Abstract
CHL27, the Arabidopsis homologue to Chlamydomonas Crd1, a plastid-localized putative diiron protein, is required for the synthesis of protochlorophyllide and therefore is a candidate subunit of the aerobic cyclase in chlorophyll biosynthesis. δ-Aminolevulinic acid-fed antisense Arabidopsis plants with reduced amounts of Crd1/CHL27 accumulate Mg-protoporphyrin IX monomethyl ester, the substrate of the cyclase reaction. Mutant plants have chlorotic leaves with reduced abundance of all chlorophyll proteins. Fractionation of Arabidopsis chloroplast membranes shows that Crd1/CHL27 is equally distributed on a membrane-weight basis in the thylakoid and inner-envelope membranes.
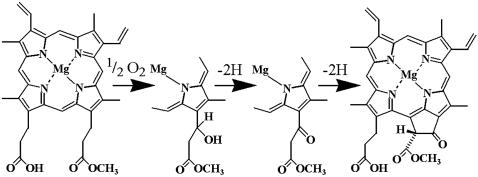
The chlorophyll (Chl) biosynthetic pathway, occurring in all photosynthetic organisms, has been described through genetic analysis of bacterial mutants and in vitro reconstitution of individual reactions (1–3). Chl production begins with the condensation of eight molecules of δ-aminolevulinic acid (ALA) to uroporphyrinogen III, the first cyclic tetrapyrrole. Uroporphyrinogen III is then converted to protoporphyrin IX, which is the branch-point intermediate to hemes and Chls. The chelation of magnesium into protoporphyrin IX results in the formation of Mg-protoporphyrin IX (MgP), which is converted to MgP monomethyl ester (MgPMME) by a methyl transferase (4). MgPMME is the substrate for the so-called cyclase reaction, which results in the formation of divinyl protochlorophyllide (Pchlide) containing the fifth ring (ring E) characteristic of all Chls (Fig. 1). In angiosperms, the subsequent steps include the extensively studied, light-dependent conversion of Pchlide to chlorophyllide a via NADPH-Pchlide oxidoreductase and the addition of a polyisoprene tail to complete Chl a production.
Fig. 1.
The oxidative cyclase reaction for the formation of the fifth ring of the Chl molecule. The conversion of MgPMME to divinyl Pchlide proceeds through three sequential two-electron oxidations. The first step requires molecular oxygen as a substrate for hydroxylation, analogous to the methane monooxygenase reaction. There is also an oxygen requirement for the third reaction.
Labeling experiments (5) suggested two different mechanisms for the cyclase reaction and presumably two different enzymes. The bchE gene product is implicated in the anaerobic reaction because bchE mutants in Rhodobacter sphaeroides accumulate pigments corresponding to MgP and MgPMME (6). The aerobic enzyme is clearly distinct. Although its genetic identity remained elusive for a long time, biochemical studies did define a reaction path and several key features of the enzyme. The aerobic cyclase reaction, an overall six-electron oxidation, is proposed to occur in three sequential steps (7) (Fig. 1). The first step is the stereospecific hydroxylation of the methyl-esterified ring C propionate followed by oxidation of the alcohol to the corresponding ketone. The now-activated methylene group reacts with the γ-meso carbon of the porphyrin nucleus in an oxidative reaction involving removal of two H to yield ring E. Both the hydroxylated and the keto compounds were suggested to be genuine intermediates, because they could function as substrates for the enzyme (8, 9). Molecular oxygen is required at the step of hydroxylation and also for the conversion of the keto intermediate to divinyl Pchlide (8).
The enzyme is iron-dependent: iron chelators inhibit the cyclase enzyme activity in vitro (10), and the substrate MgPMME accumulates in iron-deficient plants (11). The cyclase enzyme is membrane-associated and consists of multiple subunits, as indicated by the identification of multiple loci in barley that lead to accumulation of MgPMME in ALA-fed plants (2, 12). Both soluble and membrane components must be recombined to restore cyclase activity, which further supports the involvement of multiple gene products (13–15). The cyclase was suggested to have a reactive thiol, because activity is sensitive to thiol-modifying agents (13).
Crd1, a candidate diiron enzyme, is required for the accumulation of a subset of Chl proteins in Chlamydomonas (16). We previously proposed a role for Crd1 in cofactor biogenesis in the chloroplast and noted a link with iron metabolism. Crd1 is an intriguing candidate for the aerobic cyclase of chloroplasts because (i) it contains a putative diiron site, which is consistent with either of the two O2-dependent steps in cyclase chemistry, (ii) Crd1 homologues are restricted to photosynthetic organisms, suggesting a function in bacteriochlorophyll/Chl biosynthesis (17), and (iii) Crd1 is not found in photosynthetic organisms such as Chlorobium tepidum, which are strict anaerobes and hence use the anaerobic enzyme. This idea was validated by the phenotype of a Rubrivivax gelatinosus mutant carrying a loss-of-function mutation in a homologue of Crd1 called acsF (18). The authors showed that the mutant accumulated MgPMME under aerobic growth conditions, and they proposed that AcsF may be the aerobic cyclase.
Recently we showed that Crd1 may be involved also in signaling plastid iron status (19). This, coupled with the occurrence of multiple, differentially expressed isoforms, the uneven effect of crd1 alleles on specific Chl proteins, and circadian expression in vascular plants (20), argues for functional characterization of Crd1 in the eukaryotic and plastid context. The tetrapyrrole pathway is of considerable interest, not only because of the unusual chemistry of some of the enzymes but also because of the recent appreciation of its role in nucleus-organelle signaling (21–27).
Here we show that the Arabidopsis homologue of Crd1, which we now call CHL27, is required for the synthesis of Pchlide from MgPMME, consistent with a role in the cyclase reaction. We also show that CHL27 is located at both the envelope and thylakoid membranes of Arabidopsis.
Materials and Methods
Plant Material. Arabidopsis thaliana (L.) Heyn cv. Columbia was used for all the experiments except the localization studies, where cv. Wassilewskija was used. Plants were grown in peat in a controlled environment chamber (Percival AR-60L, Percival, Perry, IA) at a photosynthetic flux of 100–120 μmol of photons m-2·s-1, 20°C, and 70% relative humidity. The photoperiod was 12 h for plants used for transformation and 8 h for plants used for biochemical and physiological analysis to suppress flowering.
Vector Construction and Plant Transformation. A 630-bp fragment containing the C-terminal half of CHL27 was amplified from a full-length cDNA clone by using primers that were based on the cDNA sequence for the CHL27 gene (GenBank accession no. AF236101). The fragment was cloned in antisense orientation between the enhanced cauliflower mosaic virus 35S promoter and 35S terminator in the pPS48 vector (28, 29). Insert orientation was confirmed by DNA sequencing. Subsequently, a fragment containing the enhanced 35S promoter followed by the CHL27 gene in the antisense orientation and the 35S terminator was excised with XbaI and ligated into the binary vector pPZP111 (30). The construct was transformed into the Agrobacterium tumefaciens strain C58 (31). Arabidopsis plants were transformed by the floral dip method (32) by using Silwet L-77 (Lehle Seeds, Round Rock, TX). Control plants were generated via transformation with the vector pPZP111 alone. Seeds harvested from transformed plants were germinated on Murashige and Skoog medium (Sigma) containing 2% sucrose, 50 mg·liter-1 kanamycin sulfate, and 0.8% agar and subjected to selection for kanamycin resistance for 2 weeks. Seedlings then were transplanted to peat. A total of 87 kanamycin-resistant, green seedlings containing the antisense CHL27 construct were transferred. These are referred to as chl27-as plants. Approximately 20 control plants were transplanted also.
ALA Feeding and HPLC Pigment Analysis. Intact leaves were incubated overnight in darkness in 10 mM potassium phosphate, pH 7.0/5 mM MgCl2/10 mM 5-aminolevulinate. For pigment analysis, leaves were homogenized in liquid nitrogen and extracted with acetone/H2O/25% NH4OH (80:20:1) under dim, green light at 4°C. After centrifugation for 15 min at 20,000 × g, 4°C, the supernatant was transferred to a fresh microcentrifuge tube and extracted 3 times with hexane to remove carotenoids and Chls. Pigments were separated by chromatography on a Waters Spherisorb analytical column, ODS1 (250 × 4.6 mm i.d.; 5-μm particle size) by using a Dionex HPLC system. The mobile phase consisted of two solvents: A [0.005% (vol/vol) triethylamine in water] and B (acetonitrile). The pigments were eluted with a linear gradient from 15% A to 100% B over 10 min, followed by isocratic elution with 100% B for 5 min, and a linear gradient of 100% B to 15% A plus 85% B in 5 min. Injection volume was 60 μl, and the flow rate was 1 ml·min-1. The eluate was monitored with a photodiode array detector in the range of 290–595 nm. Pigments were identified and quantified by comparing retention times and absorption spectra with standard pigments: MgP (Frontier Science, Logan, UT), MgPMME, and Pchlide. The concentration of MgPMME and Pchlide was determined by using ε419 = 100 (mM·cm)-1 and ε626 = 30.4 (mM·cm)-1, respectively.
Isolation and Analysis of Total Leaf and Thylakoid Membrane Proteins. Plants with reduced abundance of the CHL27 protein were identified by immunoblotting. Leaf extracts were prepared and immunoblotting performed as described (33). Leaves from 8- to 10-week-old plants were used for isolation of thylakoid membranes (34). Protein concentration was determined by using the 2D Quant kit (Amersham Pharmacia) against BSA as a standard. For analysis of photosynthetic complex abundance, thylakoid membranes were analyzed by using antibodies as indicated in the figure legends. Primary antibodies were detected by using a chemiluminescent detection system (ECL, Amersham Pharmacia). The total Chl and Chl a/b ratio in thylakoids were determined in 80% acetone according to ref. 35.
Localization of CHL27. Arabidopsis chloroplasts and chloroplast subfractions were prepared and probed as described in ref. 4. Thermolysin treatment of spinach chloroplasts was performed as described in ref. 36. Treated (100 or 600 μg of thermolysin per ml) and control (no thermolysin) intact chloroplasts were recovered on a Percoll gradient, and the corresponding envelope fractions were prepared. Fractions enriched in outer- or inner-envelope membranes from spinach chloroplasts were prepared according to ref. 37. Sensitivity of CHL27 to thermolysin was determined by using solubilized envelope proteins. Envelope proteins (0.8 mg of protein per ml) were solubilized with 0.2% (wt/vol) Triton X-100 and treated with the same thermolysin concentrations used for the chloroplasts. Proteins were resolved by SDS/PAGE and transferred to nitrocellulose membrane (pore size, 0.2 μM) at 100 V for 1 h. The primary antibodies were detected by using peroxidase-conjugated AffinityPure goat anti-rabbit IgG (Jackson ImmunoResearch) and ECL reagent (Amersham Pharmacia).
Fluorescence Measurements. Fluorescence emission spectra were recorded at 77 K from 650 to 800 nm by using an excitation wavelength of 435 nm as described (33) using a spectrofluorometer (Photon Technology International, Lawrenceville, NJ). For analysis of pigments, fluorescence emission spectra were recorded at room temperature from 550 to 650 nm by using excitation wavelengths of 420 nm for detecting MgP/MgPMME and 440 nm for detecting Pchlide.
Results
Plants with Reduced CHL27 Display Chlorosis and, When Fed ALA, Accumulate MgPMME. We chose to address the function of chloroplast Crd1/CHL27 via an antisense approach in Arabidopsis, because examination of its genome revealed only one copy of the CHL27 gene. A DNA construct for strong constitutive expression of an antisense CHL27 transcript was introduced into the A. thaliana genome by transformation. Control plants carrying only the vector DNA were generated in parallel. Seeds collected from transformed plants were germinated on Murashige and Skoog medium containing sucrose and kanamycin. When the kanamycin-resistant plants had two to four leaves (≈2 weeks after germination), the seedlings were transferred to soil.
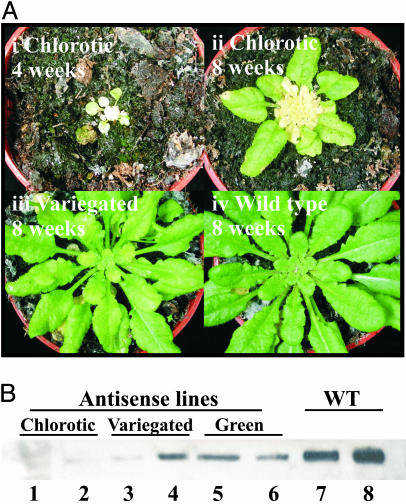
After 2–5 weeks in soil, the following varied phenotypes were observed in the chl27 antisense (chl27-as) plants: (i) Some plants died early after transfer to soil. These plants, which developed green leaves when grown on sucrose-containing plates, developed a small yellow (necrotic) inner rosette with only two to four outer leaves (Fig. 2Ai). (ii) Some plants grew on soil, developing one rosette of green leaves, and then became chlorotic with a yellow inner rosette at a later stage (Fig. 2 Aii). These plants are referred to as “chlorotic.” (iii) Some plants developed more than one rosette of uniformly green leaves before the new leaves showed chlorotic variegations (Fig. 2 Aiii). These plants, which showed significant variation in the degree of chlorosis, are referred to as “variegated” lines. (iv) Some plants were phenotypically normal with uniformly green leaves that, in color and appearance, were indistinguishable from WT plants. These plants are called “green” lines. The green and variegated lines bolted, flowered, and produced fertile seeds. However, all plants were significantly smaller and produced less seeds in comparison to typical WT plants (data not shown).
Fig. 2.
Phenotypes of chl27-as Arabidopsis plants. (Ai) A plant line (chl27-as1.7.5) that shows a severe phenotype early after transfer from sucrose-containing plates to soil. (Aii) A line (chl27-as2.38.10) that shows a severe chlorotic phenotype at a later stage of development. (Aiii) A plant line (chl27-as2.38.4) that displays a variegated phenotype. (Aiv) A WT plant transformed with pPZP111 (vector control). (B) Immunoblot detection of CHL27. Plant material was collected from chlorotic lines (1, chl27-as1.7.5; 2, chl27-as2.22.7), variegated lines (3, chl27-as2.22.10; 4, chl27-as2.22.12), green lines (5, chl27-as2.22.1; 6, chl27-as2.22.6), and WT lines (7 and 8). Proteins were resolved by SDS/PAGE, and the abundance of CHL27 was revealed by immunoblotting. Each lane contains 30 μg of total protein.
Leaves from representative plants of each phenotypic class were analyzed for the abundance of CHL27. A clear correlation between the degree of chlorosis and the abundance of CHL27 was noted (Fig. 2B), consistent with a function in Chl biosynthesis. Chlorotic plants revealed little or no CHL27 (lanes 1 and 2), whereas variegated plants accumulated intermediate amounts (lanes 3 and 4). Interestingly, the green plants, containing the chl27-as construct but exhibiting a phenotype indistinguishable from WT plants also had reduced amounts of CHL27 (lanes 5 and 6) when compared to WT plants containing just the empty transformation vector (lanes 7 and 8), suggesting that the usual amount of CHL27 in WT plants is not normally rate-limiting for Chl protein accumulation.
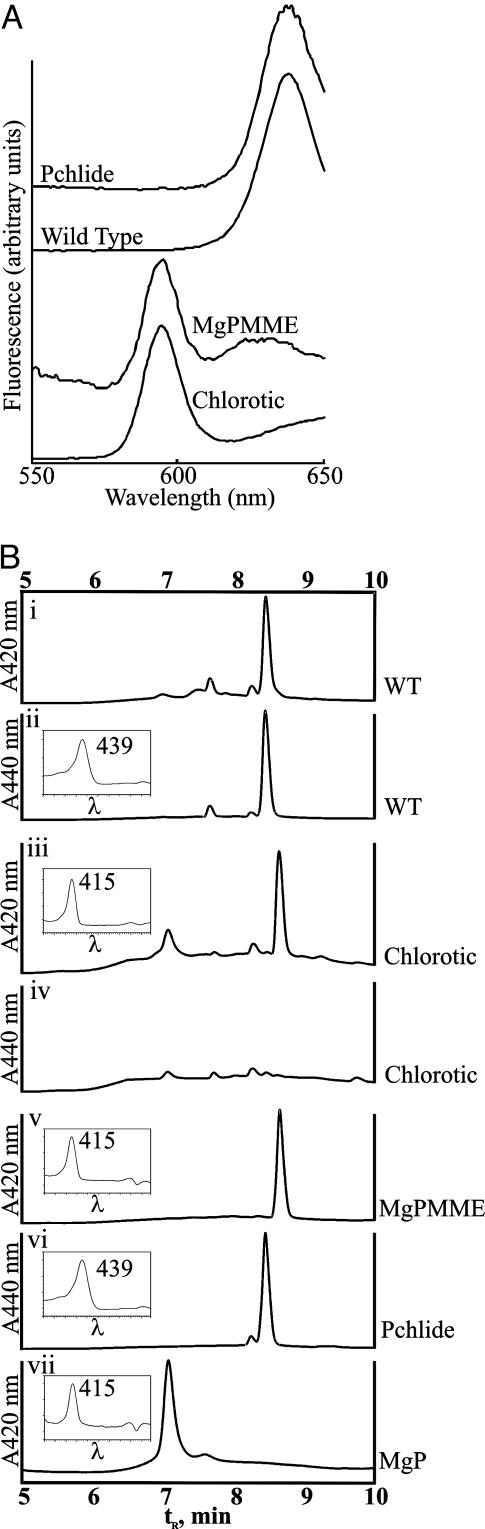
Mutants in Chl biosynthesis do not normally accumulate pathway intermediates because tetrapyrrole biosynthesis is tightly controlled (38). A key regulatory step is the synthesis of ALA. If this step is bypassed (for example, by feeding plants with ALA), precursors will accumulate. When ALA-fed angiosperms are kept in the dark, large amounts of Pchlide accumulate (39) because of the light dependency of Pchlide oxidoreductase. This experimental tool was exploited previously for classical genetic analysis of the tetrapyrrole pathway (2). Therefore, chl27-as plants showing either a severe chlorotic phenotype or a variegated phenotype and control transformed plants were fed ALA for 18 h in the dark to ascertain the location of the blockage in Chl biosynthesis. The accumulated pigments then were extracted and analyzed by using room-temperature fluorescence emission spectroscopy (Fig. 3A). The spectrum of the extracts revealed that (i) Mg-protoporphyrin or MgP accumulated in the chlorotic chl27-as plants, and (ii) this is correlated with a concurrent decrease in the accumulation of Pchlide, which is the major pigment observed in the WT plants.
Fig. 3.
Analysis of porphyrin pigments accumulating in chl27-as lines after feeding with ALA in the dark. (A) Room-temperature fluorescence emission spectra of acetone extracts of chlorotic (yellow) leaves from an antisense line (chl27-as2.38.10) and green leaves from WT plants after overnight incubation with ALA compared to purified standards. (B) HPLC traces of the extracted pigment mixtures relative to known standards. The eluate was monitored at 420 nm for optimal detection of MgPMME and MgP (i, iii, v, and vii) or 440 nm for detection of Pchlide (ii, iv, and vi). (Inset) Traces show spectral absorbance of the major eluate peak, measured from 350 to 595 nm, with the wavelength of maximum absorbance indicated.
The pigments in the extracts were separated chromatographically for their conclusive identification (Fig. 3B). The compound in the extract from the chl27-as plants had the identical retention time and absorption spectrum as standard MgPMME (Fig. 3 Biii and Bv). The major compound in extracts of WT plants was identified as Pchlide, as expected (Fig. 3 Bi and Bii). Analysis of a number of different antisense lines revealed that the extent of accumulation of MgPMME was inversely correlated with the accumulation of Pchlide and the abundance of CHL27. Extracts from the chlorotic antisense plants had no Pchlide and an abundance of MgMME (Fig. 3 Biii and Biv), whereas variegated plants accumulated both MgPMME and Pchlide (data not shown). The amounts of accumulated MgPMME and Pchlide were quantified from the chromatograms (Table 1) and clearly show that significant amounts of MgPMME accumulate in ALA-fed chl27-as plants, whereas only small or trace amounts of Pchlide are detected. We therefore conclude that the chl27-as lines are blocked at the cyclase step.
Table 1. Accumulation of MgPMME and Pchlide in leaves of Chl27-as and WT plants fed with ALA.
| ALA | n | MgPMME, nmol·(g FW)–1 | Pchlide, nmol·(g FW)–1 | |
|---|---|---|---|---|
| WT | – | 3 | 0 | 0 |
| WT | + | 4 | 0 | 405.6 ± 91.7 |
| Chl27-as-1.7 | – | 1 | Trace | 0 |
| Chl27-as-1.7 | + | 6 | 102.2 ± 14.5 | 2.2 ± 1.2 |
| Chl27-as-2.38 | – | 3 | 0 | 0 |
| Chl27-as-2.38 | + | 3 | 78.0 ± 21.6 | trace |
Analysis of MgPMME and Pchlide accumulated in selected Chl27-as and WT plants after incubation with ALA (+) or buffer only (–) for 18 hours. Porphyrins were extracted in acetone and subjected to HPLC analysis with absorbance detection as described in Materials and Methods. Data are the average of extracts from several chlorotic plants ± SE. For both pigments, the detection limit was found to be 30 nM, corresponding to 1.8 pmol per 60-μl injection. FW, fresh weight.
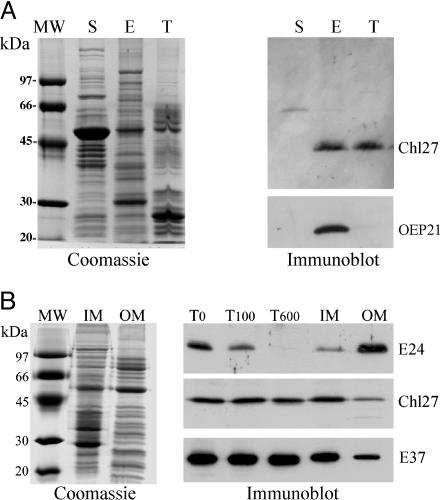
CHL27 Is Localized to the Chloroplast Inner-Envelope and Thylakoid Membranes. Because the preceding enzyme in the pathway is localized to both the envelope and thylakoid membranes (4), the location of CHL27 is clearly of interest. Previously we localized Crd1 to the chloroplast membranes of fractionated pea chloroplasts, but the distinction between envelope and thylakoid membranes was not made (40). CHL27 sublocation within the chloroplast was therefore queried after fractionation of isolated Arabidopsis chloroplasts into stroma, envelope, and thylakoid membranes. Coomassie-stained gels of the isolated fractions (Fig. 4A), Chl concentration, and immunoblot detection of envelope marker OEP21 indicated that the fractions were fully separated and that the thylakoid and envelope preparations had very little cross contamination. Immunological detection of CHL27 revealed its presence in both thylakoid and envelope membranes (Fig. 4A) similar to the localization of MgP methyl transferase (4). Only insignificant amounts of CHL27 were detected with the stromal fraction. Based on the intensity of the signal, we conclude that CHL27 is equally distributed on a membrane-weight basis between the envelope and thylakoid membranes.
Fig. 4.
Suborganellar localization of CHL27. (A) Analysis of Arabidopsis chloroplast fractions. E, envelope; T, thylakoid; S, stroma. (Left) Fractions analyzed after SDS/PAGE by Coomassie blue staining. (Right) Immunoblot detection of CHL27 and OEP21. (B) Distribution of CHL27 between envelope fractions from spinach chloroplasts. Envelope membrane fractions were enriched in inner-envelope membrane (IM) or outer-envelope membrane (OM). T0,T100, and T600, envelope fractions were obtained from chloroplasts treated without thermolysin or with 100 or 600 μg/ml thermolysin, respectively. (Left) Coomassie blue staining. (Right) Immunodetection of CHL27, E37, an inner-envelope membrane protein, and E24, an outer-envelope membrane protein. Each lane was loaded with 20 μg of protein. The membrane was incubated with anti-CHL27 (1:1,000), anti-E37 (1:10,000), and anti-E24 (1:5,000).
To distinguish CHL27 localization between the inner- vs. outer-envelope membranes, we compared the sensitivity of CHL27 to thermolysin treatment relative to E24 and E37. Thermolysin treatment degrades outer-envelope proteins that are exposed to cleavage while inner-envelope proteins remain intact. E24, an outer-membrane protein (41), is degraded rapidly by low concentrations of thermolysin (Fig. 4B), whereas E37, an inner-membrane protein (42), remains unaffected. CHL27 abundance is unchanged with increasing thermolysin concentrations, consistent with CHL27 localization to the inner-envelope membrane. CHL27 is sensitive to thermolysin if chloroplasts are solubilized in detergent (data not shown). The localization of CHL27 to the envelope inner membrane was confirmed immunologically by using isolated inner and outer chloroplast envelope fractions (Fig. 4B). Coomassie staining of resolved membrane proteins reveals the separation of the characteristic marker proteins of the inner and outer membranes, and separation is confirmed by the immunological detection of E37 and E24 in their expected locations. CHL27 is clearly present in greatest abundance within the inner-membrane fraction. The protein is also detected in the outer membrane but in quantities comparable to those observed for inner-membrane marker protein E37. We therefore conclude that in the chloroplast, CHL27 is present in thylakoids and in the inner-envelope membrane.
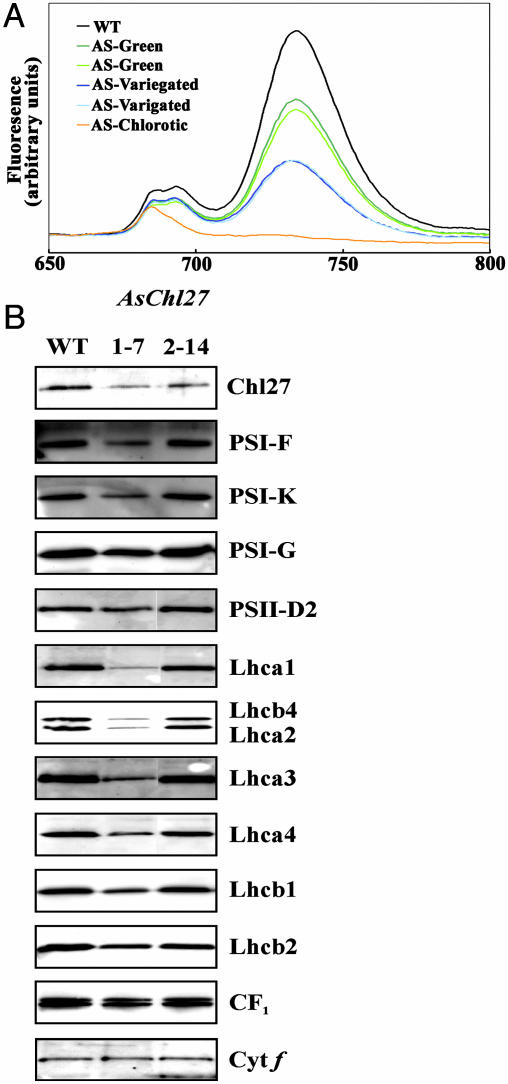
Decreased Chl-Binding Proteins in Plants with Reduced CHL27. To analyze the effect of restricted Chl availability on the two photosystems and their peripheral antennas, fluorescence emission at 77 K was monitored on leaves of WT and chl27-as plants displaying various degrees of Chl deficiency (Fig. 5A). A reduction in the far-red fluorescence emission at 734 nm was observed in all chl27-as plants assayed, when compared to WT plants. This reduction in fluorescence includes the category of plants that displayed no discernible visual phenotype but contained less CHL27 when assayed immunologically. Fluorescence emission at 734 nm is attributed to photosystem (PS)I and light-harvesting complex (LHC)I abundance (43). The degree of the decline of 734-nm fluorescence emission observed is proportional to the extent of chlorosis. Mutant plants with no visual phenotype displayed a decrease in 734-nm fluorescence to approximately half that of WT plants. Plants that contain extremely low levels of CHL27, and therefore are the most acutely chlorotic, have almost no fluorescence emission at 734 nm, consistent with the severe depletion of the LHCI and PSI core proteins. Alterations of the fluorescence at 685 and 695 nm, attributed to PSII (43), are far less pronounced than are alterations of the 734-nm peak, with no detectable decrease at 685 and 695 nm for a range of plants displaying large fluorescence decreases at 734 nm. The preferential loss of LHCI and PSI core proteins is consistent with the phenotypes of previously characterized cyanobacterial mutants that are blocked in the light-dependent Pchlide reductase (44). The most chlorotic plants do show decreased fluorescence emission at 685 and 695 nm but retain a residual fluorescence at ≈685 nm.
Fig. 5.
Reduced Chl protein abundance in chl27-as plants. (A) Fluorescence emissions (77 K) measured from 650 to 800 nm from WT (black), antisense lines showing no visible phenotype [green (chl27-as2.22.1) and light green (chl27-as2.22.6)], variegated lines [blue (chl27-as2.22.10) and light blue (chl27-as2.22.12)], and chlorotic [orange (chl27-as1.7.5)] plants. (B) Immunoblot analysis of CHL27-deficient and WT plants. Thylakoid membranes isolated from lines 2–14 (mild variegation) and 1–7 (more severe variegation) were analyzed for the abundance of the indicated thylakoid membrane protein. Each lane was loaded with 3 μg of protein.
Direct biochemical analysis of individual Chl protein complexes was undertaken by immunoblotting. Thylakoid membranes isolated from two plant lines displaying various degrees of the variegated phenotype were examined for the abundance of protein subunits from PSI, PSII, and their associated LHCs (Fig. 5B). Compared to WT Arabidopsis thylakoid membrane proteins, the antisense lines accumulated less CHL27 as expected. Correlated with the reduction of CHL27 is a widespread loss of Chl-binding proteins. Loss of the peripheral light-harvesting antenna proteins is particularly acute, as expected from the increased Chl a/b ratio. Thylakoid proteins CF1 and cytochrome f are not reduced to the same extent, revealing that the phenotype is specific for Chl containing thylakoid membrane proteins.
Discussion
CHL27 Is Involved in the Aerobic Cyclase Reaction. The phenotype of chl27-as plants is consistent with a role for CHL27 in the conversion of MgPMME to Pchlide. Specifically, we showed that chlorotic plants with reduced CHL27 cannot accumulate Pchlide when fed ALA in the dark as do WT plants, indicating a function for CHL27 in Chl biosynthesis (Fig. 3). The plants do, however, accumulate MgPMME, which, based on the phenotype of known cyclase mutants, viridis-k and xantha-l in barley (2, 12), localizes the site of CHL27 action to the cyclase. As expected, reduced Chl production results in reduced Chl protein accumulation as assessed by immunoblot analyses and low-temperature fluorescence emission spectra (Fig. 5). The strength of the phenotype is proportional to the decrease in CHL27 accumulation in individual lines; leaves containing the lowest amounts of CHL27 show the most pronounced chlorotic phenotype.
The data suggest, therefore, that CHL27 is involved in Chl biosynthesis at the step of the cyclase reaction. Formally, our data do not allow us to distinguish between a role for CHL27 in the chemistry of the reaction vs. regulation. We argue in favor of the former, because the predicted iron-containing active site and conserved cysteines of CHL27 (16) match nicely with the biochemical description of the cyclase enzyme, its cofactor and substrate requirements, and the proposed reaction sequence.
The formation of the fifth isocyclic ring in Chl is achieved via three sequential reactions (see Fig. 1 and the Introduction), of which the first and third require molecular oxygen and pyridine nucleotide cofactors (8) and are compatible with diiron chemistry. The initial hydroxylation would be analogous to the reactions of prototypical diiron enzymes, methane monooxygenase, and the related toluene monooxygenase and phenol hydroxylase (45–47). The third reaction, cyclization of the keto intermediate, is also an oxygen-dependent oxidation and could occur through radical chemistry via a diiron active site. CHL27 could be involved in one of the two O2-dependent steps or in all three reactions.
Dual Localization. Previously, the enzyme preceding the cyclase was shown to localize to both the thylakoid and envelope membranes of Arabidopsis chloroplasts (4), and interestingly, the same is true for CHL27 (Fig. 4), which supports a model where Chl is synthesized in two membrane locations. The thylakoid and envelope preparations show very little cross contamination; in particular, the envelope contamination of thylakoid fraction is very low, and on a protein basis, CHL27 is about equally distributed between the envelope and thylakoid membranes. Within the envelope membranes, CHL27 resides in the inner membrane based both on biochemical fractionation and resistance to thermolysin (Fig. 4). Because the methyltransferase and the candidate cyclase are each encoded by only one gene in Arabidopsis, their dual localization raises an interesting cell biology problem concerning the mechanism of suborganellar distribution of a single gene product.
In Chlamydomonas, the two isoforms of CHL27, originally named Crd1 and Cth1, are differentially expressed based on copper nutrition status (16, 40). We concluded that the two forms had overlapping but nonidentical functions in the biosynthesis of Chl proteins based on (i) the specific loss of PSI and LHCI proteins in copper-deficient crd1 mutants and (ii) the inability of Cth1 to fully compensate for the loss of Crd1 in both copper-replete crd1 cells or in crd1sct1 strains that display copper-independent “constitutive” expression of Cth1. Taken together with the dual localization of CHL27 in Arabidopsis, we suggest that there may be specific sites of Chl synthesis within the chloroplast and these sites may contribute Chl differentially to individual Chl proteins, perhaps depending on the developmental state of the chloroplast or environmental factors. This view is compatible with the model for different functions of some Pchlide oxidoreductase isoforms and overlapping functions of others (48, 49).
Acknowledgments
We thank Dr. Jacques Joyard and Dr. Ken Cline for stimulating discussions. MgPMME was a gift from Dr. Simon Gough (Lund University, Lund, Sweden) and Dr. David Bollivar (Illinois Wesleyan University, Bloomington), and Pchlide was a gift from Dr. Derren Heyes (University of Sheffield, Sheffield, U.K.). This work was supported by the National Institutes of Health (Grant GM42143) and the U.S. Department of Agriculture (National Research Initiative Competitive Grants Program 2002-35318-12673). S.T. was supported in part by a University of California Toxic Substances Research and Teaching Program's University of California Los Angeles/University of California, Riverside/Los Alamos National Laboratory Lead Campus Grant. Financial support from the Danish National Research Foundation (to P.E.J. and H.V.S.) is gratefully acknowledged.
Abbreviations: Chl, chlorophyll; ALA, δ-aminolevulinic acid; MgP, Mg-protoporphyrin IX; MgPMME, MgP monomethyl ester; Pchlide, protochlorophyllide; PS, photosystem; LHC, light-harvesting complex.
References
- 1.Suzuki, J. Y., Bollivar, D. W. & Bauer, C. E. (1997) Annu. Rev. Genet. 31, 61-89. [DOI] [PubMed] [Google Scholar]
- 2.von Wettstein, D., Gough, S. & Kannangara, C. G. (1995) Plant Cell 7, 1039-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beale, S. I. (1999) Photosynth. Res. 60, 43-73. [Google Scholar]
- 4.Block, M. A., Tewari, A. K., Albrieux, C., Maréchal, E. & Joyard, J. (2002) Eur. J. Biochem. 269, 240-248. [DOI] [PubMed] [Google Scholar]
- 5.Porra, R. J., Urzinger, M., Winkler, J., Bubenzer, C. & Scheer, H. (1998) Eur. J. Biochem. 257, 185-191. [DOI] [PubMed] [Google Scholar]
- 6.Yang, Z. M. & Bauer, C. E. (1990) J. Bacteriol. 172, 5001-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porra, R. J., Schäfer, W., Gad'on, N., Katheder, I., Drews, G. & Scheer, H. (1996) Eur. J. Biochem. 239, 85-92. [DOI] [PubMed] [Google Scholar]
- 8.Wong, Y.-S., Castelfranco, P. A., Goff, D. A. & Smith, K. M. (1985) Plant Physiol. 79, 725-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker, C. J., Mansfield, K. E., Rezzano, I. N., Hanamoto, C. M., Smith, K. M. & Castelfranco, P. A. (1988) Biochem. J. 255, 685-692. [PMC free article] [PubMed] [Google Scholar]
- 10.Nasrulhaq-Boyce, A., Griffiths, W. T. & Jones, O. T. (1987) Biochem. J. 243, 23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiller, S. C., Castelfranco, A. M. & Castelfranco, P. A. (1982) Plant Physiol. 69, 107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Wettstein, D., Kahn, A., Nielsen, O. F. & Gough, S. P. (1974) Science 184, 800-802. [DOI] [PubMed] [Google Scholar]
- 13.Wong, Y.-S. & Castelfranco, P. A. (1984) Plant Physiol. 75, 658-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker, C. J., Castelfranco, P. A. & Whyte, B. J. (1991) Biochem. J. 276, 691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollivar, D. W. & Beale, S. I. (1996) Plant Physiol. 112, 105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moseley, J., Quinn, J., Eriksson, M. & Merchant, S. (2000) EMBO J. 19, 2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond, J., Zhaxybayeva, O., Gogarten, J. P., Gerdes, S. Y. & Blankenship, R. E. (2002) Science 22, 1616-1620. [DOI] [PubMed] [Google Scholar]
- 18.Pinta, V., Picaud, M., Reiss-Husson, F. & Astier, C. (2002) J. Bacteriol. 184, 746-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moseley, J. L., Allinger, T., Herzog, S., Hoerth, P., Wehinger, E., Merchant, S. & Hippler, M. (2002) EMBO J. 21, 6709-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng, C. C., Porat, R., Lu, P. & O'Neill, S. D. (1998) Plant Physiol. 116, 27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kropat, J., Oster, U., Rüdiger, W. & Beck, C. F. (2000) Plant J. 24, 523-531. [DOI] [PubMed] [Google Scholar]
- 22.Papenbrock, J. & Grimm, B. (2001) Planta 213, 667-681. [DOI] [PubMed] [Google Scholar]
- 23.Strand, A., Asami, T., Alonso, J., Ecker, J. R. & Chory, J. (2003) Nature 421, 79-83. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, L. & Hach, A. (1999) Cell. Mol. Life Sci. 56, 415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surpin, M., Larkin, R. M. & Chory, M. (2002) Plant Cell 14, 327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A. & Chory, J. (2001) Proc. Natl. Acad. Sci. USA 98, 2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan, J. A. & Gray, J. C. (1999) Plant Cell. 11, 901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odell, J. T., Nagy, F. & Chua, N.-H. (1985) Nature 313, 810-812. [DOI] [PubMed] [Google Scholar]
- 29.Kay, R., Shan, A., Daly, M. & McPherson, J. (1987) Science 236, 1299-1302. [DOI] [PubMed] [Google Scholar]
- 30.Hajdukiewicz, P., Svab, Z. & Maliga, P. (1994) Plant. Mol. Biol. 25, 989-994. [DOI] [PubMed] [Google Scholar]
- 31.Zambryski, P., Joos, H., Genetello, C., Leemans, J., Van Montagu, M. & Schell, J. (1983) EMBO J. 2, 2143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 33.Jensen, P. E., Gilpin, M., Knoetzel, J. & Scheller, H. V. (2000) J. Biol. Chem. 275, 24701-24708. [DOI] [PubMed] [Google Scholar]
- 34.Haldrup, A., Naver, H. & Scheller, H. V. (1999) Plant J. 17, 689-698. [DOI] [PubMed] [Google Scholar]
- 35.Lichtenthaler, H. K. (1987) Methods Enzymol. 148, 350-382. [Google Scholar]
- 36.Joyard, J., Block, M., Pineau, B., Albrieux, C. & Douce, R. (1990) J. Biol. Chem. 265, 21820-21827. [PubMed] [Google Scholar]
- 37.Block, M. A., Dorne, A. J., Joyard, J. & Douce, R. (1983) J. Biol. Chem. 258, 13281-13286. [PubMed] [Google Scholar]
- 38.Reinbothe, S. & Reinbothe, C. (1996) Plant Physiol. 111, 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granick, S. (1959) Plant Physiol. 34, S-xviii [Google Scholar]
- 40.Moseley, J. L., Page, M. D., Alder, N. P., Eriksson, M., Quinn, J., Soto, F., Theg, S. M., Hippler, M. & Merchant, S. (2002) Plant Cell 14, 673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joyard, J., Billecocq, A., Bartlett, S. G., Block, M. A., Chua, N. H. & Douce, R. (1983) J. Biol. Chem. 25, 10000-10006. [PubMed] [Google Scholar]
- 42.Teyssier, E., Block, M. A., Douce, R. & Joyard, J. (1996) Plant J. 10, 903-912. [DOI] [PubMed] [Google Scholar]
- 43.Krause, G. H. & Weis, E. (1991) Rev. Plant Physiol. Plant Mol. Biol. 42, 313-349. [Google Scholar]
- 44.Kada, S., Koike, H., Satoh, K., Hase, T. & Fujita, Y. (2003) Plant Mol. Biol. 51, 225-235. [DOI] [PubMed] [Google Scholar]
- 45.Nordlund, P. & Eklund, H. (1995) Curr. Opin. Struct. Biol. 5, 758-766. [DOI] [PubMed] [Google Scholar]
- 46.Wallar, B. J. & Lipscomb, J. D. (1996) Chem. Rev. (Washington, D.C.) 96, 2625-2657. [DOI] [PubMed] [Google Scholar]
- 47.Merkx, M., Kopp, D. A., Sazinsky, M. H., Blazyk, J. L., Müller, J. & Lippard, S. J. (2001) Angew. Chem. Int. Ed. Engl. 40, 2782-2807. [PubMed] [Google Scholar]
- 48.Armstrong, G. A., Runge, S., Frick, G., Sperling, U. & Apel, K. (1995) Plant Physiol. 108, 1505-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frick, G., Su, Q., Apel, K. & Armstrong, G. A. (2003) Plant J. 35, 141-153. [DOI] [PubMed] [Google Scholar]