Abstract
Background
The purpose of this study was to analyze the bacterial diversity in persistent apical lesions on root-filled teeth by using culture-independent molecular methods.
Design
Twenty surgically removed apical lesions from therapy-resistant teeth were examined for the presence of bacterial DNA using PCR targeting the 16s ribosomal RNA gene, followed by cloning and sequencing.
Results
Bacterial DNA was detected in 17 of the 20 samples (85%). A total of 236 clones were analyzed. Seven different bacterial phyla were represented and a total of 75 different bacterial taxa were identified; 36% of the species have not yet been cultivated. Commonly detected bacterial species included Fusobacterium spp., Prevotella spp., Tannerella forsythia, Porphyromonas endodontalis, Treponema denticola, Bacteroidetes spp., Peptostreptococcus spp., and Streptococcus spp.
Conclusions
A wide range of bacteria was identified in periapical lesions on therapy-resistant teeth. These bacteria may contribute in the etiology of periapical infection and impede healing of these lesions.
Keywords: bacterial phyla, endodontic infection, therapy-resistant teeth, 16s ribosomal RNA, sequencing
Teeth with apical periodontitis are highly prevalent amongst adults (1). The majority of apical periodontal lesions are located in previously root-filled teeth (1–6). The proportion of root-filled teeth with apical periodontitis was 43% in a study comprising 250 35-year-old inhabitants in Norway in 2003 (7).
Bacteria play the major role in the etiology of periapical lesion formation (8, 9), resulting in bone resorption which is an active process carried out by osteoclasts. Bacteria can gain access to the dental pulp through the crown or root surfaces in association with processes such as caries, periodontal disease, or trauma. A select group of microorganisms colonizing the normally sterile root canal space can cause pulp necrosis and inflammation in the surrounding bone. In well-controlled studies, where the root canal is properly instrumented and obturated, a success rate of 80–90% has been shown (10, 11). However, if bacteria are resistant to host defense mechanisms and/or therapy, they will survive within the root canal system and periapical tissue. In such cases, healing of the periapical lesion is compromised.
Culture-independent analysis using the bacterial ribosomal RNA (rRNA) genes has provided insights into the composition of mixed microbial communities in the root canals (12–19). It has been demonstrated that several uncultivated phylotypes and newly named species are present in the root canals associated with asymptomatic primary or persistent endodontic infections (18), and these can also take part in the root canal flora associated with symptomatic endodontic infections (15).
While several studies have shown that Propionibacterium and Actinomyces spp. can survive and thrive in persistent periapical lesions (20, 21), it is controversial whether or not a consortium of different species can live in apical lesions. At present there is scarce information about the role of microorganisms in apical lesions on therapy-resistant teeth. One recent culture-independent study (22) identified bacterial DNA, including unidentified and uncultured species, in samples from extraradicular infections. Studies employing molecular techniques have shown that the bacterial diversity in most environments is strongly underestimated in surveys using cultivation-based techniques (23–25), in some cases with less than 1% of the organisms being cultivable (26). As only 50% of the oral bacteria have been cultivated, it is likely that at least some members of the uncultivable flora may contribute to disease. Molecular assays have shown that 700–1,000 species can colonize different oral biofilms, far more than detected by cultivation (25, 27). The development of 16s rRNA phylogenetic methodology has widened the scope of detectable microorganisms to include uncultivable organisms that may play a significant, as yet undefined role in the pathogenesis of oral disease (25, 27, 28).
Since tissue invasion is considered a major step in the pathogenesis and complete healing of the periapical lesion is the ultimate goal in the treatment of apical periodontitis, the understanding of endodontic disease requires knowledge on the entire bacterial population. The aim of the present study was to further investigate the bacterial diversity in persistent apical lesions on root-filled teeth by using culture-independent molecular methods.
Methods
Subjects and sample collection
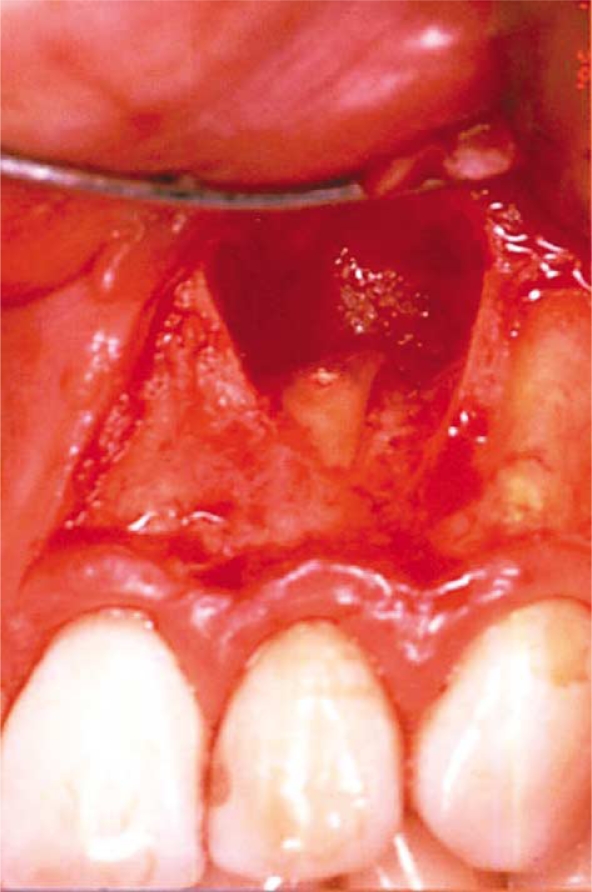
Twenty patients (age 27–93, mean age 56) with persistent apical periodontitis were surgically treated with apicectomies by the same oral surgeon. Each patient was referred to the oral surgeon for the treatment of one tooth (Fig. 1). None of the teeth had previously responded to conventional endodontic therapy. Fistulous tracts or endo-perio-like lesions related to the teeth to be treated were not present. No patients had pain at the time of surgery and none had taken antibiotics the previous two months before surgery. Radiographically, it was evident that the patients referred for treatment had root-filled teeth with periapical radiolucencies of diameters varying between 4 and 12 mm. Clinical inspection showed that all teeth had satisfactory coronal restorations.
Fig. 1.
Apical surgery with a submarginal incision to avoid contamination of the surgical area.
All patients had an oral rinse with 0.2% chlorhexidine gluconate solution for 30 sec immediately before surgery. The present investigation used a full-thickness muco-periosteal flap, reflected without involving the sulcular area of the periodontium. A submarginal incision was applied in order to avoid contamination of the periapical lesion with microorganisms present in the marginal area. Care was taken to avoid contamination from saliva. The whole lesion was enucleated before apicectomy. Samples were transferred to 200 µl of lysis buffer from the extraction kit QiaAmp® DNA Mini Kit (Qiagen, GmbH, Germany) and subsequently kept at −70°C prior to DNA extraction. The study was approved by the Norwegian Ethical Board. Informed consent was received from all patients.
Preparation of DNA and PCR amplification of 16s ribosomal RNA (rRNA) genes
DNA was extracted using the QiaAmp® DNA Mini Kit (Qiagen) according to the manufacturer's recommendation. PCR amplification of 16s rRNA gene was performed using previously published universal eubacterial primers (PA and PD) (MWG-Biotech, GmbH, Ebersberg, Germany) (29). Amplicons were purified using the QIAquick Gel Extraction Kit (Qiagen).
Cloning procedures and 16s ribosomal RNA (rRNA) sequencing
The amplicons were ligated into the plasmid vector pCR4-TOPO and transformed into Eschericia coli one Shot Top10 chemically competent cells (Invitrogen, Oslo, Norway). The transformed cells were plated onto Luria Bertoni agar plates, supplemented with kanamycin and incubated overnight at 37°C. From each sample, 12–36 clones were randomly selected to investigate bacterial species. A total of 236 clones were investigated. Correct sizes of the inserts were determined by PCR with M13 forward (−20) and M13 reverse primer (Invitrogen) followed by electrophoresis on 1% agarose gel. DNA amplicons (7 µl) were purified in a mixture containing 1 µl exonuclease and 1 µl shrimp alkaline phosphatase, and incubated for 15 min at 37°C and 15 min at 80°C, as recommended by the manufacturer (Amersham Biosciences, Cleveland, OH). The purified product was sequenced on both strands using the primers PB and PC (MWG-Biotech, GmbH) (29) and the Big-Dye Terminator mix (Applied Biosystems, Foster City, CA), according to the manufacturer's instruction. The sequencing reactions were run on an ABI 3730 DNA sequencer (Applied Biosystems).
The partial 16s rDNA sequences of approximately 500 bp were used to determine bacterial identity. For identification of closest relatives, the consensus sequences were compared with 16s rDNA sequences in GenBank databases using the Blast 2.1 program from the GenBank Online Service. Clone sequences with 98–100% identity were considered to be of the same bacterial species. The sequences were aligned with the CLUSTAL W program. A neighbor-joining phylogenetic tree was constructed using the package MEGA, version 4.1 (30). Two hundred bootstrap trees were generated, and bootstrap confidence levels were determined with the MEGA program.
Results
In the present study, no PCR product was obtained from three samples (nos. 5, 11, and 20) using the primers PA and PD while 17 of 20 (85%) lesions were positive for bacterial DNA. Bacterial species from seven different phyla were represented (Table 1). Bacteroidetes, Fusobacteria, and Firmicutes dominated, followed by Spirochaetes, Actinobacteria, Proteobacteria, and Synergistes.
Table 1.
Bacterial phyla in patients with refractory apical periodontitis
| Phylogenetic group | No. of taxa | No. of clones (%) |
|---|---|---|
| Fusobacteria | 13 | 90 (38.1) |
| Bacteroidetes | 19 | 53 (22.5) |
| Firmicutes | 19 | 32 (13.6) |
| Spirochaetes | 4 | 10 (4.2) |
| Proteobacteria | 2 | 5 (2.1) |
| Actinobacteria | 3 | 4 (1.7) |
| Synergistes | 1 | 1 (0.4) |
| Othera | 18 | 41 (17.4) |
| Total | 75 | 236 (100) |
Eighteen of the taxa were not assigned to any phylogenetic group.
All specimens harbored 1–11 different bacterial species with a mean of seven species per lesion. The bacterial profiles of patients differed in diversity and bacterial dominance (Table 2). One patient (no. 15) presented with monoculture (Fusobacterium spp.), two patients had three species (no. 3: Fusobacterium spp., Selenomonas-like spp., uncultured bacterial species; no. 4: Fusobacterium spp., Treponema spp., Propionibacterium spp.), otherwise 4–11 different species were found within the patient samples. Fusobacterium spp. were the most prevalent (38% of taxa), detected in all but two lesions (88%). Fusobacterium nucleatum was the only bacterium that could be identified to species level within this genus. Several other strains of Fusobacterium were also found, including yet uncultivable strains. Bacteroidetes was the second most prevalent phylum with Prevotella as the most commonly detected genus (present in 35% of the lesions). Other well-known endodontic bacterial species such as Porphyromonas endodontalis, Bacteroidetes spp., and Tannerella forsythia were also identified. Peptostreptococcus sp. oral clone FG014 and Streptococcus spp. were the most prevalent within Firmicutes (41 and 29% of lesions, respectively). Additionally, Selenomonas spp., Eubacterium sp., and Solobacterium moorei were identified. Amongst Spirochaetes, Treponema denticola and strains that could not be identified at species level were found. Campylobacter rectus and the less commonly encountered bacterial species Bradyrhizobium spp. and Synergistes-like sp. oral clone BH007 were detected in two lesions. No association of bacterial phyla or taxa could be observed using the chi-square test (Fisher's exact test, data not shown).
Table 2.
Distribution of bacterial taxa detected in the apical lesions
| Subject no. | Bacteriaa (no. of clones if >1) |
|---|---|
| 1 | Fusobacterium sp. (2) |
| Prevotella sp. (2) | |
| Porphyromonas sp. (3) | |
| Bacteroidetes sp. (3) | |
| Uncultured Bacteroidetes | |
| Uncultured bacterium | |
| 2 | Fusobacterium sp. (3) |
| Prevotella sp. (3) | |
| Treponema sp. | |
| Campylobacter rectus (2) | |
| Synergistes-like sp. | |
| Uncultured bacterium | |
| Unidentified oral bacterium | |
| 3 | Fusobacterium sp. (6) |
| Selenomonas-like sp. | |
| Uncultured bacteria (3) | |
| 4 | Fusobacterium sp. (6) |
| Treponema sp. (4) | |
| Propionibacterium sp. (2) | |
| 6 | Fusobacterium sp. (4) |
| Peptostreptococcus sp. (4) | |
| Streptococcus sp. | |
| Uncultured bacteria (2) | |
| Unidentified oral bacterium | |
| 7 | Fusobacterium sp. (11) |
| Porphyromonas-like sp. (2) | |
| Tannerella sp. (4) | |
| Peptostreptococcus sp. | |
| Campylobacter sp. | |
| Eubacterium sp. | |
| Uncultured bacterium | |
| 8 | Prevotella sp. (5) |
| Porphyromonas-like sp. | |
| Treponema sp. (5) | |
| Peptococcus sp. (2) | |
| Eubacterium sp. | |
| Unidentified oral bacteria (2) | |
| 9 | Uncultured bacteria (4) |
| Unidentified oral bacteria (6) | |
| 10 | Fusobacterium sp (2) |
| Uncultured Propionibacterineae | |
| Streptococcus sp. | |
| Uncultured Streptococcus sp. (3) | |
| Corynebacterium sp. | |
| Uncultured bacteria (3) | |
| Uncultured Bradyrhizobium sp. | |
| 12 | Fusobacterium sp. (2) |
| Prevotella sp. | |
| Porphyromonas sp. (2) | |
| Porphyromonas-like sp. (3) | |
| Bacteroidetes sp. (3) | |
| Tannerella sp. | |
| Unidentified oral bacteria (3) | |
| 13 | Fusobacterium sp. (8) |
| Porphyromonas sp. (2) | |
| Peptostreptococcus sp. | |
| Uncultured Veillonella sp. (2) | |
| 14 | Fusobacterium sp. (2) |
| Prevotella sp.(6) | |
| Peptostreptococcus sp. | |
| Streptococcus intermedius (2) | |
| Streptococcus sp. | |
| Uncultured bacterium | |
| 15 | Fusobacterium sp. (28) |
| 16 | Fusobacterium sp. (8) |
| Peptostreptococcus sp. | |
| Uncultured Bradyrhizobium sp. | |
| Uncultured bacterium | |
| 17 | Fusobacterium sp. (3) |
| Prevotella sp. | |
| Selenomonas sp. | |
| Solobacterium moorei strain N407 | |
| Uncultured bacteria (6) | |
| Unidentified oral bacterium | |
| 18 | Fusobacterium sp. (2) |
| Prevotella sp. | |
| Porphyromonas sp. (5) | |
| Bacteroidales genomosp. | |
| Streptococcus sp. | |
| Uncultured bacterium clone bE15-57 | |
| Unidentified oral bacteria (2) | |
| 19 | Fusobacterium sp. (2) |
| Tannerella sp. (2) | |
| Peptostreptococcus sp. | |
| Streptococcus sp. (2) | |
| Uncultured Streptococcus sp. | |
| Solobacterium sp. | |
| Uncultured bacteria (2) | |
| Unidentified oral bacterium |
Species name given only when 100% homology was seen.
A high bacterial diversity, with 75 different bacterial taxa, were identified amongst the 236 clones investigated (Fig. 2). All 16s rDNA sequences, except for three, showed 98–100% similarity to sequences deposited to GenBank. Three isolates (uncultured bacterium clone 2:3 [FJ472593], uncultured bacterium clone 7:8 [FJ472594], and uncultured bacterium clone 14:12 [FJ472595]) were not closely related to any sequence previously in the GenBank database and, thus, might represent new phylotypes. Notably, 36% of the taxa represented yet uncultivable/unidentified species and these were present in 88% of the lesions comprising 23% of the total clones investigated.
Fig. 2.
Phylogenetic tree of bacterial phylotypes detected in persistent periapical lesions. The marker bar represents a 5% difference in nucleotide sequences.
Discussion
The present study assessed the occurrence of cultivable and also uncultivated phylotypes in persistent apical lesions on root-filled teeth using culture-independent molecular methods. Most studies of the bacterial etiology of periapical lesions have used either culture-based or targeted DNA approaches (31–33), and it is therefore likely that pathogens remain undiscovered. Sampling of microorganisms in the periapical area during surgery is difficult because the samples are easily contaminated with microorganisms from the marginal area and the indigenous oral microbiota unless care is taken to avoid such contamination (31–33). During surgery, a submarginal incision was performed in order to avoid contamination in the surgical field. It was, therefore, assumed that the bacterial species recovered in this study were present in the refractory lesions before surgery. Microbiological profiling has previously indicated that following a marginal incision, bacteria from the periodontal pocket reach the underlying tissues by surgeon-released bacteremia or direct translocation (32).
In the present study, three periapical samples were negative for bacterial DNA, thus suggesting that in few cases the bacteria causing the lesions may be present exclusively in the root canal system. Alternatively, sometimes DNA extractions and subsequent PCR simply do not work. Eighty-eight percent of the lesions harbored uncultivable/unidentified bacterial species, constituting 34% of the taxa identified. These findings clearly indicate that previously unrecognized bacterial species are present in the apical lesions and may participate in persistent apical periodontitis. This is in concordance with the findings of Noguchi et al. (22) who detected unidentified and uncultivable bacterial species in 11 of 14 samples from extraradicular biofilm. The pathogenicity of the uncultivable microbiota in endodontics is not known. However, it is reasonable that at least some members of this part of the microbiota are pathogenic and may contribute in the disease process.
Many of the species identified in the present study were the same that have been reported in extraradicular disease based on cultivation and DNA–DNA hybridization techniques. Fusobacterium is commonly isolated from root canal infections, and in the present study all except one lesion where bacterial DNA was detected harbored this organism. F. nucleatum was recently found in high proportions and levels in necrotic root canals with multiple displacement amplification, checkerboard DNA–DNA hybridization, and reverse-capture checkerboard hybridization (17, 34, 35). Fusobacterium species are known to co-aggregate with many species of oral bacteria and they are common participants in oral disease (36). To our knowledge, the study by Noguchi et al. (22) is the only published of extraradicular infection using PCR-based 16s rRNA gene assay. They identified bacterial DNA in 14 of 20 lesions and 113 bacterial species were isolated. F. nucleatum (14 of 14) and Porphyromonas gingivalis (12 of 14) were frequently detected. However, some teeth were extracted before examination which can give false positive results from the periodontal or indigenous oral microbiota. This might explain the high levels of the periodontopathogen, P. gingivalis, as opposed to our study where it was absent. However, P. gingivalis has been detected in small areas of periapical lesions by fluorescence in situ hybridization and DNA–DNA hybridization (31, 37).
Organisms belonging to Porphyromonas spp., Treponema spp., and T. forsythia are well-known pathogens in periodontal diseases. In the recent years, these species have also been detected in periapical tissue by DNA–DNA and in situ hybridization techniques (31, 33, 37). T. forsythia is difficult to culture and has not yet been recovered from root canals or periapical tissue with cultivation techniques. This organism is, however, frequently detected in necrotic canals with molecular methods (34, 38, 39). Although some authors have indicated that treponemes are important pathogens in endodontic disease (40–43), their fastidious growth has led these organisms to be underestimated in endodontic disease. It is only recently that molecular genetic analyses confirm their presence in infected root canals (14, 34, 44, 45) and periapical tissue (22, 31, 33, 37). Based on their frequent detection by molecular techniques, they might be potential endodontic pathogens. Prevotella spp. have frequently been found in different types of endodontic infections (46, 47), but have only been detected in the apical tissue with DNA–DNA hybridization and in situ hybridization (31, 37). A Synergistes phylotype was detected in one lesion in the present study. This phylotype has recently been shown to be a frequent member of the endodontic microbiota (18). Most isolates of Solobacterium are found in the oral cavity, particularly in periodontitis (16, 48, 49) and in patients with halitosis (50).
A remarkably high diversity of species was identified in the periapical lesions. Although traditional views suggest that organisms surviving root canal treatment comprise a select group of the most robust organisms, the application of ecological parameters indicates that bacterial survival after root canal treatment will depend not only on the robustness of the organisms, but on how good an adaptor the organism is to the new limiting factors in their corresponding niches. Surface adherence by bacteria to form biofilms is an example of bacterial adaptation. Increasing amount of information is now available on the existence of polymicrobial biofilm communities on root tips (51) and also in the apical tissue (37). The foundation for this ecological approach to endodontic infections suggests disease is caused by a polymicrobial microbiota that undergoes physiological and genetic changes triggered by changes in the root canal environment. The frequent recovery of Enterococcus faecalis in root canals, associated with persistent infections, brought an intense research interest in this bacterium. Actually, E. faecalis is seldom identified in persistent apical lesions (22), and was not recovered in any lesion in the present study, indicating that this organism may not be as important when it comes to invading the apical area.
Molecular methods have expanded the list of endodontic pathogens by inclusion of some fastidious bacterial species and even uncultivated bacterial species that have never previously been found in endodontic infections by cultivation procedures. It has been shown that bacteria form biofilms in the root canal and extraradicular area, both on root tips, extruded gutta percha points, and directly in the periapical tissue (37, 51, 52). It is reasonable that extraradicular infections are caused during the acute phase of apical periodontitis or because of overinstrumentation by the dentist during root canal therapy. However, from a biofilm infectious viewpoint, it is possible that extraradicular sites are infected both during the acute or chronic phase of apical periodontitis. Bacterial species in the infected root canals may invade extraradicular sites and form biofilm which in turn can function as a focus of acute infections.
In conclusion, a wide variety of bacterial species, including a high percentage of uncultured/unidentified bacteria were found to colonize periapical lesions. Since endodontic infections develop in a previously sterile place which does not contain a normal microbiota, every bacterial species present in the mixed consortium has the potential to play a role in the infectious disease process. The present study has expanded the list of bacterial species present in the extraradicular area suggesting that the microbial etiology of periradicular disease is far more complex than previously anticipated.
Acknowledgements
The technical assistance of Jan Oksnes and Anne-Marie Klem (Norwegian Institute of Public Health, Oslo, Norway) is kindly acknowledged. The study was supported by the Faculty of Dentistry, University of Oslo, Oslo, Norway.
Conflict of interest and funding
The study was supported by the Faculty of Dentistry, University of Oslo, Oslo, Norway. There is no conflict of interest in the present study for any of the authors.
References
- 1.Eriksen HM, Kirkevang LL, Petersson K. Endodontic epidemiology and treatment outcome: general considerations. Endod Topics. 2002;2:1–9. [Google Scholar]
- 2.Bergström J, Eliasson S, Ahlberg KF. Periapical status in subjects with regular dental care habits. Comm Dent Oral Epidemiol. 1987;15:236–9. doi: 10.1111/j.1600-0528.1987.tb00528.x. [DOI] [PubMed] [Google Scholar]
- 3.Eriksen HM, Bjertness E. Prevalence of apical periodontitis and results of endodontic treatment in middle-aged adults in Norway. Endod Dent Traumatol. 1991;7:1–4. doi: 10.1111/j.1600-9657.1991.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 4.Kirkevang LL, Hörsted-Bindslev P, Ørstavik D, Wenzel A. Frequency and distribution of endodontically treated teeth and apical periodontitis in an urban Danish population. Int Endod J. 2001;34:198–205. doi: 10.1046/j.1365-2591.2001.00370.x. [DOI] [PubMed] [Google Scholar]
- 5.Petersson K. Endodontic status of mandibular premolars and molars in an adult Swedish population. A longitudinal study 1974–1985. Endod Dent Traumatol. 1993;9:13–28. doi: 10.1111/j.1600-9657.1993.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 6.Sidaravicius B, Aleksejuniene J, Eriksen HM. Endodontic treatment and prevalence of apical periodontitis in an adult population of Vilnius, Lithuania. Endod Dent Traumatol. 1999;15:210–5. doi: 10.1111/j.1600-9657.1999.tb00776.x. [DOI] [PubMed] [Google Scholar]
- 7.Skudutyte-Rysstad R, Eriksen HM. Endodontic status amongst 35-year-old Oslo citizens and changes over a 30-year period. Int Endod J. 2006;39:637–42. doi: 10.1111/j.1365-2591.2006.01129.x. [DOI] [PubMed] [Google Scholar]
- 8.Kakehashi S, Stanley HR, Fitzgerald RJ. The effect of surgical exposures of dental pulps in germ-free and conventional laboratory rats. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1965;20:340–9. doi: 10.1016/0030-4220(65)90166-0. [DOI] [PubMed] [Google Scholar]
- 9.Sundqvist G. Odontological Dissertations No. 7, Umeå University, Umeå, Sweden. 1976. Bacteriological studies of necrotic dental pulps. [Google Scholar]
- 10.Byström A, Happonen RP, Sjögren U, Sundqvist G. Healing of periapical lesions of pulpless teeth after endodontic treatment with controlled asepsis. Endod Dent Traumatol. 1987;3:58–63. doi: 10.1111/j.1600-9657.1987.tb00543.x. [DOI] [PubMed] [Google Scholar]
- 11.Tronstad L, Barnett F, Riso K, Slots J. Extraradicular endodontic infections. Endod Dent Traumatol. 1987;3:86–90. doi: 10.1111/j.1600-9657.1987.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 12.Dymock D, Weightman A, Scully C, Wade W. Molecular analysis of microflora associated with dentoalveolar abscesses. J Clin Microbiol. 1996;34:537–42. doi: 10.1128/jcm.34.3.537-542.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouad AF, Barry J, Caimano M, Clawson M, Zhu Q, Carver R, et al. PCR-based identification of bacteria associated with endodontic infections. J Clin Microbiol. 2002;40:3223–31. doi: 10.1128/JCM.40.9.3223-3231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung IY, Choi BK, Kum KY, Roh BD, Lee SJ, Lee CY, et al. Molecular epidemiology and association of putative pathogens in root canal infection. J Endod. 2000;26:599–604. doi: 10.1097/00004770-200010000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Rôças IN, Siqueira JF. Detection of novel oral species and phylotypes in symptomatic endodontic infections including abscesses. FEMS Microbiol Lett. 2005;15:279–85. doi: 10.1016/j.femsle.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Rolph HJ, Lennon A, Riggio MP, Saunders WP, MacKenzie D, Coldero L, et al. Molecular identification of microorganisms from endodontic infections. J Clin Microbiol. 2001;39:3282–9. doi: 10.1128/JCM.39.9.3282-3289.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakamoto M, Rôças IN, Siqueira JF, Benno Y. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol Immunol. 2003;21:112–22. doi: 10.1111/j.1399-302X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 18.Siqueira JF, Rôças IN. Uncultivated phylotypes and newly named species associated with primary and persistent endodontic infections. J Clin Microbiol. 2005;43:3314–9. doi: 10.1128/JCM.43.7.3314-3319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade WG, Spratt DA, Dymock D, Weigthman AJ. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin Infect Dis. 1997;25:235–6. doi: 10.1086/516215. [DOI] [PubMed] [Google Scholar]
- 20.Happonen RP, Söderling E, Viander M, Linko-Kettunen L, Pelliniemi LJ. Immunocytochemical demonstration of Actinomyces species and Arachnia propionica in periapical infections. J Oral Pathol. 1985;14:405–13. doi: 10.1111/j.1600-0714.1985.tb00512.x. [DOI] [PubMed] [Google Scholar]
- 21.Nair PNR. Light and electron microscopic studies of root canal flora and periapical lesions. J Endod. 1987;13:29–39. doi: 10.1016/S0099-2399(87)80089-4. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi N, Noiri Y, Narimatsu N, Ebisu S. Identification and localization of extraradicular biofilm-forming bacteria associated with refractory endodontic pathogens. Appl Environ Microbiol. 2005;71:8738–43. doi: 10.1128/AEM.71.12.8738-8743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Ecol. 1995;59:143–69. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–76. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paster BJ, Boches SK, Gavin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pace NR, Stahl DA, Lane DJ, Olsen GJ. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 27.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroes I, Lepp PW, Relman DA. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–52. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valheim M, Djønne B, Heiene R, Caugant DA. Disseminated Mycobacterium celatum (type 3) infection in a domestic ferret (Mustela putoris furo) Vet Pathol. 2001;38:460–3. doi: 10.1354/vp.38-4-460. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 31.Gatti JJ, Dobeck JM, Smith C, White RR, Socransky SS, Skobe Z. Bacteria of asymptomatic periradicular endodontic lesions identified by DNA-DNA hybridization. Endod Dent Traumatol. 2000;16:197–204. doi: 10.1034/j.1600-9657.2000.016005197.x. [DOI] [PubMed] [Google Scholar]
- 32.Sunde PT, Olsen I, Lind PO, Tronstad L. Extraradicular infection: a methodological study. Endod Dent Traumatol. 2000;16:84–90. doi: 10.1034/j.1600-9657.2000.016002084.x. [DOI] [PubMed] [Google Scholar]
- 33.Sunde PT, Tronstad L, Eribe ER, Lind PO, Olsen I. Assessment of periradicular microbiota by DNA-DNA hybridization. Endod Dent Traumatol. 2000;16:191–6. doi: 10.1034/j.1600-9657.2000.016005191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brito LCN, Teles FR, França EC, Ribeiro-Sobrinho AP, Haffajee AD, Socransky SS. The use of multiple displacement amplification and checkerboard DNA-DNA hybridization to examine the microbiota of endodontic infections. J Clin Microbiol. 2007;45:3039–49. doi: 10.1128/JCM.02618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rpôças IN, Siqueira JF. Root canal microbiota of teeth with chronic apical periodontitis. J Clin Microbiol. 2008;46:3599–606. doi: 10.1128/JCM.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. Coaggregation: specific adherence among human oral plaque bacteria. Fed Am Soc Exper Biol J. 1993;7:406–13. doi: 10.1096/fasebj.7.5.8462782. [DOI] [PubMed] [Google Scholar]
- 37.Sunde PT, Olsen I, Göbel UB, Theegarten D, Winter S, Debelian GJ, et al. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiol. 2003;149:1095–102. doi: 10.1099/mic.0.26077-0. [DOI] [PubMed] [Google Scholar]
- 38.Conrads G, Gharbia SE, Gulabivala K, Lampert F, Shah HN. The use of a 16S rDNA directed PCR for the detection of endodontopathogenic bacteria. J Endod. 1997;23:433–8. doi: 10.1016/S0099-2399(97)80297-X. [DOI] [PubMed] [Google Scholar]
- 39.Siqueira JF, Roças IN. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2004;97:85–94. doi: 10.1016/s1079-2104(03)00353-6. [DOI] [PubMed] [Google Scholar]
- 40.Dahle UR, Olsen I, Tronstad L, Caugant DA. Population genetic analysis of oral treponemes by multilocus enzyme electrophoresis. Oral Microbiol Immunol. 1995;5:265–70. doi: 10.1111/j.1399-302x.1995.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 41.Dahle UR, Tronstad L, Olsen I, Cooper P, Smibert RM. A new method for routine isolation of oral treponemes using U-tubes and pectin medium. Anaerobe. 1995;1:315–9. doi: 10.1006/anae.1995.1033. [DOI] [PubMed] [Google Scholar]
- 42.Kerekes K, Olsen I. Similarities in the microfloras of root canals and deep periodontal pockets. Endod Dent Traumatol. 1990;6:1–5. doi: 10.1111/j.1600-9657.1990.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 43.Trope M, Tronstad L, Rosenberg ES, Listgarten M. Darkfield microscopy as a diagnostic aid in differentiating exudates from endodontic and periodontal abscesses. J Endod. 1988;14:35–8. doi: 10.1016/S0099-2399(88)80239-5. [DOI] [PubMed] [Google Scholar]
- 44.Jung IY, Choi B, Kum KY, Yoo YJ, Yoon TC, Lee SJ, et al. Identification of oral spirochetes at the species level and their association with other bacteria in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;92:329–34. doi: 10.1067/moe.2001.117263. [DOI] [PubMed] [Google Scholar]
- 45.Siqueira JF, Rôças IN, Favieri A, Santos KR. Detection of Treponema denticola in endodontic infections by 16S rRNA gene-directed polymerase chain reaction. Oral Microbiol Immunol. 2000;15:335–7. doi: 10.1034/j.1399-302x.2000.150512.x. [DOI] [PubMed] [Google Scholar]
- 46.Baumgartner JC, Watkins BJ, Bae KS, Xia T. Association of black-pigmented bacteria with endodontic infections. J Endod. 1999;25:413–5. doi: 10.1016/S0099-2399(99)80268-4. [DOI] [PubMed] [Google Scholar]
- 47.Haapasalo M. Bacteroides spp. in dental root canal infections. Endod Dent Traumatol. 1989;5:1–10. doi: 10.1111/j.1600-9657.1989.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 48.Downes BV, Van Der Berg J, Wade WG. Gram-positive anaerobic bacilli in human periodontal disease. J Periodont Res. 2004;39:213–20. doi: 10.1111/j.1600-0765.2004.00726.x. [DOI] [PubMed] [Google Scholar]
- 49.Harper-Owen R, Dymock D, Booth V, Weightman AJ, Wade WG. Detection of unculturable bacteria in periodontal health and disease by PCR. J Clin Microbiol. 1999;37:1469–73. doi: 10.1128/jcm.37.5.1469-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41:558–63. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tronstad L, Cervone F, Barnett F. Periapical bacterial plaque in teeth refractory to endodontic treatment. Endod Dent Traumatol. 1990;6:73–7. doi: 10.1111/j.1600-9657.1990.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 52.Takemura N, Noiri Y, Ehara A, Kawahara T, Noguchi N, Ebisu S. Single species biofilm-forming ability of root canal isolates on gutta-percha points. Eur J Oral Sci. 2004;112:523–9. doi: 10.1111/j.1600-0722.2004.00165.x. [DOI] [PubMed] [Google Scholar]