Abstract
Light-dependent regulation of a growing number of chloroplast enzymatic activities has been found to occur through the reversible reduction of intra- or intermolecular disulphides by thioredoxins. In cyanobacteria, despite their similarity to chloroplasts, no proteins have hitherto been shown to interact with thioredoxins, and the role of the cyanobacterial ferredoxin/thioredoxin system has remained obscure. By using an immobilized cysteine 35-to-serine site-directed mutant of the Synechocystis sp. PCC 6803 thioredoxin TrxA as bait, we screened the Synechocystis cytosolic and peripheral membrane protein complements for proteins interacting with TrxA. The covalent bond between the isolated target proteins and mutated TrxA was confirmed by nonreducing/reducing two-dimensional SDS/PAGE. Thus, we have identified 18 cytosolic proteins and 8 membrane-associated proteins as candidate thioredoxin substrates. Twenty of these proteins have not previously been associated with thioredoxin-mediated regulation. Phosphoglucomutase, one of the previously uncharacterized thioredoxin-linked enzymes, has not earlier been considered a target for metabolic control through disulphide reduction. In this article, we show that phosphoglucomutase is inhibited under oxidizing conditions and activated by DTT and reduced wild-type TrxA in vitro. The results imply that thioredoxin-mediated redox regulation is as extensive in cyanobacteria as in chloroplasts but that the subjects of regulation are largely different.
Dithiol/disulphide exchange catalyzed by thioredoxin (Trx) forms the molecular basis for light-dependent regulation of many enzymes in the chloroplasts of higher plants and algae (1-3). Ferredoxin receives reducing equivalents from the photosynthetic electron transport in the light and reduces Trx m and f by means of ferredoxin-Trx reductase. The Trxs, in turn, convert disulphides to dithiols in their respective target enzymes, thereby modulating their activities. The earliest discovered targets for Trx-mediated regulation belong to the Calvin cycle of CO2 assimilation (1). Since the initial discoveries, several more chloroplast enzymes have been recognized as substrates for Trx (3). A breakthrough in the investigation of chloroplast redox regulation came with the introduction of a new method to isolate Trx target proteins (4-6). The method involves mutation of the buried redox-active cysteine of Trx, which favors the formation of stable mixed disulphides with its target proteins. This experimental approach confirmed the interaction between Trx and its known targets and more than doubled the number of potential Trx-regulated proteins (6).
Cyanobacteria are oxygenic photosynthetic prokaryotes that probably share a common ancestor with the chloroplast. The complete sequence of the cyanobacterium Synechocystis sp. PCC 6803 genome (Cyanobase, www.kazusa.or.jp/cyano/cyano.html) reveals that this organism, hereafter referred to as Synechocystis, contains ferredoxin-Trx reductase and at least four different Trxs. Nevertheless, attempts to demonstrate light-dependent redox regulation of three cyanobacterial enzymes of the Calvin cycle (phosphoribulokinase, fructose-1,6-bisphosphatase, and glyceraldehyde-3-phosphate dehydrogenase) were unsuccessful (7), although purified phosphoribulokinase from Synechococcus sp. PCC 7942 could be activated by DTT in vitro (8). The stark contrast between the growing number of chloroplast Trx targets and the lack of data indicating Trx function in cyanobacteria might lead to the conclusion that light-induced Trx-mediated regulation evolved with the need to coordinate chloroplastic and extrachloroplastic metabolism in photosynthetic eukaryotes. Surprisingly, Synechocystis TrxA was found essential for survival under photoautotrophic as well as heterotrophic growth conditions (9). Among the diverse Synechocystis Trxs, TrxA is the one that most resembles the chloroplast m-type Trx.
Aiming at clarifying the roles of TrxA in Synechocystis, we have screened the Synechocystis cytosolic and peripheral membrane protein complements for proteins interacting with TrxA. To this end, we used a strategy similar to that described in refs. 5 and 6. Thus, we have identified 18 cytosolic proteins and 8 membrane-associated proteins as candidate substrates for TrxA. One of these targets, phosphoglucomutase (PGM), which represents a metabolic branch point between storage and utilization of carbohydrates, was shown to be activated by TrxA in vitro.
Materials and Methods
Materials. DEAE Sephacel matrix, His-Bind resin, and Superdex 75 were purchased from Sigma, Novagen, and Amersham Pharmacia Biosciences, respectively. DTT, Triton X-100, and NAD+ were from Sigma. Other chemicals were of the highest grade commercially available.
Mutagenesis, Expression, and Purification of TrxA. The trxA gene (ORF slr0623, Cyanobase) was mutated, exchanging cysteine 35 for a serine. The mutated TrxA (TrxA35) was expressed with a C-terminal His-tag by using the pET22b vector in Escherichia coli BL21 DE3 and purified to homogeneity by anion-exchange chromatography on DEAE Sephacel matrix, Ni-affinity chromatography with the His-Bind resin, and gel filtration on Superdex 75. The gel-filtration step was preceded by a 1-h incubation with 20 mM DTT on ice. Pure TrxA35 was eluted in 25 mM Tris·HCl (pH 7.9) and 150 mM KCl.
Preparation of Cytosolic and Peripheral Membrane Protein Fractions. Synechocystis cultures were grown photoautotrophically at 30°C in BG11 medium (10) including 1 g/l NaHCO3 and bubbled with 1% (vol/vol) CO2 in air under continuous illumination with white light at an intensity of 50 μmol photons·m-2·s-1. Cells were harvested and broken with glass beads as described in ref. 11 with buffer A (25 mM Hepes·NaOH, pH 7.0/15 mM CaCl2/5 mM MgCl2/15% (vol/vol) glycerol/1 mM PMSF). After removal of unbroken cells by centrifugation at 2,300 × g for 5 min, total membranes were pelleted by centrifugation at 16,000 × g for 20 min, yielding a supernatant containing cytosolic proteins. Membranes were washed three times by resuspension in buffer A and centrifugation. Peripheral membrane proteins were extracted by 15-min incubation on ice with buffer A supplemented with 1 M NaCl and 0.02% (vol/vol) Triton X-100 followed by centrifugation at 16,000 × g for 30 min. The supernatant was concentrated, and NaCl and Triton X-100 were eliminated by centrifugation filtration in Ultrafree-MC (Millipore), 3-kDa cut-off, and successive replacement with buffer A. Protein concentrations were measured as described (12).
Isolation of TrxA Target Proteins. We added 1.2 mg of pure TrxA35 to 150 μl of His-Bind resin in a 1.5-ml microcentrifuge tube and incubated it at 4°C under gentle agitation for 1 h. The resin was washed twice with 1 ml of buffer A after removal of the supernatant. Then 2.8 mg of cytosolic protein or 1.8 mg of peripheral membrane protein was added followed by a 16-h incubation at 4°C under gentle agitation. Unbound proteins were removed, and the resin was washed four times in 1 ml of buffer B (20 mM Tris·HCl, pH 7.9/0.5 M NaCl) with the addition of 60 mM imidazole. TrxA35-target protein complexes were released by addition of 120 μl of buffer B including 1 M imidazole, and the eluates were recovered by centrifugation for 10 sec at 2,000 × g.
Nonreducing/Reducing Gel Electrophoresis. Eluates were electrophoresed under nonreducing conditions in the first dimension, maintaining the TrxA35-target protein complexes intact, and reducing conditions in the second dimension, releasing TrxA35 from its targets. Seventy microliters of eluate was mixed with 15 μl of See Blue prestained protein standard (Invitrogen) and 30 μl of solubilizing buffer (0.25 M Tris·HCl, pH 6.8/12.5% (vol/vol) glycerol/10% (wt/vol) SDS). After separation by SDS/PAGE on a 16 × 18 cm, 1.5-mm thick gel, the lane was excised and incubated for 1 h at 25°C in 2 ml of solubilizing buffer diluted 2.5 times with the addition of DTT to 100 mM final concentration. The lane was fixed on top of an acrylamide gel of identical size and, after separation at 40 mA, the gel was stained with Coomassie brilliant blue R-250. Acrylamide gels of 12% and 10% (wt/vol) were used for cytosolic and peripheral membrane proteins, respectively. Apparent molecular masses were determined by using one-dimensional gels, reducing conditions, and nonstained protein markers (Bio-Rad) followed by Coomassie staining for maximal accuracy.
Protein Identification. Coomassie-stained proteins were excised and digested with trypsin. Peptides were subjected to matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) analysis performed on an Ultraflex apparatus (Bruker, Billerica, MA). Proteins were identified by using the mascot peptide mass fingerprint search engine (Matrix Science, London).
Western Blot. For glutamine synthetase (GS), the antibody (13) was used at 1:5,000 dilution. For NADH-glutamate synthase (GOGAT), an antibody raised against the large subunit, GltB (F. Navarro and F.J.F., unpublished data), was used at 1:2,000 dilution. Signals were visualized with enhanced chemiluminescence Western blotting detection reagents from Amersham Pharmacia Biosciences.
Measurement of PGM Activity. The activity was measured by coupling to reduction of NAD+ catalyzed by glucose-6-phosphate dehydrogenase (G6PD) from Leuconostoc mesenteroides (Sigma). G6PD from L. mesenteroides is devoid of cysteines, and thiol reagents do not affect its activity. Each assay of 1 ml final volume contained 50 mM Tris·HCl (pH 7.6), 4 mM MgCl2, 2 mM NAD+, 40 μM α-d-glucose-1,6-diphosphate, 1.4 units G6PD, and 23 μg of Synechocystis cytosolic proteins. The reaction was started by addition of 4 mM α-d-glucose-1-phosphate, and the absorbance at 340 nm was monitored. Incubations of cytosolic extract before measurement were performed in 25 mM Tris·HCl (pH 7.6) and 2 mM MgCl2 at 25°C and at a protein concentration of 4 mg/ml with additions as specified. Wild-type TrxA was prepared as in ref. 14.
Results and Discussion
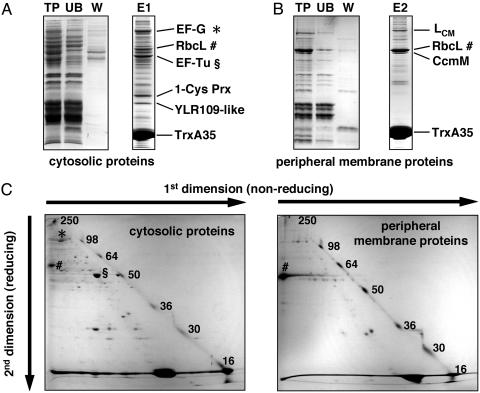
Isolation of Proteins Bound to TrxA35. Screening of the Synechocystis extracts for proteins interacting with the TrxA Cys-35-Ser mutant, TrxA35, yielded cytosolic target proteins (Fig. 1A) as well as peripheral membrane target proteins (Fig. 1B). The covalent bond between these proteins and TrxA35 was confirmed by nonreducing/reducing two-dimensional SDS/PAGE (Fig. 1C). The majority of the captured Synechocystis proteins migrate under nonreducing conditions with an apparent molecular mass that is ≈15 kDa larger than the apparent molecular mass under reducing conditions (Fig. 1C). This difference in mass corresponds to one covalently bound TrxA35 molecule, which is released after reduction. In some cases, the difference in migration between the first and second dimension is so great that it implies more than one TrxA35 molecule joined to a target and/or an oligomeric structure of the target protein mediated by intermolecular disulphides. TrxA35 contains only one cysteine, thus excluding the possibility of Trx trimers. A large amount of TrxA35 forms homodimers during the binding assay and migrates with coordinates 33 kDa/15 kDa, and some TrxA35 remains a monomer and migrates at the diagonal (Fig. 1C). The diagonal is practically devoid of Synechocystis proteins.
Fig. 1.
(A and B) Isolation of Synechocystis Trx target proteins with the His-tagged site-directed Trx mutant TrxA35 immobilized on a Ni-affinity matrix. Total protein before binding (TP), unbound proteins (UB), and proteins washed off with 60 mM imidazole (W) were separated on 12% acrylamide gels and stained with Coomassie brilliant blue. Six microliters from each fraction of cytosolic proteins and 10 μl from fractions of peripheral membrane proteins were applied on the gels, corresponding to 45 and 30 μg of initial total protein (TP), respectively. Seventy microliters each of cytosolic proteins (E1) and peripheral membrane proteins (E2) bound to TrxA35 and eluted with 1 M imidazole were separated on 12% acrylamide gels under reducing conditions. The identities of the most abundant targets are indicated. (C) Separation of Trx target proteins by nonreducing/reducing two-dimensional SDS/PAGE. Eluted proteins E1 and E2 were separated on 12% and 10% acrylamide gels, respectively. Before the second dimension, proteins were reduced with 100 mM DTT. The molecular masses of the prestained standard proteins are indicated by numbers.
Identification of Synechocystis TrxA-Linked Proteins. Twenty-six of the resolved proteins were positively identified with their corresponding ORFs in the Synechocystis genome. The 18 cytosolic proteins are listed in Table 1, and the 8 peripheral membrane proteins are listed in Table 2. One of the targets, the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) (RbcL), was found both in the cytosolic and the peripheral membrane protein fractions. The vast majority of the target proteins, 20 of 25, have not previously been associated with Trx-mediated regulation. Only two of the Synechocystis targets, translation elongation factors EF-G and EF-Tu (Table 1), have been reported earlier to interact with chloroplast Trxs (6). It is striking that there are no other common proteins between the 35 chloroplast Trx targets identified by Balmer et al. (6) and the 25 Synechocystis Trx targets reported in this study. The implications of our findings in relation to the current knowledge about the target enzymes and the metabolic processes in which they participate are discussed below.
Table 1. Synechocystis cytosolic TrxA-linked proteins.
| ORF | Gene product | Apparent molecular mass, kDa | Theoretical molecular mass, kDa | Number of cysteines conserved* |
|---|---|---|---|---|
| sll1499 | Ferredoxin-GOGAT (GlsF) | 200 | 169.4 | 16 (15) |
| sll1502 | NADH-GOGAT (GltB) | 186 | 169.0 | 10 (10) |
| slr0557 | Valyl-tRNA synthetase (ValS) | 105 | 102.7 | 5 (3) |
| slr1463 | Elongation factor EF-G | 90 | 76.7 | 2 (2) |
| sll0158 | Glucan branching enzyme (GlgB) | 84 | 89.5 | 6 (0) |
| slr2076 | 60-kDa chaperonin GroEL | 63 | 57.5 | 1 (1) |
| sll0726 | Phosphoglucomutase | 58 | 61.1 | 1 (0) |
| slr0009 | Rubisco large subunit (RbcL) | 57 | 52.5 | 9 (8) |
| sll1393 | Glycogen synthase (Glg2) | 55 | 56.2 | 10 (4) |
| slr1176 | ADPglucose pyrophosphorylase | 54 | 49.3 | 4 (3) |
| slr0585 | Argininosuccinate synthetase | 53 | 44.5 | 2 (1) |
| sll1099 | Elongation factor EF-Tu | 51 | 43.7 | 1 (1) |
| sll1212 | GDP-mannose dehydratase | 45 | 41.3 | 2 |
| sll1994 | Porphobilinogen synthase | 42 | 36.1 | 4 (0) |
| sll0576 | Sugar-nucleotide epimerase | 35 | 34.9 | 1 (1) |
| slr1198 | 1-Cys peroxiredoxin | 29 | 23.5 | 1 |
| sll1621 | YLR109-homologue | 25 | 21.2 | 2 (2) |
| ssr3383 | Phycobilisome linker (Lc) | 13 | 7.8 | 1 |
Total number of conserved cysteines in homologues from any organism and then, in parentheses, the number of cysteines conserved in plant chloroplast homologues
Table 2. Synechocystis peripheral membrane TrxA-linked proteins.
| ORF | Gene product | Apparent molecular mass, kDa | Theoretical molecular mass, kDa | Number of cysteines conserved* |
|---|---|---|---|---|
| sll1789 | RNA polymerase β′ subunit | 191 | 144.7 | 6 (5) |
| sll1787 | RNA polymerase β subunit | 152 | 123.3 | 3 (3) |
| slr0335 | Phycobilisome linker (LCM) | 92 | 100.2 | 3 |
| slr0963 | Ferredoxin-sulfite reductase | 67 | 71.4 | 6 (5) |
| slr0009 | Rubisco large subunit (RbcL) | 56 | 52.5 | 9 (8) |
| sll1031 | Carboxysomal protein (CcmM) | 54 | 73.1 (56.9)† | 2 |
| slr1165 | Sulfate adenylyltransferase | 43 | 43.7 | 4 (0) |
| sll1804 | 30S ribosomal protein S3 | 32 | 27.1 | 1 (0) |
Carbon dioxide fixation. Cytosolic (Table 1) and membrane-associated (Table 2) forms of RbcL were found to be targets for Trx. In both cases, RbcL migrated with much too high apparent molecular mass under nonreducing conditions to be compatible with an RbcL-TrxA35 heterodimer (Fig. 1C). Cysteine 247 of spinach RbcL, which corresponds to Synechocystis Cys-242, is known to mediate dimer formation between neighboring RbcL subunits (15). Next to the active site of RbcL are two other conserved cysteines whose redox state affects enzyme activity in Synechocystis (16). A bond between TrxA35 and one of these cysteines in addition to a disulphide between two Cys-242 residues would yield an RbcL2-TrxA352 heterotetramer. Such a polypeptide with a complex branched structure could be more retarded on SDS/PAGE than would be expected from the sum of the molecular masses of the individual subunits. There is as yet no evidence for Trx-mediated regulation of RbcL in any organism, but the small subunit of Rubisco in chloroplasts has been identified as a Trx-target (5, 6). CcmM (Table 2) is a carbon dioxide-concentrating mechanism protein, which is required for growth at air levels of CO2 and is probably a constituent of the carboxysome (17). We identified CcmM as a protein migrating on SDS/PAGE with an apparent molecular mass of 54 kDa (Fig. 1B), which is in agreement with Ogawa et al. (17) but at variance with Cyanobase, where it is reported as a 73-kDa protein. No structural carboxysomal proteins have so far been reported to interact with Trxs, but a chloroplast carbonic anhydrase was recently discovered as a target for Trx (6).
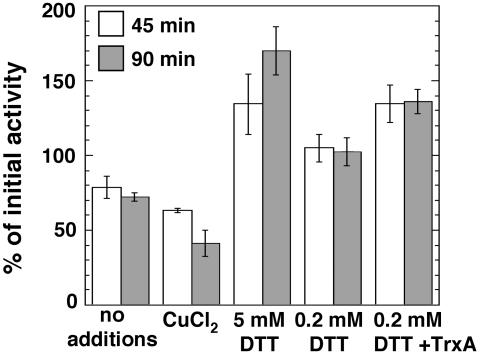
PGM. The interconversion of glucose-1-phosphate and glucose-6-phosphate catalyzed by PGM (Table 1) bridges glycolysis and glycogen metabolism. Although PGM is recognized to play a key role in sugar metabolism, its activity has not been considered a target for metabolic control. PGM activity in intact chloroplasts was interestingly found to be up-regulated in the light, but preincubation with 10 mM DTT in the dark had no effect on this activity (18). To test the possibility of redox regulation of PGM in Synechocystis, we examined the effects of DTT and TrxA on PGM activity. Incubation of cytosol with 5 mM DTT for 45 or 90 min before measurement led to a substantial increase in PGM activity (Fig. 2). In contrast, control incubation without additions resulted in loss of activity. Further loss was observed when preincubation was performed in the presence of a low concentration of CuCl2, which is known to induce the formation of disulphide bonds (5). Addition of DTT at low (0.2 mM) concentration maintained the activity constant throughout the incubation and, finally, addition of 2.5 μM purified Synechocystis wild-type TrxA in the presence of 0.2 mM DTT caused a significant increase in PGM activity (Fig. 2). The fact that 0.2 mM DTT alone prevented loss of activity may be attributable to the presence of endogenous Trxs in the cytosolic extract. Synechocystis PGM contains a single cysteine residue, which implies that the regulatory disulphide is intermolecular.
Fig. 2.
Activity of Synechocystis PGM. Conversion of glucose-1-phosphate to glucose-6-phosphate in cytosolic extracts was measured before and after incubation at 25°C for 45 and 90 min with the additions specified. Concentrations of CuCl2 and TrxA were 25 and 2.5 μM, respectively. The values are the means of three different experiments, and SDs are presented as error bars. The initial activity before incubation was 0.38 μmol·mg of protein-1·min-1.
Glycogen synthesis. ADPglucose pyrophosphorylase (Table 1) catalyzes the synthesis of ADPglucose from glucose-1-phosphate and ATP. This reversible reaction constitutes the first step of glycogen synthesis in bacteria and starch synthesis in the plastids of plants and algae (19). The higher plant enzyme is a heterotetramer consisting of two large subunits and two small subunits. Experiments in vitro (20, 21) and in vivo (22) have shown that the N-terminal cysteine (Cys-12) of the small subunits may form an intermolecular disulphide bond between them, thereby inactivating the enzyme. The oxidized, inactive enzyme is slowly reduced and, thus, reactivated by DTT (20) or Trx (21) in vitro. This mechanism of reductive activation has been proposed to operate in all dicot plants, because Cys-12 is conserved in these plants (20). In cyanobacteria, ADPglucose pyrophosphorylase is encoded by a single gene whose product forms homotetramers (23). The cyanobacterial enzyme closely resembles the small subunit of higher plants.
Glycogen synthases in bacteria and starch synthases in plant plastids catalyze the transfer of ADPglucose to the nonreducing end of a growing α-1,4-linked glucan. The Synechocystis glycogen synthase (Glg2) (Table 1) is similar to the soluble starch synthases from plants and algae (24). Our finding that Glg2 is a potential target for Trx is an indication of a posttranslational regulation of this enzyme in photosynthetic organisms. The glycogen-branching enzyme (Table 1) catalyzes the final step in glycogen synthesis, transferring a segment of a preexisting α-1,4-linked glucan in α-1,6 position (19). Thus, the complete pathway for glycogen synthesis seems to be linked to Trx in Synechocystis.
Sugar-nucleotide metabolism. Neither the putative sugar-nucleotide epimerase/dehydratase nor GDP-mannose dehydratase (Table 1) previously has been linked to Trx. The latter catalyzes the first step of the pathway that leads to synthesis of GDP-l-fucose, a component of, e.g., bacterial capsular polysaccharides.
Sulfur metabolism. The first step of reductive sulfate assimilation is the activation of sulfate catalyzed by sulfate adenylyltransferase. We found this enzyme among the peripheral membrane proteins bound to TrxA35 (Table 2). 3′-Phosphoadenylylsulfate (PAPS) reductase, which was not found among the Trx targets in this study, reduces activated sulfate to sulfite and is one of the best known acceptors of reducing equivalents from Trx in E. coli (25). Instead, we found the next enzyme of the pathway, ferredoxin-dependent sulfite reductase (Table 2), which is homologous to the higher plant plastid ferredoxin-sulfite reductase (26). Cysteine synthase, the last enzyme of the pathway, recently has been identified as a target for chloroplast Trxs (6).
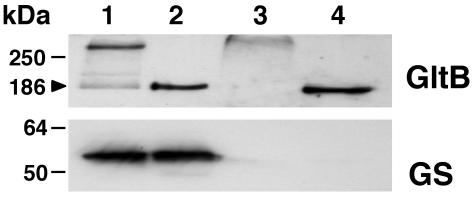
Nitrogen metabolism. Ammonium assimilation occurs through a cyclic pathway involving two enzymes, GS and GOGAT. In this pathway, GS has been considered the exclusive target for regulation at both transcriptional and posttranslational levels in cyanobacteria (27). Surprisingly, both ferredoxin-dependent GOGAT and the large subunit of NADH-dependent GOGAT (GltB) were captured by TrxA35 (Table 1), suggesting that glutamate synthesis may be redox-regulated. Western blot using antibodies against GltB confirms that this protein is present in eluate E1, which contains cytosolic proteins bound to TrxA35 (Fig. 3). In addition, we found that in vivo most of GltB participates in high-molecular weight complexes crosslinked by disulphides (Fig. 3). The apparent molecular mass of these complexes cannot be determined, because it exceeds that of the largest marker protein of 250 kDa, but it is possible that it corresponds to a dimer of the 186-kDa GltB subunits. However, it can be concluded that TrxA35 does not interact with the particular cysteine involved in complex formation, because that would produce a GltB-TrxA35 heterodimer of ≈200 kDa molecular mass (Fig. 3, lane 3). Chlamydomonas reinhardtii GS (28) and plant GS (29) are activated on reduction by Trx or DTT. In both refs. 5 and 6, GS is reported as a target for chloroplast Trxs. Synechocystis GS (type I) has never been found to respond to thiol reagents but instead is known to be regulated through the reversible binding of two small proteins (27). In this article, we show that Synechocystis GS is absent from eluate E1 despite its abundance in the cytosolic extract (Fig. 3).
Fig. 3.
Examination of NADH-GOGAT and GS by Western blot analysis. Lanes 1 and 2 contain 15 μg of cytosolic Synechocystis proteins; lanes 3 and 4 contain 5 μl of eluate E1 (cytosolic proteins bound to TrxA35). Lanes 1 and 3 do not include reducing agents; lanes 2 and 4 include 10 mM DTT. Proteins separated by SDS/PAGE were analyzed by using antibodies against GS and the large subunit of NADH-GOGAT (GltB). Molecular mass markers of 250, 64, and 50 kDa are indicated, as is the 186-kDa apparent molecular mass of the monomeric reduced GltB.
Argininosuccinate synthetase (Table 1) is the rate-limiting enzyme of both the urea- and arginine-citrulline cycles. Like most cyanobacteria, Synechocystis stores excess reduced nitrogen in the form of cyanophycin, a nonribosomally synthesized polymer of aspartate and arginine (30). Thus, the synthesis of arginine is analogous to the synthesis of activated glucose for storage of reduced carbon as glycogen.
Transcription. RNA polymerase subunits β and β′ were found to react with TrxA35 (Table 2). Interestingly, in vitro transcription with purified RNA polymerase from the cyanobacterium Anabaena sp. PCC 7120 requires the presence of thiol-reducing agents (31). Thus, it is possible that polymerase activity in the dark is low because of oxidation of regulatory thiols and that activity is recovered in the light after reduction by Trx. Indeed, cyanobacterial overall gene expression decreases drastically after a shift from light to darkness and returns to initial levels after the onset of light (32).
Protein synthesis and folding. A yeast homologue of valyl-tRNA synthetase (Table 1) already is known to undergo reductive activation in a reaction that involves thiols (33). The translation elongation factors EF-G and EF-Tu (Table 1) were both isolated as targets of chloroplast Trxs (6), and EF-Tu possesses protein disulphide isomerase activity in E. coli (34). However, it can be questioned whether the 30S ribosomal protein S3 (Table 2) is a true target, because its single cysteine is conserved in only Cyanophora paradoxa but no other species. The 60-kDa chaperonin GroEL (Table 1) closely resembles the chloroplast Rubisco-binding protein subunits α and β, which were reported earlier as chloroplast Trx targets (6). Furthermore, the Hsp70 chaperone is regulated by a Trx-like protein in plants (35).
Tetrapyrrole biosynthesis. It has been demonstrated that synthesis of protoporphyrin IX from 5-aminolevulinic acid is stimulated by DTT in chloroplast extracts (36). Three enzymes from the chloroplast heme and chlorophyll synthesis pathway recently were found to interact with Trxs (6). In this article, we report another enzyme from the same pathway, porphobilinogen synthase (Table 1).
Oxidative stress response. Peroxiredoxins (Prxs) constitute an ubiquitous family of thiol-specific peroxidases (37). A light-dependent, thiol-specific peroxidase activity was discovered in a catalase deletion mutant of Synechocystis (38), suggesting the presence of Prx. Among our Trx targets we found a 1-Cys Prx and a homologue of the yeast peroxidase YLR109 (Table 1). Yeast YLR109 has been shown to interact with an Arabidopsis thaliana Trx h (4). Trxs, or Trx-like proteins, are known to reduce chloroplast 2-Cys Prx (6, 39) and Prx-Q (5).
Light harvesting. The phycobilisome core linker polypeptide (LC) (Table 1) links three allophycocyanin heterodimers, thereby forming the core of the phycobilisome (40). The Synechocystis LC contains a single cysteine residue, which is conserved in several cyanobacterial species, such as Synechococcus sp. PCC 6301, but not in Anabaena sp. PCC 7120. The phycobilisome core-membrane linker polypeptide (LCM) (Table 2) anchors the entire phycobilisome to the membrane (40). One of the three absolutely conserved cysteines in the Synechocystis LCM polypeptide, Cys-190, is probably involved in chromophore binding (40). The presence of redox-active cysteines in both LC and LCM suggests that they may bind each other when joined under oxidizing conditions. This finding means that the phycobilisome could be covalently associated to the membrane under light-limiting conditions by a disulphide that is broken by Trx after an increase in light intensity, ensuring greater flexibility in light harvesting at higher light intensities. Interestingly, treatment of Synechococcus with N-ethylmaleimide disrupts energy transfer from phycobilisomes to photosystem I without perturbing the integrity of either phycobilisomes or photosystems I or II (41).
Conservation of Cysteines in Synechocystis TrxA-Linked Proteins. Considering the possibility of conservation of Trx-related regulatory mechanisms through evolution of photosynthetic organisms, we examined the presence of conserved cysteines in the Synechocystis amino acid sequences (see Table 3, which is published as supporting information on the PNAS web site). All TrxA-linked Synechocystis proteins have at least one cysteine conserved in homologous proteins from some other organism (Tables 1 and 2), and 17 of these Synechocystis TrxA targets have at least one cysteine conserved in chloroplast homologues (Tables 1 and 2). Three proteins with no cysteines conserved between Synechocystis and Arabidopsis chloroplast counterparts [glucan branching enzyme, PGM, and sulfate adenylyltransferase (Tables 1 and 2)] display very low overall similarities to the chloroplast proteins. A closer look at these chloroplast enzymes reveals that they are much more similar to eukaryotic proteins (data not shown). It must therefore be concluded that a possible Trx-mediated regulation has not been conserved through chloroplast evolution in these cases. Porphobilinogen synthase in bacteria, including cyanobacteria, fungi, and animals, is a zinc-dependent enzyme that uses three vicinal cysteines to coordinate the zinc ion (42). In plant chloroplasts and some bacteria, these cysteines have been replaced with aspartates that participate in coordinating a magnesium ion. The purified zinc-containing enzyme depends on exogenous thiols for maintaining the bound zinc and, consequently, for its activity (43), whereas the plant enzyme does not (44). Thus, Trx may function as an antioxidant, enabling zinc binding in vivo and so controlling the activity. This could be an example of a function for Trx lost in one branch through evolution. Synechocystis ADPglucose pyrophosphorylase shares three cysteines (Cys-55, -325, and -330) with its Arabidopsis chloroplast homologue. More interesting, however, are the two N-terminal cysteines (Cys-2 and -3) that are not strictly conserved but appear only 2 aa upstream of the chloroplast Cys-12, which has been implied in redox regulation of this enzyme. Hence, there is a possibility that cysteines could be functionally equivalent without being conserved in the sequence.
Concluding Remarks. The putative Synechocystis TrxA target proteins identified in this study are mainly enzymes participating in anabolic processes. A picture begins to emerge in which the cyanobacterial processes of assimilation and storage of carbon, sulfur, and nitrogen in glycogen, proteins, and cyanophycin, respectively, are all possible targets for light-induced redox regulation by means of Trx. Based on the knowledge of Trx action in chloroplasts (1-3), it is expected that reduced Trx has a stimulatory effect on these anabolic processes, thus signaling the availability of light energy. However, the identities and relative abundance of Trx targets are likely to change depending on growth conditions.
The new potential TrxA target proteins point to differences in the Trx-mediated regulation between cyanobacteria and chloroplasts. In contrast to plants, cyanobacteria depend on the carbon dioxide-concentrating mechanism for a functioning CO2 assimilation and growth at physiological levels of CO2 (45). The finding that CcmM interacts with TrxA suggests that the carbon dioxide-concentrating mechanism is subject to Trx-mediated redox regulation in cyanobacteria, whereas the Calvin cycle has become a prime Trx target process in chloroplasts. Each Synechocystis enzyme of the complete pathway of glycogen synthesis was found to interact with Trx and not only the rate-limiting enzyme, ADPglucose pyrophosphorylase, as seems to be the case in plastids (21). Synechocystis GOGAT, rather than GS, were found to interact with TrxA, implying a shift of the focus for redox regulation of ammonium assimilation between cyanobacteria and chloroplasts.
This study paves the way for future work on regulation mediated by Trx in cyanobacteria, which has long been considered insignificant. It also gives an interesting perspective to the molecular evolution of susceptibility toward reduction by Trx.
Supplementary Material
Acknowledgments
This work was financed by Grant BMC 2001-2635 from the Spanish Ministry of Science and Technology (MCYT). M.L. is a holder of a Ramon-y-Cajal contract from the MCYT.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Trx, thioredoxin; GS, glutamine synthetase; GOGAT, glutamate synthase; Rubisco, ribulose 1,5-bisphosphate carboxylase/oxygenase; RbcL, large subunit of Rubisco; PGM, phosphoglucomutase; Prx, peroxiredoxin.
References
- 1.Buchanan, B. B. (1980) Annu. Rev. Plant Physiol. 31, 341-374. [Google Scholar]
- 2.Jacquot, J.-P., Lancelin, J.-M. & Meyer, Y. (1997) New Phytol. 136, 543-570. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan, B. B., Schürmann, P., Wolosiuk, R. A. & Jacquot, J.-P. (2002) Photosynth. Res. 73, 215-222. [DOI] [PubMed] [Google Scholar]
- 4.Verdoucq, L., Vignols, F., Jacquot, J.-P., Chartier, Y. & Meyer, Y. (1999) J. Biol. Chem. 274, 19714-19722. [DOI] [PubMed] [Google Scholar]
- 5.Motohashi, K., Kondoh, A., Stumpp, M. T. & Hisabori, T. (2001) Proc. Natl. Acad. Sci. USA 98, 11224-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balmer, Y., Koller, A., del Val, G., Manieri, W., Schürmann, P. & Buchanan, B. B. (2003) Proc. Natl. Acad. Sci. USA 100, 370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamoi, M., Murakami, A., Takeda, T. & Shigeoka, S. (1998) Biosci. Biotechnol. Biochem. 62, 374-376. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi, D., Tamoi, M., Iwaki, T., Shigeoka, S. & Wadano, A. (2003) Plant Cell Physiol. 44, 269-276. [DOI] [PubMed] [Google Scholar]
- 9.Navarro, F. & Florencio, F. J. (1996) Plant Physiol. 111, 1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rippka, R., Deruelles, J., Waterbury, J. B., Herman, M. & Stanier, R. Y. (1979) J. Gen. Microbiol. 111, 1-61. [Google Scholar]
- 11.Reyes, J. C. & Florencio, F. J. (1995) Plant Mol. Biol. 27, 789-799. [DOI] [PubMed] [Google Scholar]
- 12.Markwell, M. A., Haas, S. M., Bieber, L. L. & Tolbert, N. E. (1978) Anal. Biochem. 87, 206-210. [DOI] [PubMed] [Google Scholar]
- 13.Marqués, S., Mérida, A., Candau, P. & Florencio, F. J. (1992) Planta 187, 247-253. [DOI] [PubMed] [Google Scholar]
- 14.Navarro, F., Martín-Figueroa, E. & Florencio, F. J. (2000) Plant Mol. Biol. 43, 23-32. [DOI] [PubMed] [Google Scholar]
- 15.Ranty, B., Lorimer, G. & Gutteridge, S. (1991) Eur. J. Biochem. 200, 353-358. [DOI] [PubMed] [Google Scholar]
- 16.Marcus, Y., Altman-Gueta, H., Finkler, A. & Gurewitz, M. (2003) J. Bacteriol. 185, 1509-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa, T., Amichay, D. & Gurewitz, M. (1994) Photosynth. Res. 39, 183-190. [DOI] [PubMed] [Google Scholar]
- 18.Sicher, R. C. (1989) Plant Physiol. 89, 557-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball, S. G. & Morell, M. K. (2003) Annu. Rev. Plant Biol. 54, 207-233. [DOI] [PubMed] [Google Scholar]
- 20.Fu, Y., Ballicora, M. A., Leykam, J. F. & Preiss, J. (1998) J. Biol. Chem. 273, 25045-25052. [DOI] [PubMed] [Google Scholar]
- 21.Ballicora, M. A., Frueauf, J. B., Fu, Y., Schürmann, P. & Preiss, J. (2000) J. Biol. Chem. 275, 1315-1320. [DOI] [PubMed] [Google Scholar]
- 22.Tiessen, A., Hendriks, J. H. M., Stitt, M., Branscheid, A., Gibon, Y., Farré, E. M. & Geigenberger, P. (2002) Plant Cell 14, 2191-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iglesias, A. A., Kakefuda, G. & Preiss, J. (1991) Plant Physiol. 97, 1187-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cid, E., Geremia, R. A., Guinovart, J. J. & Ferrer, J. C. (2002) FEBS Lett. 528, 5-11. [DOI] [PubMed] [Google Scholar]
- 25.Gleason, F. K. & Holmgren, A. (1988) FEMS Microbiol. Rev. 4, 271-297. [DOI] [PubMed] [Google Scholar]
- 26.Bruhl, A., Haverkamp, T., Gisselmann, G. & Schwenn, J. D. (1996) Biochim. Biophys. Acta 1295, 119-124. [DOI] [PubMed] [Google Scholar]
- 27.Muro-Pastor, M. I. & Florencio, F. J. (2003) Plant Physiol. Biochem. 41, 595-603. [Google Scholar]
- 28.Florencio, F. J., Gadal, P. & Buchanan, B. B. (1993) Plant Physiol. Biochem. 31, 649-655. [Google Scholar]
- 29.Choi, Y. A., Kim, S. G. & Kwon, Y. M. (1999) Plant Sci. 149, 175-182. [Google Scholar]
- 30.Ziegler, K., Diener, A., Herpin, C., Richter, R., Deutzmann, R. & Lockau, W. (1998) Eur. J. Biochem. 254, 154-159. [DOI] [PubMed] [Google Scholar]
- 31.Schneider, G. J., Tumer, N. E., Richaud, C., Borbely, G. & Haselkorn, R. (1987) J. Biol. Chem. 262, 14633-14639. [PubMed] [Google Scholar]
- 32.Singer, R. A. & Doolittle, W. F. (1975) Nature 253, 650-651. [DOI] [PubMed] [Google Scholar]
- 33.Black, S. (1986) Science 234, 1111-1114. [DOI] [PubMed] [Google Scholar]
- 34.Richarme, G. (1998) Biochem. Biophys. Res. Commun. 252, 156-161. [DOI] [PubMed] [Google Scholar]
- 35.Vignols, F., Mouaheb, N., Thomas, D. & Meyer, Y. (2003) J. Biol. Chem. 278, 4516-4523. [DOI] [PubMed] [Google Scholar]
- 36.Manohara, M. S. & Tripathy, B. C. (2000) Planta 212, 52-59. [DOI] [PubMed] [Google Scholar]
- 37.Wood, Z. A., Schröder, E., Harris, J. R. & Poole, L. B. (2003) Trends Biochem. Sci. 28, 32-40. [DOI] [PubMed] [Google Scholar]
- 38.Tichy, M. & Vermaas, W. (1999) J. Bacteriol. 181, 1875-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broin, M., Cuiné, S., Eymery, F. & Rey, P. (2002) Plant Cell 14, 1417-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidler, W. A. (1994) in The Molecular Biology of Cyanobacteria, ed. Bryant, D. A. (Kluwer, Dordrecht, The Netherlands), pp. 139-216.
- 41.Glazer, A. N., Gindt, Y. M., Chan, C. F. & Sauer, K. (1994) Photosynth. Res. 40, 167-173. [DOI] [PubMed] [Google Scholar]
- 42.Shoolingin-Jordan, P. M., Spencer, P., Sarwar, M., Erskine, P. E., Cheung, K.-M., Cooper, J. B. & Norton, E. B. (2002) Biochem. Soc. Trans. 30, 584-590. [DOI] [PubMed] [Google Scholar]
- 43.Spencer, P. & Jordan, P. M. (1993) Biochem. J. 290, 279-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liedgens, W., Lütz, C. & Schneider, H. A. W. (1983) Eur. J. Biochem. 135, 75-79. [DOI] [PubMed] [Google Scholar]
- 45.Badger, M. R. & Price, G. D. (2003) J. Exp. Bot. 54, 609-622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.