptive immune evasion
When the innate immune system confronts a pathogen, it becomes activated and prepares the adaptive arm of the immune system to respond appropriately. The adaptive immune system consists of two branches: the humoral immune response arm (production of antibodies by B cells) and the cellular immune response arm (activities carried out by cytotoxic CD4+ and CD8+ T cells). Both typically require antigen presentation in conjunction with MHC and a co-stimulatory signal for full activation. Accordingly, the antigen-processing machinery, particularly the MHC class I pathway has been targeted by many viruses, including KSHV.
Humoral immune response
Antibodies (e.g., IgG, IgM and IgA) are produced by plasma cells, the final stage of B-cell development after B cells have recognized an antigen and have been stimulated by CD4+ T cells and T-cell-derived cytokines. Unlike T cells, B cells can recognize antigen in its native form. In the lymph nodes, naive B cells recognize cognate antigen by their surface antibodies, become activated, switch from IgM to IgG production (class-switch), increase their immunoglobulin specificity and affinity, and differentiate into plasma or memory B cells as the cell continues to divide in the presence of cytokines. As a natural host defense mechanism, antibodies can directly neutralize viruses by sterically hindering the receptor–virus ligand interaction or by inducing conformational changes in viral receptor ligands. Other indirect effects caused by antibodies include the recruitment or activation of the innate immune effector system, such as antibody-dependent cell cytotoxicity, engulfment of antibody-coated (opsonized) viruses by phagocytes and complement activation. B cells are the likely cellular reservoir of KSHV infection [1,165]; therefore, KSHV seems to influence several aspects of B-cell biology through the modulation of the humoral immune response.
Recently, several groups have shown that a B-cell terminal differentiation factor, X-box-binding protein-1 (XBP-1), can effectively initiate KSHV reactivation by activating the RTA promoter, thus providing a link between B-cell development and KSHV pathogenesis [166–168]. XBP-1 has two major functions: it is triggered by a variety of different cellular stresses as part of the induction of unfolded protein response [169]; and participates in the terminal differentiation of B cells to plasma cells [170]. Interestingly, PEL cells predominantly express the inactive form of XBP-1, XBP-1u, however, when the active form, XBP-1s, is induced, the KSHV lytic cycle is activated by XBP-1s [171]. This raises the possibility that in KSHV-infected B cells, latency is maintained until plasma cell differentiation occurs.
The study of HIV-1 Vpu recently led to discovery of the IFN-induced transmembrane protein, tetherin (BST-2, CD317), as a novel component of host innate defense against enveloped viruses. To date, six viral proteins have been reported to counteract tetherin: HIV-1 Vpu, HIV-2 and SIV Env, SIV Nef, Ebola glycoprotein and KSHV K5 [172–178]. KSHV K5 induces the proteasomal degradation of tetherin during primary infection and upon reactivation from latency in endothelial cells. Furthermore, tetherin reduced KSHV release upon inhibition of K5 expression by siRNA, suggesting that tetherin is part of the IFN-induced innate immune response against KSHV [175].
Cellular immune response
Besides effective host humoral immunity, the activation of cellular immunity is usually required for clearance of established infection. The recognition of viral peptides presented by MHC class I molecules on cytotoxic T lymphocytes (CTLs) is a key event in the elimination of cells producing abnormal or foreign proteins, specifically during a virus infection, and CTLs thus play a critical role in the control of a viral infection, especially as a long-term immune surveillance effector that can react more quickly against the same virus after a primary infection [179]. Hence CTL evasion is a prerequisite for the replication of persistent viruses, particularly in the case of herpesviruses, which must establish a persistent, latent infection and must then reactivate in immunologically primed hosts to shed infectious virions. Indeed, all herpesviruses probably implement some strategy of CTL evasion during their lytic cycle (reviewed in [180,181]). Two proteins encoded by KSHV K3 and K5 efficiently downregulate the expression of MHC class I molecules and a large number of supplementary membrane receptors on the surfaces of infected cells, thereby preventing antiviral CTL responses [182,183].
Inhibition of MHC class I antigen presentation
The K3 and K5 proteins, also known as modulator of immune recognition (MIR)1 and MIR2, are highly homologous and encode a RING-CH domain with a nonclassical C4HC3 configuration of the cysteines and histidine residues [184,185], making them prototypes of a novel membrane-associated RING-CH (MARCH) family of ubiquitin E3 ligases. They interact with MHC class I molecules through transmembrane interactions, and trigger endocytosis and proteasomal degradation of MHC class I molecules by ubiquitinating its cytoplasmic tail, but without affecting their assembly or transport [186]. Interestingly, K3 downregulates the expression of both canonical and noncanonical MHC class I molecules in humans (HLA-A, -B, -C and -E), whereas K5 downregulates only HLA-A and -B alleles as substrate specificity is dictated by transmembrane interactions [182,183,187,188]. Additionally, vIRF1 has been suggested to interact with p300 to prevent basal transcription of MHC class I molecules. Thus, KSHV can also inhibit MHC class I transcription as another way of down-regulating MHC class I molecules on the cell surface of infected cells [189]. Notably, from the viral perspective, the virus must thwart both CTLs and NK cells because the downregulation of only MHC class I molecules renders infected cells sensitive to NK cell-mediated lysis. NK cells recognize infected cells in an antigen-independent manner, subsequently destroying such cells via their cytotoxic activities. Furthermore, they can rapidly produce large amounts of IFN-γ, which induce diverse antiviral immune consequences upon receptor ligation, including the augmentation of antigen presentation and activation/polarization of CD4+ T cells and macrophage [190]. Hence, K3 and K5 also target the IFN-γ receptor-1 (IFN-γR1) by inducing its ubiquitination, endocytosis and degradation, ultimately resulting in the inhibition of IFN-γR1-mediated activation of transcription factors [191]. Overall, it is suggested that the downregulation of surface MHC class I molecules and IFN- γR1 by K3- and K5-mediated ubiquitylation and lysosomal degradation inhibits primary host immunity against viral infection.
Inhibition of co-stimulation
The generation of a robust adaptive immune response requires the engagement of T cells with professional antigen-presenting cells such as DCs, macrophages and B cells. To sufficiently activate T cells, co-stimulatory signals, such as the interaction between CD28 and its ligands, B7-1 and B7-2 on antigen-presenting cell surfaces [192,193], and between ICAM1 and lymphocyte function-associated antigen-1 are essential [194]. In addition to lowering the level of MHC class I molecules, K5 also downregulates other components of the immune synapse, such as ICAM (CD54) and B7-2 (CD86), two co-activating molecules for NK cell activation, on the B-cell surface by inducing their endocytosis and degradation [183,195].
Taken together, K3 seems to specifically target MHC class I molecules, whereas K5 shares this function in addition to targeting multiple receptors, including MHC class I, CD86 and ICAM, with the overall effect of preventing detection by CTL and NK cells. Even in a single gene overexpression system, the resultant downregulation is sufficient to confer resistance to immune cells in culture, and therefore may protect KSHV-infected cells undergoing lytic replication in vivo. However, the contribution of K3 and K5 to immune evasion by KSHV during latency is less clear. Their influence appears to be most important in the early stages of KSHV infection, when diminished detection by T cells can result in reduced antiviral cytokine responses and an impaired production of CTLs, thereby allowing the virus to establish a persistent infection.
Conclusion
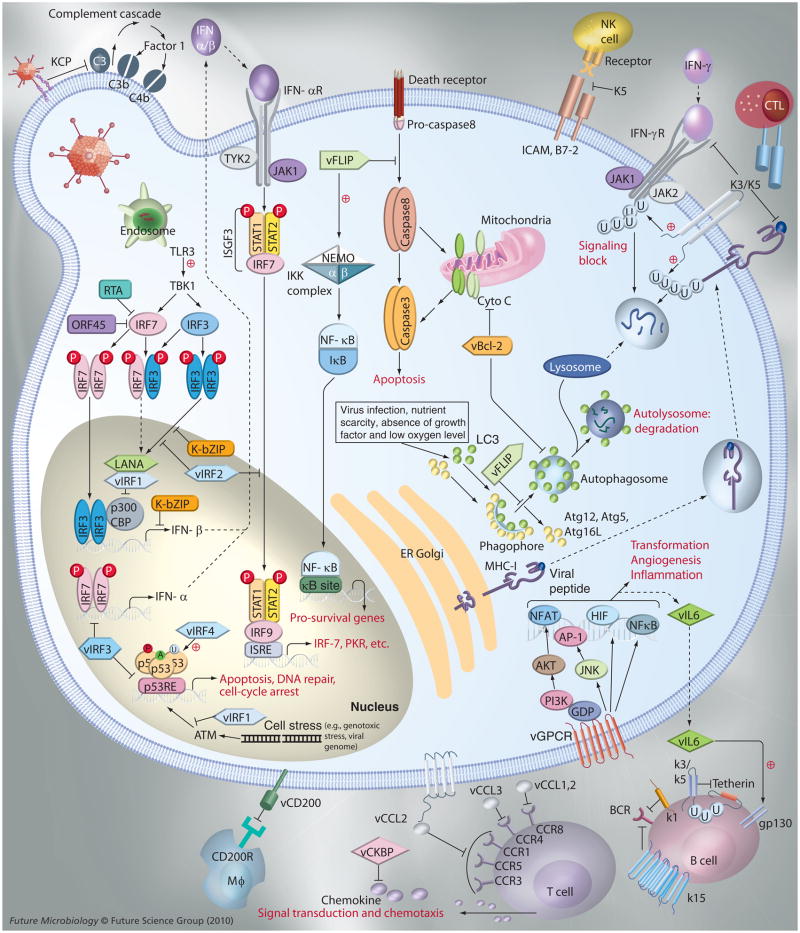
As previously described, KSHV harbors a surprising number of viral genes either pirated or modified from host genes, which contribute to immune evasion and set the foundation for a persistent viral infection and pathogenesis. Based on these, we briefly summarize the most prominent immune evasion mechanisms utilized by unique KSHV viral proteins, homologs of cellular proteins which function in a coordinated manner to antagonize host innate and adaptive immunity (Figure 1 & Table 1). Given, KSHV’s success at establishing a life-long persistence, it seems safe to say that these immunomodulation activities are quite effective. Furthermore, KSHV has multiple tropisms for different cell types, such that a certain subset of viral genes may help support virus survival in different cellular environments, securing its place as a parasite in the host. However, due to the lack of a permissive cell culture and an in vivo model system for KSHV, there remains the great task of integrating our observations regarding KSHV persistent infection with the sophisticated molecular mechanisms used by each viral immunomodulatory protein.
Figure 1. Kaposi’s sarcoma-associated herpesvirus immune evasion strategies.
CCR: CC-chemokine receptor; GPCR: G-protein-coupled chemokine receptor; LANA: Latency-associated nuclear antigen; NK: Natural killer; RTA: Replication and transcription activator; TLR: Toll-like receptor.
Table 1.
Immune evasion strategies of Kaposi’s sarcoma-associated herpesvirus.
| Strategy | Gene product | Function |
|---|---|---|
| Innate immune evasion | ||
| Inhibit complement | ORF4 (KCP) | Inhibit complement activation |
| Inhibit IFN | ORF45 | Prevent IRF7 activation |
| ORF50 (RTA) | Promotes IRF7 degradation | |
| vIRF1 | Inhibition of IRF3-mediated transcription | |
| vIRF2 | Suppression of IRF1 and IRF3 | |
| vIRF3 | Inhibition of IRF7 DNA binding activity | |
| LANA | Competing with IRF3 | |
| K8 (K-bZIP) | Impede IRF3 binding on IFN-β | |
| Inhibit cytokines/chemokines | K6 (vCCL1) | CCR8 agonist and Th2 chemoattractant |
| K4 (vCCL2) | CCR3 and CCR8 agonist, as well as a C-, CC-, CXC- and CX3C-chemokine antagonist | |
| K4.1 (vCCL3) | CCR4 agonist and Th2 chemoattractant | |
| K14 (vCD200) | Inhibition of myeloid cell activation, reduction of Th1-associated cytokine production | |
| K2 (vIL-6) | B-cell growth factor | |
| ORF74 (vGPCR) | Homolog of the cellular IL-8 receptor | |
| Inhibit apoptosis and autophagy | K13 (vFLIP) | Inhibition of caspase-8 activity and prevention of Atg3 interaction |
| ORF16 (vBcl-2) | Bind proapoptotic Bak and Bax proteins and direct bind with Beclin-1 | |
| Adaptive immune evasion | ||
| Inhibit humoral immune response | K5 | Induces tetherin degradation |
| Inhibit MHC class I antigen presentation | K3 | Downregulates MHC class I molecules as well as CD1d |
| K5 | Downregulates HLA-A and -B as well as CD1d | |
| Inhibit co-stimulation | K5 | Downregulates ICAM1 and B7-2 |
KCP: KSHV complement control protein; LANA: Latency-associated nuclear antigen.
Future perspective
Kaposi’s sarcoma-associated herpesvirus is linked to several malignancies in the human population and accounts for a large proportion of cancer deaths in Africa where the AIDS pandemic continues to fuel the incidence of KSHV infection. Notwithstanding these pressing human health problems, attempts to study KSHV infection are greatly hampered by the lack of cell culture and animal models. Although numerous immune evasion mechanisms employed by KSHV seem to be well-understood, it is too early to translate the knowledge we have obtained from basic science research into developing more effective clinical management and therapies because we can only speculate on how latent and lytic genes function within the context of an intact host immune system. To address this serious necessity, a variety of animal models including macaques, NOD-SCID mice and humanized SCID mice were tested for KSHV infection, yet none of them led to the appearance of virus-associated diseases upon infection [196–199]. However, Mutlu et al. recently reported that engraftments of mouse bone marrow endothelial-lineage cells that had previously been transfected with a KSHVBac36 induce an angiogenic phenotype and a KS-like, KSHV-dependent tumorigenicity [200]. Despite this significant advance, this model, like others, fails to truly recapitulate the overall aspects of the KSHV lifecycle in a living subject. Fortunately, we have recently provided the first animal model of KSHV persistent infection established via a zoonotic transmission of KSHV into common marmosets (Callithrix jacchus), a New World primate [201]. In this study rKSHV.219 infection of the common marmoset was shown to result in sustained serological responses, latent infection of PBMCs, virus persistence and KS-like skin lesion development, making this primate model highly analogous to what is seen in human infection [201]. Thus, this model is expected to not only help advance future research efforts studying KSHV pathobiology, but also greatly improve our chances to design and develop more effective antiviral therapies and vaccines against KSHV.
Acknowledgments
We apologize to our colleagues whose work could not be cited because of space constraints.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was partly supported by CA082057, CA31363, CA115284, CA147868, CA148616, AI073099, AI083025, DE019085, Hastings Foundation, Fletcher Jones Foundation, and Korean GRL and KRIBB grants. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1▪.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. The first descovery of the human rhadinovirus Kaposi’s sarcoma-associated herpesvirus (KSHV) [DOI] [PubMed] [Google Scholar]
- 2.Jung JU, Choi JK, Ensser A, Biesinger B. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin Cancer Biol. 1999;9(3):231–239. doi: 10.1006/scbi.1998.0115. [DOI] [PubMed] [Google Scholar]
- 3.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74(7):3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo JJ, Bohenzky RA, Chien MC, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neipel F, Albrecht JC, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71(6):4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albrecht JC, Nicholas J, Biller D, et al. Primary structure of the Herpesvirus saimiri genome. J Virol. 1992;66(8):5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang FZ, Akula SM, Sharma-Walia N, Zeng L, Chandran B. Human herpesvirus 8 envelope glycoprotein B mediates cell adhesion via its RGD sequence. J Virol. 2003;77(5):3131–3147. doi: 10.1128/JVI.77.5.3131-3147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akula SM, Wang FZ, Vieira J, Chandran B. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology. 2001;282(2):245–255. doi: 10.1006/viro.2000.0851. [DOI] [PubMed] [Google Scholar]
- 9.Birkmann A, Mahr K, Ensser A, et al. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J Virol. 2001;75(23):11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pertel PE. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J Virol. 2002;76(9):4390–4400. doi: 10.1128/JVI.76.9.4390-4400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪.Kaleeba JA, Berger EA. Kaposi’s sarcoma-associated herpesvirus fusion-entry receptor: cystine transporter xCT. Science. 2006;311(5769):1921–1924. doi: 10.1126/science.1120878. Identified xCT as another KSHV entry receptor. [DOI] [PubMed] [Google Scholar]
- 12.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95(18):10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradoville L, Gerlach J, Grogan E, et al. Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74(13):6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252(2):304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 15.Renne R, Zhong W, Herndier B, et al. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2(3):342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 16.Duprez R, Lacoste V, Briere J, et al. Evidence for a multiclonal origin of multicentric advanced lesions of Kaposi sarcoma. J Natl Cancer Inst. 2007;99(14):1086–1094. doi: 10.1093/jnci/djm045. [DOI] [PubMed] [Google Scholar]
- 17.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79(14):9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102(15):5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA. 2006;12(5):733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 21.Cai X, Cullen BR. Transcriptional origin of Kaposi’s sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80(5):2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 2010;84(6):2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei X, Bai Z, Ye F, et al. Regulation of NF-κB inhibitor IκBα and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12(2):193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6(6):570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25▪.Marshall V, Parks T, Bagni R, et al. Conservation of virally encoded microRNAs in Kaposi sarcoma--associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis. 2007;195(5):645–659. doi: 10.1086/511434. References [19–25] describe the identification of miRNAs in KSHV. These miRNAs are essential for controlling KSHV replication and latency through different mechanisms. [DOI] [PubMed] [Google Scholar]
- 26.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93(21):11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad NK, Steitz JA. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24(10):1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan BP, Marchbank KJ, Longhi MP, Harris CL, Gallimore AM. Complement: central to innate immunity and bridging to adaptive responses. Immunol Lett. 2005;97(2):171–179. doi: 10.1016/j.imlet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20(1):34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 30.Blom AM. Strategies developed by bacteria and virus for protection from the human complement system. Scand J Clin Lab Invest. 2004;64(5):479–496. doi: 10.1080/00365510410002904. [DOI] [PubMed] [Google Scholar]
- 31▪.Spiller OB, Robinson M, O’Donnell E, et al. Complement regulation by Kaposi’s sarcoma-associated herpesvirus ORF4 protein. J Virol. 2003;77(1):592–599. doi: 10.1128/JVI.77.1.592-599.2003. Describes the first characterization of KSHV ORF4 transcription and protein function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fodor WL, Rollins SA, Bianco-Caron S, et al. The complement control protein homolog of Herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J Virol. 1995;69(6):3889–3892. doi: 10.1128/jvi.69.6.3889-3892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapadia SB, Molina H, van Berkel V, Speck SH, Virgin HW., IV Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J Virol. 1999;73(9):7658–7670. doi: 10.1128/jvi.73.9.7658-7670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okroj M, Mark L, Stokowska A, et al. Characterization of the complement inhibitory function of rhesus rhadinovirus complement control protein (RCP) J Biol Chem. 2009;284(1):505–514. doi: 10.1074/jbc.M806669200. [DOI] [PubMed] [Google Scholar]
- 35.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147(927):258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 36.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 37.Muller U, Steinhoff U, Reis LF, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 38.Weber F, Kochs G, Haller O. Inverse interference: how viruses fight the interferon system. Viral Immunol. 2004;17(4):498–515. doi: 10.1089/vim.2004.17.498. [DOI] [PubMed] [Google Scholar]
- 39.Dupuis S, Jouanguy E, Al-Hajjar S, et al. Impaired response to interferon-α/β and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 40.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41▪▪.Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274(5293):1739–1744. doi: 10.1126/science.274.5293.1739. Demonstrates the functions of KSHV-encoded cytokines. [DOI] [PubMed] [Google Scholar]
- 42.Jenner RG, Alba MM, Boshoff C, Kellam P. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol. 2001;75(2):891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parravicini C, Chandran B, Corbellino M, et al. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am J Pathol. 2000;156(3):743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dittmer DP. Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 2003;63(9):2010–2015. [PubMed] [Google Scholar]
- 45.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol. 2001;75(1):429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham C, Barnard S, Blackbourn DJ, Davison AJ. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J Gen Virol. 2003;84(Pt 6):1471–1483. doi: 10.1099/vir.0.19015-0. [DOI] [PubMed] [Google Scholar]
- 47.Lee HR, Kim MH, Lee JS, Liang C, Jung JU. Viral interferon regulatory factors. J Interferon Cytokine Res. 2009;29(9):621–627. doi: 10.1089/jir.2009.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takaoka A, Tamura T, Taniguchi T. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci. 2008;99(3):467–478. doi: 10.1111/j.1349-7006.2007.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flowers CC, Flowers SP, Nabel GJ. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor confers resistance to the antiproliferative effect of interferon-α. Mol Med. 1998;4(6):402–412. [PMC free article] [PubMed] [Google Scholar]
- 50.Zimring JC, Goodbourn S, Offermann MK. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72(1):701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 52.Park J, Lee MS, Yoo SM, et al. Identification of the DNA sequence interacting with Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 1. J Virol. 2007;81(22):12680–12684. doi: 10.1128/JVI.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burysek L, Yeow WS, Lubyova B, et al. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73(9):7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Lee H, Guo J, et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72(7):5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung JU. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol. 2000;20(21):8254–8263. doi: 10.1128/mcb.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin R, Genin P, Mamane Y, et al. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene. 2001;20(7):800–811. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- 57.Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ. Inhibition of interferon signaling by the Kaposi’s sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol. 2006;80(6):3092–3097. doi: 10.1128/JVI.80.6.3092-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58▪.Areste C, Mutocheluh M, Blackbourn DJ. Identification of caspase-mediated decay of interferon regulatory factor-3, exploited by a Kaposi sarcoma-associated herpesvirus immunoregulatory protein. J Biol Chem. 2009;284(35):23272–23285. doi: 10.1074/jbc.M109.033290. Reports that the KSHV vIRF2 protein facilitates caspase-3-mediated decay of IRF3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burysek L, Pitha PM. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J Virol. 2001;75(5):2345–2352. doi: 10.1128/JVI.75.5.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burysek L, Yeow WS, Pitha PM. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2) J Hum Virol. 1999;2(1):19–32. [PubMed] [Google Scholar]
- 61.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi’s sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol. 2007;81(15):8282–8292. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wies E, Hahn AS, Schmidt K, et al. The Kaposi’s Sarcoma-associated herpesvirus-encoded vIRF-3 inhibits cellular IRF-5. J Biol Chem. 2009;284(13):8525–8538. doi: 10.1074/jbc.M809252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wies E, Mori Y, Hahn A, et al. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood. 2008;111(1):320–327. doi: 10.1182/blood-2007-05-092288. [DOI] [PubMed] [Google Scholar]
- 64.Cloutier N, Flamand L. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) β expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J Biol Chem. 2010;285(10):7208–7221. doi: 10.1074/jbc.M109.018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol. 2007;81(20):10950–10960. doi: 10.1128/JVI.00183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci USA. 2002;99(8):5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67▪.Yu Y, Wang SE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity. 2005;22(1):59–70. doi: 10.1016/j.immuni.2004.11.011. Identification that another function of replication and transcription activator in host immune evasion. [DOI] [PubMed] [Google Scholar]
- 68.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 69.Fernandez EJ, Lolis E. Structure, function, and inhibition of chemokines. Annu Rev Pharmacol Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- 70.Ransohoff RM. Chemokines and chemokine receptors: standing at the crossroads of immunobiology and neurobiology. Immunity. 2009;31(5):711–721. doi: 10.1016/j.immuni.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laing KJ, Secombes CJ. Chemokines. Dev Comp Immunol. 2004;28(5):443–460. doi: 10.1016/j.dci.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 72.McFadden G, Murphy PM. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr Opin Microbiol. 2000;3(4):371–378. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 73.Murphy PM. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat Immunol. 2001;2(2):116–122. doi: 10.1038/84214. [DOI] [PubMed] [Google Scholar]
- 74.Sozzani S, Luini W, Bianchi G, et al. The viral chemokine macrophage inflammatory protein-II is a selective Th2 chemoattractant. Blood. 1998;92(11):4036–4039. [PubMed] [Google Scholar]
- 75.Stine JT, Wood C, Hill M, et al. KSHV-encoded CC chemokine vMIP-III is a CCR4 agonist, stimulates angiogenesis, and selectively chemoattracts TH2 cells. Blood. 2000;95(4):1151–1157. [PubMed] [Google Scholar]
- 76.Dairaghi DJ, Fan RA, McMaster BE, Hanley MR, Schall TJ. HHV8-encoded vMIP-I selectively engages chemokine receptor CCR8 Agonist and antagonist profiles of viral chemokines. J Biol Chem. 1999;274(31):21569–21574. doi: 10.1074/jbc.274.31.21569. [DOI] [PubMed] [Google Scholar]
- 77▪.Iellem A, Mariani M, Lang R, et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2001;194(6):847–853. doi: 10.1084/jem.194.6.847. Provides the first evidence that CD4+CD25+ cells exhibit a distinctive Treg chemotactic response profile and chemokine receptor expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kledal TN, Rosenkilde MM, Coulin F, et al. A broad-spectrum chemokine antagonist encoded by Kaposi’s sarcoma-associated herpesvirus. Science. 1997;277(5332):1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 79.Weber KS, Grone HJ, Rocken M, et al. Selective recruitment of Th2-type cells and evasion from a cytotoxic immune response mediated by viral macrophage inhibitory protein-II. Eur J Immunol. 2001;31(8):2458–2466. doi: 10.1002/1521-4141(200108)31:8<2458::aid-immu2458>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 80.Chen S, Bacon KB, Li L, et al. In vivo inhibition of CC and CX3C chemokine-induced leukocyte infiltration and attenuation of glomerulonephritis in Wistar-Kyoto (WKY) rats by vMIP-II. J Exp Med. 1998;188(1):193–198. doi: 10.1084/jem.188.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C, Okruzhnov Y, Li H, Nicholas J. Human herpesvirus 8 (HHV-8)-encoded cytokines induce expression of and autocrine signaling by vascular endothelial growth factor (VEGF) in HHV-8-infected primary-effusion lymphoma cell lines and mediate VEGF-independent antiapoptotic effects. J Virol. 2001;75(22):10933–10940. doi: 10.1128/JVI.75.22.10933-10940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi YB, Nicholas J. Induction of angiogenic chemokine CCL2 by human herpesvirus 8 chemokine receptor. Virology. 2010;397(2):369–378. doi: 10.1016/j.virol.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chung YH, Means RE, Choi JK, Lee BS, Jung JU. Kaposi’s sarcoma-associated herpesvirus OX2 glycoprotein activates myeloid-lineage cells to induce inflammatory cytokine production. J Virol. 2002;76(10):4688–4698. doi: 10.1128/JVI.76.10.4688-4698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright GJ, Cherwinski H, Foster-Cuevas M, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol. 2003;171(6):3034–3046. doi: 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- 85.Rijkers ES, de Ruiter T, Baridi A, Veninga H, Hoek RM, Meyaard L. The inhibitory CD200R is differentially expressed on human and mouse T and B lymphocytes. Mol Immunol. 2008;45(4):1126–1135. doi: 10.1016/j.molimm.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 86.Minas K, Liversidge J. Is the CD200/CD200 receptor interaction more than just a myeloid cell inhibitory signal? Crit Rev Immunol. 2006;26(3):213–230. doi: 10.1615/critrevimmunol.v26.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87▪.Foster-Cuevas M, Wright GJ, Puklavec MJ, Brown MH, Barclay AN. Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor. J Virol. 2004;78(14):7667–7676. doi: 10.1128/JVI.78.14.7667-7676.2004. K14 (vCD200) interacts with CD200-R, and consequently inhibits secretion by activated macrophage of proinflammatory cytokines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rezaee SA, Gracie JA, McInnes IB, Blackbourn DJ. Inhibition of neutrophil function by the Kaposi’s sarcoma-associated herpesvirus vOX2 protein. AIDS. 2005;19(16):1907–1910. doi: 10.1097/01.aids.0000189849.75699.46. [DOI] [PubMed] [Google Scholar]
- 89▪.Neipel F, Albrecht JC, Ensser A, et al. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71(1):839–842. doi: 10.1128/jvi.71.1.839-842.1997. Reports the identification of vIL-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nishimoto N, Kishimoto T. Interleukin 6: from bench to bedside. Nat Clin Pract Rheumatol. 2006;2(11):619–626. doi: 10.1038/ncprheum0338. [DOI] [PubMed] [Google Scholar]
- 91.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem. 1997;272(31):19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 92.Breen EC, Gage JR, Guo B, et al. Viral interleukin 6 stimulates human peripheral blood B cells that are unresponsive to human interleukin 6. Cell Immunol. 2001;212(2):118–125. doi: 10.1006/cimm.2001.1852. [DOI] [PubMed] [Google Scholar]
- 93.Staskus KA, Sun R, Miller G, et al. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J Virol. 1999;73(5):4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brousset P, Cesarman E, Meggetto F, Lamant L, Delsol G. Colocalization of the viral interleukin-6 with latent nuclear antigen-1 of human herpesvirus-8 in endothelial spindle cells of Kaposi’s sarcoma and lymphoid cells of multicentric Castleman’s disease. Hum Pathol. 2001;32(1):95–100. doi: 10.1053/hupa.2001.21131. [DOI] [PubMed] [Google Scholar]
- 95.Cannon JS, Nicholas J, Orenstein JM, et al. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J Infect Dis. 1999;180(3):824–828. doi: 10.1086/314956. [DOI] [PubMed] [Google Scholar]
- 96.Nicholas J, Ruvolo V, Zong J, et al. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71(3):1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97▪.Chen D, Sandford G, Nicholas J. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J Virol. 2009;83(2):722–733. doi: 10.1128/JVI.01517-08. Investigation of the role of endogenously produced vIL-6 in primary effusion lymphoma cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aoki Y, Jaffe ES, Chang Y, et al. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93(12):4034–4043. [PubMed] [Google Scholar]
- 99.Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science. 2002;298(5597):1432–1435. doi: 10.1126/science.1074883. [DOI] [PubMed] [Google Scholar]
- 100.Rosenkilde MM, Kledal TN, Brauner-Osborne H, Schwartz TW. Agonists and inverse agonists for the herpesvirus 8-encoded constitutively active seven-transmembrane oncogene product, ORF-74. J Biol Chem. 1999;274(2):956–961. doi: 10.1074/jbc.274.2.956. [DOI] [PubMed] [Google Scholar]
- 101▪▪.Bais C, Santomasso B, Coso O, et al. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391(6662):86–89. doi: 10.1038/34193. Describes the biological effects of G-protein-coupled chemokine receptors in KSHV-mediated oncogenesis. [DOI] [PubMed] [Google Scholar]
- 102.Sodhi A, Montaner S, Patel V, et al. The Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 2000;60(17):4873–4880. [PubMed] [Google Scholar]
- 103.Montaner S, Sodhi A, Pece S, Mesri EA, Gutkind JS. The Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor promotes endothelial cell survival through the activation of Akt/protein kinase B. Cancer Res. 2001;61(6):2641–2648. [PubMed] [Google Scholar]
- 104.Bais C, Van Geelen A, Eroles P, et al. Kaposi’s sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/KDR. Cancer Cell. 2003;3(2):131–143. doi: 10.1016/s1535-6108(03)00024-2. [DOI] [PubMed] [Google Scholar]
- 105.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn MC, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385(6614):347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 106.Virgin HW, Latreille P, Wamsley P, et al. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71(8):5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paulsen SJ, Rosenkilde MM, Eugen-Olsen J, Kledal TN. Epstein–Barr virus-encoded BILF1 is a constitutively active G protein-coupled receptor. J Virol. 2005;79(1):536–546. doi: 10.1128/JVI.79.1.536-546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davis-Poynter NJ, Lynch DM, Vally H, et al. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol. 1997;71(2):1521–1529. doi: 10.1128/jvi.71.2.1521-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paulose-Murphy M, Ha NK, Xiang C, et al. Transcription program of human herpesvirus 8 (kaposi’s sarcoma-associated herpesvirus) J Virol. 2001;75(10):4843–4853. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Beisser PS, Grauls G, Bruggeman CA, Vink C. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J Virol. 1999;73(9):7218–7230. doi: 10.1128/jvi.73.9.7218-7230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McFadden G, Lalani A, Everett H, Nash P, Xu X. Virus-encoded receptors for cytokines and chemokines. Semin Cell Dev Biol. 1998;9(3):359–368. doi: 10.1006/scdb.1998.0245. [DOI] [PubMed] [Google Scholar]
- 112.Parry CM, Simas JP, Smith VP, et al. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J Exp Med. 2000;191(3):573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Webb LM, Alcami A. Virally encoded chemokine binding proteins. Mini Rev Med Chem. 2005;5(9):833–848. doi: 10.2174/1389557054867110. [DOI] [PubMed] [Google Scholar]
- 114.Seet BT, McFadden G. Viral chemokine-binding proteins. J Leukoc Biol. 2002;72(1):24–34. [PubMed] [Google Scholar]
- 115.Carfi A, Smith CA, Smolak PJ, McGrew J, Wiley DC. Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus. Proc Natl Acad Sci USA. 1999;96(22):12379–12383. doi: 10.1073/pnas.96.22.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 117.Green DR. Apoptotic pathways: ten minutes to dead. Cell. 2005;121(5):671–674. doi: 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 118▪▪.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741–752. doi: 10.1038/nrm2239. Describes the multiple connections between autophagy and apoptosis. [DOI] [PubMed] [Google Scholar]
- 119.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115(10):2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120▪.Thorburn J, Moore F, Rao A, et al. Selective inactivation of a Fas-associated death domain protein (FADD)-dependent apoptosis and autophagy pathway in immortal epithelial cells. Mol Biol Cell. 2005;16(3):1189–1199. doi: 10.1091/mbc.E04-10-0906. Identified a novel cell death pathway that combines apoptosis and autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mills KR, Reginato M, Debnath J, Queenan B, Brugge JS. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc Natl Acad Sci USA. 2004;101(10):3438–3443. doi: 10.1073/pnas.0400443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 123.Arico S, Petiot A, Bauvy C, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276(38):35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 124.Jia L, Dourmashkin RR, Allen PD, Gray AB, Newland AC, Kelsey SM. Inhibition of autophagy abrogates tumour necrosis factor α induced apoptosis in human T-lymphoblastic leukaemic cells. Br J Haematol. 1997;98(3):673–685. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 125.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 126.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102(23):8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 128.Fakhari FD, Dittmer DP. Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol. 2002;76(12):6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72(10):8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2001;75(4):1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bieleski L, Talbot SJ. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol. 2001;75(4):1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Low W, Harries M, Ye H, Du MQ, Boshoff C, Collins M. Internal ribosome entry site regulates translation of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol. 2001;75(6):2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Belanger C, Gravel A, Tomoiu A, et al. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J Hum Virol. 2001;4(2):62–73. [PubMed] [Google Scholar]
- 134.Chaudhary PM, Jasmin A, Eby MT, Hood L. Modulation of the NF-κ B pathway by virally encoded death effector domains-containing proteins. Oncogene. 1999;18(42):5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 135.Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the IκB kinase complex. J Biol Chem. 2002;277(16):13745–13751. doi: 10.1074/jbc.M110480200. [DOI] [PubMed] [Google Scholar]
- 136.Field N, Low W, Daniels M, et al. KSHV vFLIP binds to IKK-γ to activate IKK. J Cell Sci. 2003;116(Pt 18):3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 137▪.Chugh P, Matta H, Schamus S, et al. Constitutive NF-κB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc Natl Acad Sci USA. 2005;102(36):12885–12890. doi: 10.1073/pnas.0408577102. Provides in vivo evidence of K13’s (vFLIP) abilities in K13 transgenic mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Grossmann C, Podgrabinska S, Skobe M, Ganem D. Activation of NF-κB by the latent vFLIP gene of Kaposi’s sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J Virol. 2006;80(14):7179–7185. doi: 10.1128/JVI.01603-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sun Q, Matta H, Lu G, Chaudhary PM. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene. 2006;25(19):2717–2726. doi: 10.1038/sj.onc.1209298. [DOI] [PubMed] [Google Scholar]
- 140.Ye FC, Zhou FC, Xie JP, et al. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-κB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J Virol. 2008;82(9):4235–4249. doi: 10.1128/JVI.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao J, Punj V, Matta H, et al. K13 blocks KSHV lytic replication and deregulates vIL6 and hIL6 expression: a model of lytic replication induced clonal selection in viral oncogenesis. PLoS One. 2007;2(10):E1067. doi: 10.1371/journal.pone.0001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sun Q, Zachariah S, Chaudhary PM. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-κB activation. J Biol Chem. 2003;278(52):52437–52445. doi: 10.1074/jbc.M304199200. [DOI] [PubMed] [Google Scholar]
- 143▪.Lee JS, Li Q, Lee JY, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11(11):1355–1362. doi: 10.1038/ncb1980. Describes how the antiapoptotic protein vFLIP act as a negative regulator of autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Korsmeyer SJ. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 1999;59(7 Suppl):S1693–S1700. [PubMed] [Google Scholar]
- 145.Cheng EH, Nicholas J, Bellows DS, et al. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94(2):690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3(3):293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 147.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc Natl Acad Sci USA. 2002;99(6):3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Polster BM, Pevsner J, Hardwick JM. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim Biophys Acta. 2004;1644(2–3):211–227. doi: 10.1016/j.bbamcr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 149.Cuconati A, White E. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 2002;16(19):2465–2478. doi: 10.1101/gad.1012702. [DOI] [PubMed] [Google Scholar]
- 150.Bellows DS, Chau BN, Lee P, Lazebnik Y, Burns WH, Hardwick JM. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J Virol. 2000;74(11):5024–5031. doi: 10.1128/jvi.74.11.5024-5031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Loh J, Huang Q, Petros AM, et al. A surface groove essential for viral Bcl-2 function during chronic infection in vivo. PLoS Pathog. 2005;1(1):E10. doi: 10.1371/journal.ppat.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang GH, Garvey TL, Cohen JI. The murine γherpesvirus-68 M11 protein inhibits Fas- and TNF-induced apoptosis. J Gen Virol. 1999;80 (Pt 10):2737–2740. doi: 10.1099/0022-1317-80-10-2737. [DOI] [PubMed] [Google Scholar]
- 153.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 154.Pattingre S, Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res. 2006;66(6):2885–2888. doi: 10.1158/0008-5472.CAN-05-4412. [DOI] [PubMed] [Google Scholar]
- 155.Ku B, Woo JS, Liang C, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine γ-herpesvirus 68. PLoS Pathog. 2008;4(2):E25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xiaofei E, Hwang S, Oh S, et al. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009;5(10):E1000609. doi: 10.1371/journal.ppat.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 158.Beutler BA. TLRs and innate immunity. Blood. 2009;113(7):1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 160.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19(1):24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 161.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388(4):621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 162.West J, Damania B. Upregulation of the TLR3 pathway by Kaposi’s sarcoma-associated herpesvirus during primary infection. J Virol. 2008;82(11):5440–5449. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lagos D, Vart RJ, Gratrix F, et al. Toll-like receptor 4 mediates innate immunity to Kaposi sarcoma herpesvirus. Cell Host Microbe. 2008;4(5):470–483. doi: 10.1016/j.chom.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc Natl Acad Sci USA. 2009;106(28):11725–11730. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ambroziak JA, Blackbourn DJ, Herndier BG, et al. Herpes-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268(5210):582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 166.Wilson SJ, Tsao EH, Webb BL, et al. X box binding protein XBP-1s transactivates the Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF50 promoter, linking plasma cell differentiation to KSHV reactivation from latency. J Virol. 2007;81(24):13578–13586. doi: 10.1128/JVI.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu F, Feng J, Harada JN, Chanda SK, Kenney SC, Sun R. B cell terminal differentiation factor XBP-1 induces reactivation of Kaposi’s sarcoma-associated herpesvirus. FEBS Lett. 2007;581(18):3485–3488. doi: 10.1016/j.febslet.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 168.Dalton-Griffin L, Wilson SJ, Kellam P. X-box binding protein 1 contributes to induction of the Kaposi’s sarcoma-associated herpesvirus lytic cycle under hypoxic conditions. J Virol. 2009;83(14):7202–7209. doi: 10.1128/JVI.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 170.Reimold AM, Ponath PD, Li YS, et al. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183(2):393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jenner RG, Maillard K, Cattini N, et al. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc Natl Acad Sci USA. 2003;100(18):10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci USA. 2009;106(8):2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Van Damme N, Goff D, Katsura C, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174▪.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. Inhibition of HIV-1 release by tetherin is antagonized by Vpu. [DOI] [PubMed] [Google Scholar]
- 175.Mansouri M, Viswanathan K, Douglas JL, et al. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2009;83(19):9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jia B, Serra-Moreno R, Neidermyer W, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5(5):E1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang F, Wilson SJ, Landford WC, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6(1):54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Le Tortorec A, Neil SJ. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol. 2009;83(22):11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Micheletti F, Monini P, Fortini C, et al. Identification of cytotoxic T lymphocyte epitopes of human herpesvirus 8. Immunology. 2002;106(3):395–403. doi: 10.1046/j.1365-2567.2002.01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Alcami A, Koszinowski UH. Viral mechanisms of immune evasion. Trends Microbiol. 2000;8(9):410–418. doi: 10.1016/S0966-842X(00)01830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ambagala AP, Solheim JC, Srikumaran S. Viral interference with MHC class I antigen presentation pathway: the battle continues. Vet Immunol Immunopathol. 2005;107(1–2):1–15. doi: 10.1016/j.vetimm.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 182▪.Coscoy L, Ganem D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA. 2000;97(14):8051–8056. doi: 10.1073/pnas.140129797. Along with [183] describe the identification of K3 and K5 as proteins that downregulate MHC class I molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183▪.Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74(11):5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. Along with [182] describe the identification of K3 and K5 as proteins that downregulate MHC class I molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev. 2005;207:112–125. doi: 10.1111/j.0105-2896.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 185.Coscoy L. Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Nat Rev Immunol. 2007;7(5):391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 186.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155(7):1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sanchez DJ, Coscoy L, Ganem D. Functional organization of MIR2, a novel viral regulator of selective endocytosis. J Biol Chem. 2002;277(8):6124–6130. doi: 10.1074/jbc.M110265200. [DOI] [PubMed] [Google Scholar]
- 188.Wang X, Lybarger L, Connors R, Harris MR, Hansen TH. Model for the interaction of γherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J Virol. 2004;78(16):8673–8686. doi: 10.1128/JVI.78.16.8673-8686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lagos D, Trotter MW, Vart RJ, et al. Kaposi sarcoma herpesvirus-encoded vFLIP and vIRF1 regulate antigen presentation in lymphatic endothelial cells. Blood. 2007;109(4):1550–1558. doi: 10.1182/blood-2006-05-024034. [DOI] [PubMed] [Google Scholar]
- 190.Goodbourn S, Didcock L, Randall RE. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol. 2000;81(Pt 10):2341–2364. doi: 10.1099/0022-1317-81-10-2341. [DOI] [PubMed] [Google Scholar]
- 191▪▪.Li Q, Means R, Lang S, Jung JU. Downregulation of γ interferon receptor 1 by Kaposi’s sarcoma-associated herpesvirus K3 and K5. J Virol. 2007;81(5):2117–2127. doi: 10.1128/JVI.01961-06. Demonstrates that KSHV K3 and K5 proteins shut down type II IFN signaling as well as MHC class I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chambers CA, Allison JP. Costimulatory regulation of T cell function. Curr Opin Cell Biol. 1999;11(2):203–210. doi: 10.1016/s0955-0674(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 193.Sperling AI, Bluestone JA. The complexities of T-cell co-stimulation: CD28 and beyond. Immunol Rev. 1996;153:155–182. doi: 10.1111/j.1600-065x.1996.tb00924.x. [DOI] [PubMed] [Google Scholar]
- 194.Dustin ML, Springer TA. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 1989;341(6243):619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 195.Coscoy L, Ganem D. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T cell costimulation. J Clin Invest. 2001;107(12):1599–1606. doi: 10.1172/JCI12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Renne R, Dittmer D, Kedes D, et al. Experimental transmission of Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) to SIV-positive and SIV-negative rhesus macaques. J Med Primatol. 2004;33(1):1–9. doi: 10.1046/j.1600-0684.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 197▪.Parsons CH, Adang LA, Overdevest J, et al. KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice. J Clin Invest. 2006;116(7):1963–1973. doi: 10.1172/JCI27249. First useful animal model for KSHV infection using NOD/SCID mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dittmer D, Stoddart C, Renne R, et al. Experimental transmission of Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J Exp Med. 1999;190(12):1857–1868. doi: 10.1084/jem.190.12.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Foreman KE, Friborg J, Chandran B, et al. Injection of human herpesvirus-8 in human skin engrafted on SCID mice induces Kaposi’s sarcoma-like lesions. J Dermatol Sci. 2001;26(3):182–193. doi: 10.1016/s0923-1811(01)00087-1. [DOI] [PubMed] [Google Scholar]
- 200.Mutlu AD, Cavallin LE, Vincent L, et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV a cell and animal model of virally induced Kaposi’s sarcoma. Cancer Cell. 2007;11(3):245–258. doi: 10.1016/j.ccr.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201▪▪.Chang H, Wachtman LM, Pearson CB, et al. Non-human primate model of Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2009;5(10):E1000606. doi: 10.1371/journal.ppat.1000606. Provides an appropriate animal model for KSHV infection, which demonstrates seroconversion and persistent viral infection with the hallmark latency. [DOI] [PMC free article] [PubMed] [Google Scholar]