Abstract
Among commonly applied molecular markers, simple sequence repeats (SSRs, or microsatellites) possess advantages such as a high level of polymorphism and codominant pattern of inheritance at individual loci. To facilitate systematic and rapid genetic mapping in soybean, we designed a genotyping panel comprised 304 SSR markers selected for allelic diversity and chromosomal location so as to provide wide coverage. Most primer pairs for the markers in the panel were redesigned to yield amplicons of 80–600 bp in multiplex polymerase chain reaction (PCR) and fluorescence-based sequencer analysis, and they were labelled with one of four different fluorescent dyes. Multiplex PCR with sets of six to eight primer pairs per reaction generated allelic data for 283 of the 304 SSR loci in three different mapping populations, with the loci mapping to the same positions as previously determined. Four SSRs on each chromosome were analysed for allelic diversity in 87 diverse soybean germplasms with four-plex PCR. These 80 loci showed an average allele number and polymorphic information content value of 14.8 and 0.78, respectively. The high level of polymorphism, ease of analysis, and high accuracy of the SSR genotyping panel should render it widely applicable to soybean genetics and breeding.
Keywords: SSR marker, fluorescent primer, multiplex PCR, polymorphic information content, high-throughput genotyping
1. Introduction
A comprehensive genetic linkage map is fundamental to modern plant genetics and breeding because it allows the identification and utilization of agronomic trait loci, such as qualitative and quantitative trait loci, as well as the evaluation of genetic diversity and genomic structure of genetic resources. It also serves as a scaffold for construction of a physical map. In the case of soybean [Glycine max (L.) Merrill], a grain legume of global importance, many useful agronomic trait loci associated with growth, product quality, tolerance to biotic and abiotic stresses, and other characteristics have been identified in genetic resources and deposited in public databases such as Soybase (http://soybase.org). The first soybean linkage map was constructed on the basis of phenotypic traits,1 but it did not contain sufficient information for application to the above-mentioned purposes. Since the 1990s, various types of molecular markers, including restriction fragment length polymorphism, random amplification of polymorphic DNA, amplified fragment length polymorphism, simple sequence repeat (SSR, or microsatellite), and single nucleotide polymorphism (SNP) markers, have been developed and applied to soybean, and enriched marker information has enabled genetic analyses of qualitative and quantitative traits.2–4
Among such molecular markers, SSRs in particular have contributed to the construction of a genome-wide linkage map for soybean with a converged linkage group (LG) number equal to the chromosome number. As previously described,5 these microsatellite markers have several advantages, including (i) a codominant manner of inheritance at each locus, (ii) a high level of polymorphism in the form of multiple alleles, (iii) a non-biased distribution in the genome, and (iv) ease of detection of polymorphism by the polymerase chain reaction (PCR) and subsequent electrophoresis.6–9 It is thus possible to detect allelic differences at highly polymorphic loci among genetic resources as well as among many segregating populations derived from the hybridization of any given genotypes.10–12 Furthermore, given that most PCR primer pairs for SSR markers were designed to yield a single amplification product for each allele in spite of the complex chromosomal structure of soybean,13–15 each SSR marker localizes a definite site in the genome, unlike other molecular markers. These features also allow the simultaneous detection of multiple SSR loci with the use of multiplex PCR analysis.11 In addition, high-throughput and repetitive genotyping can be performed by semiautomated methods with a combination of fluorescently labelled SSR markers.11,12,16 This strategy has been successfully adapted and improved for analysis of genetic diversity and high-throughput mapping in various species, including human,17,18 mouse,19 rat,20 rice,21,22 and sunflower.23 However, in spite of the public availability of many SSR markers for soybean,24 no comprehensive SSR genotyping panel has yet been developed for whole-genome coverage. A high-throughput genotyping system for soybean that is based on an SNP array capable of high multiplexing and which discriminates up to 384 or 1536 mapped SNPs in one reaction has been described.25,26 Although the high multiplexing capacity and continued improvement of the SNP array may make this a standard technique in the foreseeable future, the quantity and quality of SNP loci in soybean are still not sufficient for application of this system to many genotypes. Moreover, the present panel system for SNPs is not sufficiently flexible for modification of marker selection and is not a cost-effective solution.
In the latest extensive molecular linkage map, nearly 2000 SSR markers were mapped to the 20 consensus soybean LGs without any large gaps with the exception of one region in LG C1 (chromosome 4).5 In addition, allelic variation at a large number of SSR loci was examined in 23 different soybean genotypes. The availability of this large amount of information prompted us to develop a whole-genome SSR panel for high-throughput genotyping in soybean. We selected SSR markers on the basis of their polymorphism and chromosomal location, and then redesigned them for adaptation to multiplex PCR. The resulting SSR panel system was applied to construct linkage maps of three segregating populations with different genetic backgrounds, and the positions of the redesigned SSR markers were confirmed.
2. Materials and methods
2.1. Design of a whole-genome SSR panel
On the basis of their positions and polymorphism information content (PIC) values previously described,5,27 we initially selected 322 SSR markers from the mapped SSR loci. The selected markers in principle had a PIC value of >0.5 and were separated from each other by a distance of <20 cM. They consisted of 263 SSR markers developed by USDA-ARS (United States Department of Agriculture, Agriculture Research Service),27,28 15 SSR markers developed by Chiba University,29 and 44 expressed sequence tag (EST)-derived SSR markers developed by Kazusa DNA Research Institute.30 For adaptation to multiplex PCR, most of the primer pairs for the SSR markers developed by USDA were redesigned with the use of the Primer 3 program on the basis of the following parameters: a melting temperature of 50°–60°C (optimum, 55°C); a product size of 80–600 bp; and all other items set to default.31 One of the four different fluorophores (6-FAM, VIC, NED, or PET) was attached to each forward primer, and a 5′ tail was added to the reverse primer in order to promote full adenylation (Applied Biosystems, Foster City, CA, USA). Each SSR marker was arranged to eliminate overlap with other SSR markers labelled with the same fluorophore in each multiplex set. Details of the markers and primers are provided in Supplementary Table S1.
2.2. PCR conditions and SSR fragment detection
PCR was performed in a reaction mixture of 5.5 µl containing 50 nM of each primer, 5 ng of total genomic DNA, and 2.5 µl of 2× Qiagen Multiplex PCR Master Mix (Qiagen, Hilden, Germany) and with the use of a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems). Total plant DNA was extracted from 10 mg of seed powder with the use of an automated purification system (BioSprint 96 DNA Plant Kit, Qiagen). The amplification protocol comprised an initial denaturation for 15 min at 95°C; 35 cycles of denaturation for 30 s at 95°C, annealing for 90 s at 50°C, and extension for 90 s at 60°C; and a final extension for 30 min at 60°C. A portion (1 µl) of the amplification products was then mixed with 0.3 µl of GeneScan 600 LIZ Size Standard (Applied Biosystems) and 10 µl of Hi-Di Formamide (Applied Biosystems), and the resulting mixture was heated for 3 min at 95°C and then cooled rapidly to 4°C in the thermal cycler. Sizing of SSR fragments was performed with a 3730 DNA Analyzer equipped with POP-7 Polymer and a 36-cm Capillary Array (Applied Biosystems), and allele calling and binning were performed with GeneMapper software v4.0 (Applied Biosystems).
2.3. SSR mapping
Three mapping populations were used to position fluorescently labelled SSR markers. (i) The JI population, which consists of 94 F2 individuals originating from a cross between the Japanese wild accession JP-36121 and the Japanese landrace Ibarakimame 7. (ii) The OA population, which is derived from a cross between the Japanese cultivar Osuzu and the USA cultivar Athow and comprises 225 F6 recombinant inbred lines (RILs). (iii) The IF population, which consists of 143 F7 RILs derived from a cross between the Japanese landrace Ippon_sangoh and a leading Japanese cultivar, Fukuyutaka. On the basis of allelic information for the SSR markers of the whole-genome SSR panel, individual linkage maps of the three populations were constructed with the use of MAPMAKER/EXP 3.0b.32 Recombination values were converted to genetic distance (cM) with the use of the Kosambi mapping function.33 Each map of the 20 LGs was visualized graphically with MapChart.34 The nomenclature of LGs was as described previously,27 based on the common markers mapped.
2.4. Diversity analysis
Four SSR markers labelled with different fluorophores were chosen for even distribution on each chromosome on the basis of previous results.12 Allelic diversity for the total of 80 selected markers was assessed among 87 soybean accessions for which diversity has been previously characterized on the basis of analysis by 3% agarose gel electrophoresis.12 The 87 diverse genotypes consisted of 37 modern cultivars and 15 landraces collected from various regions in Japan, 27 cultivars derived from countries other than Japan (mostly those in East Asia and the USA), and 8 wild accessions. The PIC value for each SSR marker locus i was calculated according to the formula previously described:2
where Pij is the frequency of the jth allele of the ith SSR locus and the summation extends over n alleles. Allele number and PIC value determined with the present system were compared with those determined in the previous study.12
3. Results and discussion
3.1. Detail of the designed whole-genome SSR panel
A whole-genome SSR panel was designed predominantly on the basis of SSR markers developed by USDA-ARS, since these markers have been well documented.27 The markers were selected according to their map position and polymorphic information.5 However, given that the USDA markers alone did not cover the entire soybean genome with an even distribution, we selected additional SSR markers that were in the public domain. The selected markers were designed according to different strategies and could not be amplified under the same conditions.28–30 In particular, primer pairs for the USDA markers were characterized by a relatively low annealing temperature (∼47°C) and the presence of a GCG-clamp near the 5′ end of each primer. Most (255 out of 263) of the primer pairs for the USDA SSR markers were therefore redesigned to enhance their suitability for multiplex PCR analysis and to yield PCR products within the detection range of the DNA analyser. All of the 322 fluorescent SSR markers initially constituting the whole-genome SSR panel were individually assessed with the six parents of the three mapping populations as well as with two standard cultivars, Enrei and Williams 82, that have been used for genomic studies in Japan and the USA, respectively. Eighteen SSR primer pairs yielded no amplicons by singleplex PCR with the eight test genotypes (Supplementary Table S1), likely because of the primer modification for multiplex PCR and fluorescence labelling. The remaining 304 primer pairs, which generated unambiguous and stable PCR amplicons, were therefore selected and assembled into 41 multiplex sets of six to eight markers each (Supplementary Table S1). The SSR primer pairs in each multiplex set were assigned one of the four different fluorescent labels (6-FAM, VIC, NED, or PET) in such a manner that no two markers with the same fluorescent dye showed overlap in allele size range. The 304 SSR markers were estimated to cover 2306 cM, or 95%, of the previous integrated linkage map,5 and were separated by an average interval of 7.6 cM (Table 1). Each chromosome (Chr) was covered with 9 (Chr 16) to 21 (Chr 7) SSR makers. There remained a large gap of 35.3 cM between Sat_235 and GMES1325 on Chr 4, but no public SSR markers have been described for this region.5 A gap of 33.9 cM existed between GMES2057 and GMES1693 on Chr 9, because the selected marker Satt539 gave no amplicons. An additional eight regions on different chromosomes showed a gap of >20 cM between proximal markers (Supplementary Table S1). No confident SSR markers were selected in these regions. The longest and shortest average distances between markers were on Chr 4 (9.6 cM) and Chr 7 (5.9 cM), respectively, indicating that the SSR panel was capable of genotyping with a relatively uniform marker density over the entire soybean genome, with only a few exceptions.
Table 1.
Assignment of SSR markers designed for the whole-genome SSR panel system
| Chromosome | Linkage group | No. of SSR marker loci | Coveragea (cM) | Marker densityb (cM) |
|---|---|---|---|---|
| 1 | D1a | 14 | 113 | 8.1 |
| 2 | D1b | 16 | 138 | 8.6 |
| 3 | N | 15 | 106 | 7.1 |
| 4 | C1 | 12 | 116 | 9.6 |
| 5 | A1 | 14 | 122 | 8.7 |
| 6 | C2 | 19 | 140 | 7.4 |
| 7 | M | 21 | 124 | 5.9 |
| 8 | A2 | 20 | 155 | 7.8 |
| 9 | K | 13 | 111 | 8.5 |
| 10 | O | 15 | 142 | 9.5 |
| 11 | B1 | 18 | 128 | 7.1 |
| 12 | H | 15 | 106 | 7.0 |
| 13 | F | 15 | 122 | 8.1 |
| 14 | B2 | 12 | 97 | 8.1 |
| 15 | E | 15 | 103 | 6.8 |
| 16 | J | 9 | 66 | 7.3 |
| 17 | D2 | 15 | 96 | 6.4 |
| 18 | G | 14 | 120 | 8.5 |
| 19 | L | 17 | 102 | 6.0 |
| 20 | I | 15 | 100 | 6.7 |
| Total | 304 | 2306 | ||
| Average | 15.2 | 115.3 | 7.6 |
aThe coverage (cM) of the SSR marker loci in each LG is based on information for the previous integrated linkage map.5
bCoverage/marker number.
3.2. Genetic mapping of SSR loci in three mapping populations
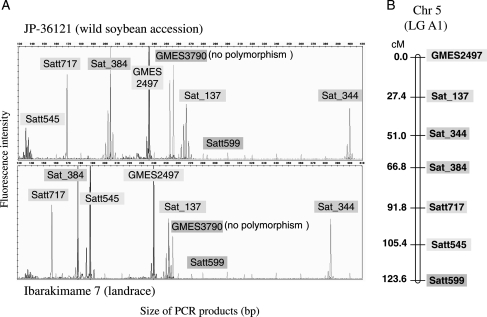
Linkage analyses were performed with the three mapping populations (JI, OA, and IF) to assess the efficacy of the whole-genome SSR panel. At the same time, we verified whether the fluorescent SSR markers, including those that had been redesigned, were located at the same positions as those originally determined. Among the three populations, the JI population, comprising 94 F2 individuals, was assumed to be diverse at many SSR loci because it was developed from a cross between a wild soybean (JP-36121) and a Japanese landrace (Ibarakimame 7).35 For each multiplex set, the allele products for each SSR locus were readily distinguished from each other as well as from those of the other loci represented (Fig. 1A). One multiplex set built a rough map of Chr 5 (Fig. 1B), whereas the two multiplex sets for Chr 5 made up a linkage frame spanning 142.1 cM with 11 polymorphic markers (Fig. 2). Given that the other three markers selected for Chr 5 in the SSR panel did not exhibit polymorphism between the parents of the JI population, the polymorphic ratio for Chr 5 was calculated at 0.786 (Table 2). This process was applied sequentially to all 20 chromosomes, with the 41 multiplex PCRs achieving genotyping of the 304 SSR markers. All the primer pairs yielded amplicons from at least one of the parents of the JI population, although 55 primer pairs showed no polymorphism between the parents. A linkage map covering 2864 cM was thus constructed from 249 markers with an average interval of 11.5 cM (Fig. 2, Table 2). All of the mapped markers were arranged in the same order and with similar genetic intervals compared with previous studies.5,27–30 Our whole-genome SSR panel thus appears to achieve highly accurate and efficient genotyping with sufficient marker coverage.
Figure 1.
Representative genotyping and mapping of eight SSR marker loci on soybean chromosome 5 (LG A1). (A) Multiplex PCR analysis of the eight SSR marker loci with primer sets labelled with four different fluorophores: 6-FAM (Sat_137 and Satt717), VIC (GMES3790 and Satt599), NED (GMES2497 and Satt545), and PET (Sat_344 and Sat_384). The separation and detection of PCR products corresponding to alleles of the eight marker loci with a fluorescence-based sequencer and GeneMapper 4.0 software are shown. (B) Genetic linkage map constructed with MAPMAKER/EXP 3.0b for the polymorphic SSR marker loci and the 94 F2 individuals of the JI population derived from a cross between JP-36121 (wild soybean accession) and Ibarakimame 7 (landrace).
Figure 2.
Molecular linkage map constructed with the whole-genome SSR panel system for the JI population. A total of 304 SSR marker loci were analysed with the 41 multiplex PCR sets of six to eight primer pairs, resulting in the generation of a linkage map consisting of 249 SSR marker loci covering 2864 cM of the soybean genome.
Table 2.
Summary of molecular linkage maps constructed for three mapping populations with the whole-genome SSR panel system
| Chromosome | Linkage group | No. of SSR marker loci | Polymorphic markers in each mapping population |
No. of non-polymorphic markers | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JI (2864 cM)a |
OA (2613 cM)b |
IF (2470 cM)c |
Total | |||||||
| No. | Ratiod | No. | Ratiod | No. | Ratiod | No. | ||||
| 1 | D1a | 14 | 12 | 0.857 | 7 | 0.500 | 5 | 0.357 | 13 | 1 |
| 2 | D1b | 16 | 13 | 0.813 | 9 | 0.563 | 8 | 0.500 | 15 | 1 |
| 3 | N | 15 | 13 | 0.867 | 14 | 0.933 | 10 | 0.667 | 15 | 0 |
| 4 | C1 | 12 | 10 | 0.833 | 9 | 0.750 | 9 | 0.750 | 12 | 0 |
| 5 | A1 | 14 | 11 | 0.786 | 9 | 0.643 | 5 | 0.357 | 12 | 2 |
| 6 | C2 | 19 | 16 | 0.842 | 9 | 0.474 | 7 | 0.368 | 18 | 1 |
| 7 | M | 21 | 20 | 0.952 | 11 | 0.524 | 13 | 0.619 | 21 | 0 |
| 8 | A2 | 20 | 16 | 0.800 | 10 | 0.500 | 12 | 0.600 | 17 | 3 |
| 9 | K | 13 | 10 | 0.769 | 9 | 0.692 | 10 | 0.769 | 13 | 0 |
| 10 | O | 15 | 10 | 0.667 | 11 | 0.733 | 8 | 0.533 | 12 | 3 |
| 11 | B1 | 18 | 16 | 0.889 | 8 | 0.444 | 8 | 0.444 | 16 | 2 |
| 12 | H | 15 | 11 | 0.733 | 13 | 0.867 | 8 | 0.533 | 13 | 2 |
| 13 | F | 15 | 13 | 0.867 | 9 | 0.600 | 6 | 0.400 | 15 | 0 |
| 14 | B2 | 12 | 12 | 1.000 | 7 | 0.583 | 8 | 0.667 | 12 | 0 |
| 15 | E | 15 | 11 | 0.733 | 5 | 0.333 | 6 | 0.400 | 12 | 3 |
| 16 | J | 9 | 5 | 0.556 | 6 | 0.667 | 7 | 0.778 | 8 | 1 |
| 17 | D2 | 15 | 13 | 0.867 | 13 | 0.867 | 11 | 0.733 | 15 | 0 |
| 18 | G | 14 | 10 | 0.714 | 11 | 0.786 | 9 | 0.643 | 13 | 1 |
| 19 | L | 17 | 16 | 0.941 | 8 | 0.471 | 7 | 0.412 | 16 | 1 |
| 20 | I | 15 | 11 | 0.733 | 9 | 0.600 | 10 | 0.667 | 15 | 0 |
| Total | 304 | 249 | 0.819 | 187 | 0.615 | 167 | 0.549 | 283 | 21 | |
| Average | 15.2 | 12.5 | 9.4 | 8.4 | 14.2 | 1.1 | ||||
aA population of 94 F2 plants derived from a cross between JP-36121 and Ibarakimame 7 was used for construction of a linkage map spanning 2864 cM.
bA population of 225 RILs derived from a cross between Osuzu and Athow was used for construction of a linkage map spanning 2613 cM.
cA population of 143 RILs derived from a cross between Ippon_sangoh and Fukuyutaka was used for construction of a linkage map spanning 2470 cM.
dPolymorphic ratio = number of polymorphic marker loci/number of analysed marker loci.
Wild soybeans including JP-36121 are thought to be more genetically diverse than cultivated soybeans, with the result that the genome panel was able effectively to build a linkage map of the JI population. However, molecular linkage maps are frequently constructed from segregating populations with relatively similar genetic backgrounds in order to analyse important agricultural traits such as maturity and yield performance.36 We therefore next applied the SSR genome panel to the analysis of two mapping populations, OA and IF, derived from crosses between soybean cultivars. The polymorphic ratio (0.549) of the IF population, which is derived from Japanese germplasm, was found to be much lower than that (0.819) of the JI population, but it was similar to that (0.615) of the OA population (Table 2). A total of 187 SSR markers showed polymorphism between the Japanese cultivar Osuzu and the US cultivar Athow, which are the parents of the OA RILs, and provided a linkage map covering 2613 cM with an average marker interval of 14.0 cM. The Japanese landrace Ippon_sangoh and the elite Japanese cultivar Fukuyutaka, which are the parents of the IF population of 143 F7 RILs, exhibited allelic variation at 167 SSR loci, and the genotype data for these RILs allowed the construction of a linkage map covering 2470 cM with an average interval of 14.8 cM. The SSR genome panel system thus achieved the construction of linkage maps with sufficient marker coverage in the three mapping populations, and it was less labour-intensive and more rapid than the precedent methods by singleplex PCRs and gel electrophoreses. The molecular linkage maps obtained provide a basis for genetic analysis of specific traits such as deficiency of group A acetylated saponins.35
The polymorphic markers were evenly distributed among the 20 LGs, with 201 (66.1%) of the 304 SSR loci being mapped in at least two populations. We thus verified that the SSR genome panel system was well designed to minimize bias of marker location and to include highly polymorphic markers. A total of 21 markers were not polymorphic in the three mapping populations; however, their locations were not confirmed. Although these markers may show polymorphic alleles in other genetic backgrounds, they should possibly be excluded from the genome panel. Many of the markers exhibiting no polymorphism were selected from the EST-derived SSR markers, which were previously found to be less diverse, with fewer alleles, than the USDA SSR markers.5,30 Large marker intervals of >30 cM were observed between Satt634 and Satt600 on Chr 2 and between Sat_039 and Satt663 on Chr 13 in all three mapping populations. The addition of new highly polymorphic markers to these two regions and other relatively sparse regions would enhance the capability of the SSR genome panel system. We are currently in the process of arranging additional SSR markers for these genomic regions based on the recently completed whole-genome sequence of soybean and the abundant SSR motifs identified in the genome.24,37
3.3. Diversity analysis of 87 soybean genotypes with the whole-genome SSR panel
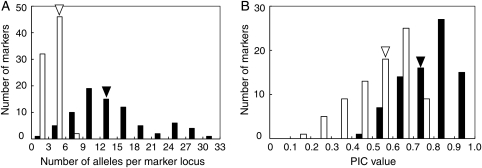
We next examined allelic diversity of selected SSR markers in 87 soybean genotypes, comprising 8 wild accessions and 79 cultivars, including Japanese elite cultivars, that we had analysed previously.12 From the 304 SSR loci in the whole-genome SSR panel, we selected four markers for each chromosome, to give a total of 80 SSR loci, on the basis of their map position and polymorphism (Supplementary Table S2). The previous analysis by PCR and 3% agarose gel electrophoresis12 revealed that the allele number for the 80 SSR loci ranged from 2 to 7, with an average of 4.0 (Fig. 3A). The most frequent allele number was 3, which accounted for 31.3% of all the marker loci. The PIC value, a measure of allelic diversity at a locus, ranged from 0.12 (Satt568) to 0.80 (Sat_115), and the average PIC value was 0.55 (Fig. 3B). In contrast, in our panel system with the selected 80 SSR marker set, the number of alleles for each locus ranged from 3 (Satt307) to 32 (Satt263), with an average of 14.8, and the PIC value ranged from 0.45 (Satt568) to 0.96 (Sat_115), with a mean of 0.78 for all the marker loci. This level of polymorphism was thus substantially higher than that obtained previously by analysis with agarose or polyacrylamide gel electrophoresis,5,12 likely reflecting the fact that a capillary sequencer is able to determine the size of repetitive motifs with higher resolution and greater accuracy. The mean PIC value and the average allele number obtained in the present study are similar to those determined in previous studies with high-resolution analysis systems, although different diverse materials were used in each study.10,11,38 However, the PIC value and allele number for each SSR locus in the present study did not correlate well with our previous results,12 indicating that allelic diversity at each SSR locus includes small differences that are difficult to determine by 3% agarose gel electrophoresis. The high level of diversity of the SSR loci will allow wide application of the whole-genome SSR panel to detailed genotype identification as well as to mapping phenotypic and quantitative trait loci in various segregating populations.
Figure 3.
Distribution of allele numbers (A) and PIC values (B) for 80 SSR marker loci in 87 soybean cultivars or wild accessions as determined by multiplex PCR and a fluorescence-based sequencer in the present study (solid bars) or by singleplex PCR and 3% agarose gel electrophoresis in a previous study12 (open bars). Arrowheads indicate the corresponding averages for allele number and PIC value determined by the two methods.
3.4. Conclusion
Thousands of microsatellite markers in soybean have been developed over the last two decades, with >2000 SSR loci having been integrated into a common linkage map.5,27–30 The abundant and well-documented SSR loci allow the relatively unlimited selection of SSR markers on the basis of their diverse length polymorphism and map location. We selected 304 SSR markers and assembled them into 41 multiplex PCR sets to give a whole-genome SSR panel. This genome panel system has been applied to linkage analysis of >10 segregating populations in addition to the three populations analysed in the present study, and it has successfully built the framework of each linkage map (data not shown). The results indicate the wide applicability of the genome panel system to various combinations of soybean genotypes. The combination of multiplex PCR and the high-resolution detection system is also applicable to other genotyping studies such as marker-assisted selection and the fine mapping of qualitative and quantitative traits. The flexibility of our genome panel system should improve the efficiency of molecular marker genotyping by reducing the labour intensity and time required for this process, although this system requires a fluorescent sequencer and hundreds of custom-labelled primers with fluorescent dye.
The recent breakthrough of high multiplexing has moved the SNP array into the mainstream of genotyping technology.39 Many thousands of SNPs have been identified in soybean and have been used to design the GoldenGate assay, which discriminates up to 1536 SNPs in one reaction.4,25,26 On the basis of its high multiplexing capacity, automation, and continued improvement, the SNP array is becoming one of most reliable methods for whole-genome genotyping. However, the quantity and quality of SNP loci in the entire soybean genome are currently not sufficient for application of this system to many genotypes. In addition, the GoldenGate assay is not flexible for modification of SNP marker selection and is expensive for each reaction. Although the multiplex PCR system coupled with high-resolution detection is capable of analysing at most 20 SSR loci at a time, it is possible to combine in a flexible manner the mapped SSR markers and SSR motifs that are scattered throughout the soybean genome.24
Supplementary data: Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This research was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan [Genomics for Agricultural Innovation (DD-3260)].
Supplementary Material
References
- 1.Palmer R.G., Kilen T.C. Qualitative genetics and cytogenetics. In: Wilcox J.R., editor. Soybeans: Improvement, Production, and Uses. 2nd edition. Madison, Wisconsin: Agronomy Monograph; 1987. pp. 135–209. [Google Scholar]
- 2.Keim P., Beavis W., Schupp J., Freestone R. Evaluation of soybean RFLP marker diversity in adapted germplasm. Theor. Appl. Genet. 1992;85:205–12. doi: 10.1007/BF00222861. [DOI] [PubMed] [Google Scholar]
- 3.Williams J.G.K., Kubelik A.R., Livak K.J., Rafalski J.A., Tingey S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–5. doi: 10.1093/nar/18.22.6531. doi:10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi I.Y., Hyten D.L., Matukumalli L.K., et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–96. doi: 10.1534/genetics.107.070821. doi:10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang T.Y., Sayama T., Takahashi M., et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–25. doi: 10.1093/dnares/dsp010. doi:10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akkaya M.S., Bhagwat A.A., Cregan P.B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics. 1992;132:1131–9. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akkaya M.S., Shoemaker R.C., Specht J.E., Bhagwat A.A., Cregan P.B. Integration of simple sequence repeat DNA markers into a soybean linkage map. Crop Sci. 1995;35:1439–45. doi:10.2135/cropsci1995.0011183X003500050030x. [Google Scholar]
- 8.Morgante M., Olivieri A.M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 2008;3:175–82. [PubMed] [Google Scholar]
- 9.Maughan P.J., Saghi Maroof M.A., Buss G.R. Microsatellite and amplified sequence length polymorphisms in cultivated and wild soybean. Genome. 1995;38:715–23. doi: 10.1139/g95-090. doi:10.1139/g95-090. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Wang D., Arelli P., Ebrahimi M., Nelson R.L. Molecular marker diversity of SCN-resistant sources in soybean. Genome. 2006;49:938–49. doi: 10.1139/g06-057. doi:10.1139/G06-057. [DOI] [PubMed] [Google Scholar]
- 11.Diwan N., Cregan P.B. Automated sizing of fluorescent labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor. Appl. Genet. 1997;95:723–33. doi:10.1007/s001220050618. [Google Scholar]
- 12.Hwang T.Y., Nakamoto Y., Kono I., et al. Genetic diversity of cultivated and wild soybean lines including Japanese elite cultivars as revealed by length polymorphism of SSR markers. Breed. Sci. 2008;58:315–23. doi:10.1270/jsbbs.58.315. [Google Scholar]
- 13.Cregan P.B., Bhagwat A.A., Akkaya M.S., Rongwen J. Microsatellite fingerprinting and mapping of soybean. Methods Mol. Cell Biol. 1994;5:49–61. [Google Scholar]
- 14.Shoemaker R.C., Polzin K., Labate J., et al. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–38. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill N., Findley S., Walling J.G., et al. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol. 2009;151:1167–74. doi: 10.1104/pp.109.137935. doi:10.1104/pp.109.137935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn J.R., Temnykh S.V., Paul E.M., McCouch S.R. Design and application of microsatellite marker panels for semiautomated genotyping of rice (Oryza sativa L.) Crop Sci. 2002;42:2092–9. doi:10.2135/cropsci2002.2092. [Google Scholar]
- 17.Doxiadis G.G.M., De Groot N., Claas F.H.J., Doxiadis I.I.N., Van Rood J.J., Bontrop R.E. A highly divergent microsatellite facilitating fast and accurate DRB haplotyping in humans and rhesus macaques. Proc. Natl Acad. Sci. USA. 2007;104:8907–12. doi: 10.1073/pnas.0702964104. doi:10.1073/pnas.0702964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindqvist A.K., Magnusson P.K., Balciuniene J., et al. Chromosome-specific panels of tri-and tetranucleotide microsatellite markers for multiplex fluorescent detection and automated genotyping: evaluation of their utility in pathology and forensics. Genome Res. 1996;6:1170–6. doi: 10.1101/gr.6.12.1170. doi:10.1101/gr.6.12.1170. [DOI] [PubMed] [Google Scholar]
- 19.Schalkwyk L.C., Jung M., Daser A., et al. Panel of microsatellite markers for whole-genome scans and radiation hybrid mapping and a mouse family tree. Genome Res. 1999;9:878–87. doi: 10.1101/gr.9.9.878. doi:10.1101/gr.9.9.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryda E.C., Riley L.K. Multiplex microsatellite marker panels for genetic monitoring of common rat strains. J. Am. Assoc. Lab. Anim. Sci. 2008;47:37–41. [PMC free article] [PubMed] [Google Scholar]
- 21.Pessoa-Filho M., Belo A., Alcochete A.A.N., Rangel P.H.N., Ferreira M.E. A set of multiplex panels of microsatellite markers for rapid molecular characterization of rice accessions. BMC Plant Biol. 2007;7:23. doi: 10.1186/1471-2229-7-23. doi:10.1186/1471-2229-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Oliveira Borba T.C., Brondani R.P.V., Rangel P.H.N., Brondani C. Microsatellite marker-mediated analysis of the EMBRAPA Rice Core Collection genetic diversity. Genetica. 2009;137:293–304. doi: 10.1007/s10709-009-9380-0. doi:10.1007/s10709-009-9380-0. [DOI] [PubMed] [Google Scholar]
- 23.Tang S., Kishore V.K., Knapp S.J. PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor. Appl. Genet. 2003;107:6–19. doi: 10.1007/s00122-003-1233-0. [DOI] [PubMed] [Google Scholar]
- 24.Song Q., Jia G., Zhu Y., et al. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 2010;50:1950–60. doi:10.2135/cropsci2009.10.0607. [Google Scholar]
- 25.Hyten D.L., Song Q., Choi I., et al. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor. Appl. Genet. 2008;116:945–52. doi: 10.1007/s00122-008-0726-2. doi:10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 26.Hyten D.L., Choi I.-Y., Song Q., et al. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010;50:960–8. doi:10.2135/cropsci2009.06.0360. [Google Scholar]
- 27.Song Q.J., Marek L.F., Shoemaker R.C., et al. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004;109:122–8. doi: 10.1007/s00122-004-1602-3. doi:10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 28.Cregan P.B., Jarvik T., Bush A.L., et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–90. doi:10.2135/cropsci1999.3951464x. [Google Scholar]
- 29.Xia Z., Tsubokura Y., Hoshi M., et al. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14:257–69. doi: 10.1093/dnares/dsm027. doi:10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hisano H., Sato S., Isobe S., et al. Characterization of the soybean genome using EST-derived micorsatellite markers. DNA Res. 2007;14:271–81. doi: 10.1093/dnares/dsm025. doi:10.1093/dnares/dsm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 32.Lincoln S.E., Daly M.J., Lander E.S. Constructing Genetic Maps with MAPMAKER/EXP 3.0, Whitehead Institute Technical Report. 3rd edition. Cambridge, MA: Whitehead Institute; 1992. [Google Scholar]
- 33.Kosambi D.D. The estimation of map distance from recombination values. Ann. Eugen. 1944;12:172–5. [Google Scholar]
- 34.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. doi:10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 35.Takada Y., Sayama T., Kikuchi A., et al. Genetic analysis of variation in sugar chain composition at the C-22 position of group A saponins in soybean, Glycine max (L.) Merrill. Breed. Sci. 2010;60:3–8. doi:10.1270/jsbbs.60.3. [Google Scholar]
- 36.Sayama T., Hwang T.Y., Yamazaki H., et al. Mapping and comparison of quantitative trait loci for soybean branching phenotype in two locations. Breed. Sci. 2010;60:380–9. doi:10.1270/jsbbs.60.380. [Google Scholar]
- 37.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Guan R., Zhangxiong L., Chang R., Qiu L. Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Sci. 2006;46:1032–8. doi:10.2135/cropsci2005.0051. [Google Scholar]
- 39.Fan J.B., Gunderson K.L., Bibikova M., et al. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. doi:10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.