Abstract
The androgen receptor (AR) is the principal target for treatment of non-organ confined prostate cancer (PCa). Androgen deprivation therapies (ADTs) directed against the AR ligand-binding domain do not fully inhibit androgen-dependent signaling critical for PCa progression. Thus, information that could direct the development of more effective ADTs are desired. Systems and bioinformatics approaches suggest that considerable variation exists in the mechanisms by which AR regulates expression of effector genes, pointing to a role for secondary transcription factors. A combination of microarray and in silico analyses led us to identify a 158 gene signature that relies on AR along with the transcription factor SRF, representing < 6% of androgen-dependent genes. This AR-SRF signature is sufficient to distinguish microdissected benign and malignant prostate samples, and it correlates with the presence of aggressive disease and poor outcome. Compared to other AR target gene signatures of similar size, the AR-SRF signature described here associates more strongly with biochemical failure. Further, it is enriched in malignant versus benign prostate tissues, compared to other signatures. To our knowledge, this profile represents the first demonstration of a distinct mechanism of androgen action with clinical relevance in PCa, offering a possible rationale to develop novel and more effective forms of ADT.
Keywords: prostate cancer, androgen receptor, transcription, gene expression, disease progression
Introduction
Prostate cancer (PCa) remains the most frequently diagnosed cancer of an internal organ and the second leading cause of cancer-related death in men (1). One in six American men will be confronted with PCa, which makes this disease a significant health problem. Localized PCa is treated with surgical or radiation therapies that have a curative intent (2). For patients with locally advanced PCa, metastatic disease or whose cancer recurs after initial treatment, treatment options are limited to preventing disease progression. As PCa progression depends on androgen signaling, the androgen receptor (AR) is the principal target for treating non-organ confined disease. Traditional androgen deprivation therapy (ADT) interferes with the systemic production of androgens and/or involves administration of antiandrogens (3). Initially, ADT prevents tumor growth and leads to a favorable clinical response. Unfortunately, ADT does not eradicate disease and eventually PCa recurs as castration-recurrent PCa (CRPC), which is invariably lethal. Intriguingly, the emergence of CRPC is due, at least in part, to inappropriate activation of the AR (4–7). Recent therapeutic approaches that are tailored specifically to target aberrant AR action in CRPC lead to antitumor activity in a substantial subset of patients (8–10). However, these effects are partial and temporary, which indicates that AR activity is not inhibited fully by current ADTs (11,12). Developing more effective means to interfere with AR signaling requires an in-depth understanding of the molecular mechanism(s) by which AR governs clinically relevant events in PCa.
The AR is a ligand-dependent transcription factor belonging to the nuclear receptor superfamily. Upon androgen binding, the AR translocates from the cytoplasm to the nucleus, binds as a dimer to “Androgen Response Elements” (AREs) in the regulatory regions of target genes and recruits a productive transcriptional complex (reviewed in 13). Considerable efforts have been directed towards identifying AR-dependent genes which contribute to PCa progression. Large scale gene expression profiling studies in model systems have identified dozens of androgen-regulated genes (14–19). Many of these have been proposed to play a role in differentiated cell function, cell proliferation and cell survival (20). None-the-less, correlation of androgen-responsive mRNA expression profiles obtained from in vitro model systems with transcriptomes derived from clinical PCa specimens has been challenging. To date, specific genes and transcriptional programs which are critical for PCa cell proliferation and metastasis and that are regulated by androgens remain largely elusive. These difficulties may reflect, in part, that attempts at classification have encompassed all androgen-dependent gene expression, which assumed little or no variation in the manner by which AR controls transcription of target genes. Recent systems approaches, however, suggest variability in the composition of the AR transcriptional complex at regulatory sites in effector genes, indicate that not all androgen-dependent genes are subject to direct ARE-driven mechanisms of regulation and provide evidence that AR signaling in PCa cells relies on secondary transcription factors (TFs) (21–25). Some TFs interact directly with the AR to affect its ability to bind to AREs and compete for coregulators or to cooperate in the transcription of AR target genes (13). Other TFs mediate critical effects on PCa cells by mechanisms that do not rely on physical interaction with the AR but involve androgen regulation of their activities (26,27). These “indirect” mechanisms of androgen action convey androgen responsiveness to target genes that do not contain AREs and can induce coordinated responses of genes and/or cells. Identification of TFs whose transcriptional program contributes to the development of aggressive disease may, therefore, open novel avenues to target clinically relevant androgen signaling in PCa.
We recently identified a novel indirect mechanism of androgen action in which effects of androgens on PCa cells are mediated by Serum Response Factor (SRF) (28). SRF is a MADS-box containing TF that was originally identified by its ability to convey the effects of serum to immediate early response genes (29). Since then, SRF has been shown to also control expression of genes involved in the organization of the cytoskeleton and to be critical for embryonic development, experimental metastasis and angiogenesis (30–33). Androgen exposure induces expression of Four-and-a-half-LIM domain protein 2 (FHL2) in a manner that is AR-dependent but independent of any AREs in the FHL2 gene. Instead, androgens stimulate expression of FHL2 through action of SRF on its consensus binding site (CArG-box) in the FHL2 promoter (28). In view of these cellular and physiological roles, it is tempting to speculate that androgen control over the SRF transcriptional program may have important implications for PCa progression.
Materials and methods
Cell culture
LNCaP and VCaP cells were purchased from American Type Culture Collection (Manassas, VA) and were maintained as described (28,34). Behavior of cells was monitored throughout the study by assessing overall androgen-responsiveness, morphology and transcriptional regulation, which were consistent with previous observations for these cell lines.
siRNA transfection
LNCaP and VCaP cells were seeded in 60 mm dishes at a density of 5.5 × 105 or 3 × 106 cells per dish, respectively, in antibiotic-free medium. The next day, cells were transfected with siGenome SmartPool siRNA targeting SRF (Dharmacon, Lafayette, CO) or a custom-made control SmartPool targeting luciferase (LUC condition) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. 42 hours after transfection, cells were treated with 5nM R1881 or ethanol vehicle. 3 biological triplicates were included per treatment group. 48 hours later, cells were harvested in Trizol reagent (Invitrogen).
Cell line RNA preparation and microarray analysis
RNA was isolated from cells using Trizol (Invitrogen), purified on RNeasy columns (Qiagen, Germantown, MD) and checked for integrity by Agilent testing (Affymetrix, Santa Clara, CA). cDNA was generated and hybridized to Human Genome U133 Plus 2.0 arrays (Affymetrix) according to the manufacturer’s instructions at the Mayo Clinic Advanced Genomics Technology Microarray Shared Resource core facility. The microarray datasets have been deposited in Gene Expression Omnibus under accession number GSE22606. A detailed description of the microarray data analysis can be found in the supplemental data.
Patient material and microarray data analysis
A detailed description of the patient materials and microarray dataset analysis is included in the supplemental data.
Ingenuity Pathway analysis
Ingenuity pathway analysis was performed using the 158 SRF-dependent androgen-responsive gene signature as a focus gene set and ingenuity curated knowledge base as a reference.
Real-time RT-PCR
cDNA was prepared and real time RT-PCR was performed as before (28). Primers targeting human FHL2, SRF, PSA, AR and GAPDH have been described (28). Primer sequences used to analyze SRF-dependent gene expression are listed in Table S1.
Results
Identification of an SRF-dependent androgen-responsive gene expression profile
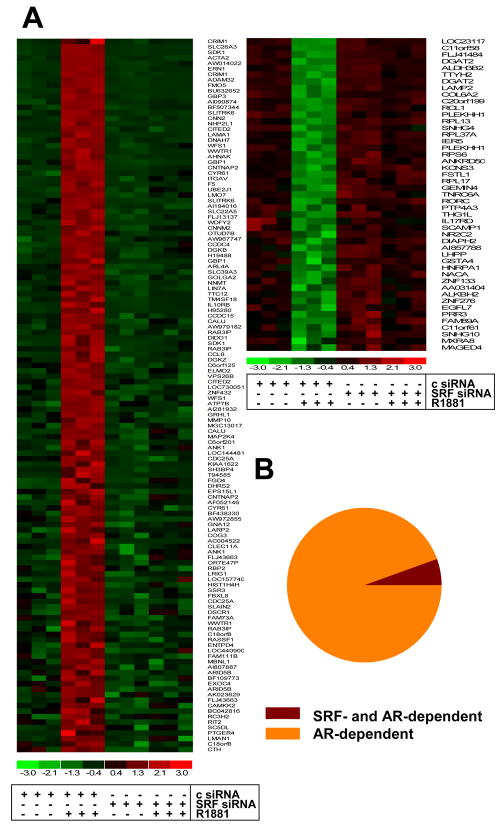
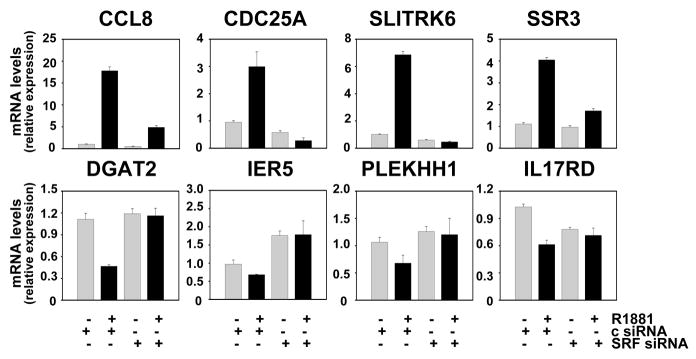
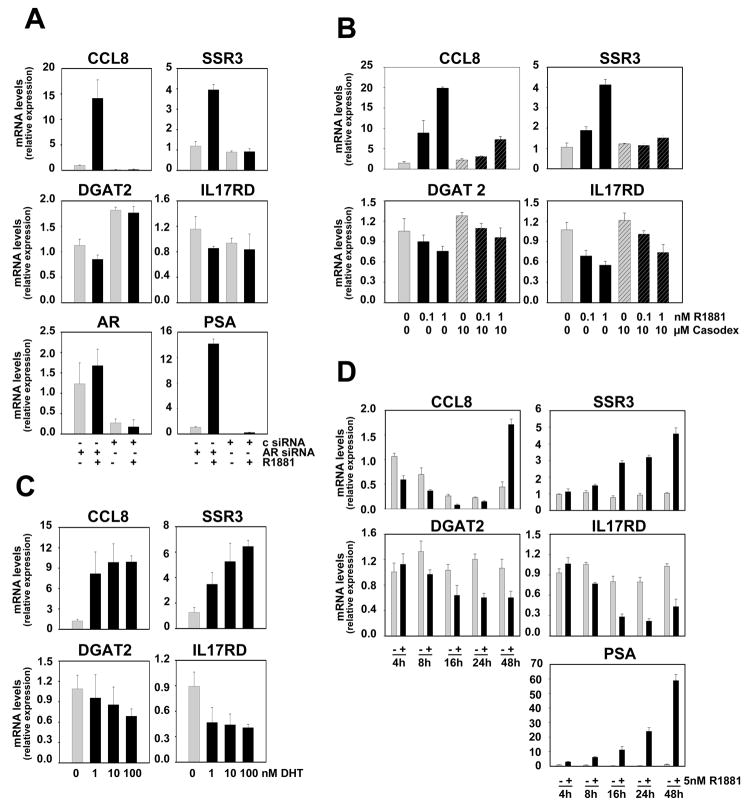
To identify and characterize genes and cellular processes that are androgen-regulated in an SRF-dependent manner in PCa, Affymetrix HG-U133 Plus 2.0 GeneChip Array analysis was performed starting from RNA obtained from LNCaP cells in which androgen stimulation was combined with siRNA-mediated SRF silencing. Biological triplicates were processed for each treatment group. The efficacy of the SRF knock-out and its effect on the expression of the positive control gene FHL2 were verified by real-time RT-PCR and are shown in Figure S1. Microarray data analysis focused on identifying genes which, for each replicate, show at least 2-fold androgen-dependent changes in expression, rely entirely on the presence of SRF for androgen-dependence and for which basal expression is not affected by loss of SRF. This approach detected 158 unique genes (178 probe sets), 113 (131 probe sets) and 45 (47 probe sets) of which were upregulated or downregulated in an SRF-dependent manner, respectively, following androgen treatment (Fig. 1A, Tables S2 and S3) (p<0.05). Overall androgen- and SRF-responsiveness of this expression profile, which represents less than 6 percent of androgen-regulated genes (Fig. 1B), was validated by real-time RT-PCR (Fig. 2) and immunoblotting (Fig. S2). AR involvement was further confirmed using AR-specific siRNA and the AR antagonist bicalutamide, and by performing dose response curves with the natural ligand dihydrotestosterone (DHT) and the synthetic androgen R1881 (Fig. 3A–C). Time course studies revealed that androgen regulation of genes belonging to the 158 gene profile requires at least 8 to 16 hours of ligand exposure, with some genes (e.g. CCL8) displaying slower androgen-dependent induction or repression. These kinetics are consistent with observations for the androgen-induced SRF-dependent gene FHL2 (28) and in contrast to rapid changes in expression of direct, ARE-driven AR target genes (e.g. PSA) (Fig. 3D). In the independent AR-positive cell line VCaP, androgen-regulation was conserved for 6 of the 7 genes (cfr Fig.2) for which expression was detectable by real-time RT-PCR. Moreover, knockdown of SRF partially or completely abolished androgen-dependence for 4 out of these 6 genes, which indicates that SRF-dependent androgen action is a common event in PCa cells (Fig. S3). Basal expression of 1,420 probe sets, which do not overlap with the 178 SRF- and AR-dependent probe sets, was affected upon loss of SRF, whereas overexpression of SRF did not alter expression of these genes (Fig. S4).
Figure 1. Identification of a SRF-dependent androgen-responsive expression profile.
LNCaP cells were transfected with siRNA targeting SRF or control siRNA (c) as described. RNA was isolated, column purified and processed for hybridization to Human Genome Affymetrix U133 Plus 2.0 arrays as described. Microarray raw data was analyzed and heatmaps were generated as described. 113 unique androgen-induced genes (131 probe sets) (left panel) and 45 unique androgen-repressed genes (47 probe sets) (right panel) were identified. Each row on the heatmap represents a probe set and each column represents an individual replicate. Gene expression on each probe set was standardized on the mean of samples where red color is higher than the mean and green color is lower than the mean (A). The 158 gene signature which relies on SRF for full androgen regulation represents 5.5 percent of all genes which are at least 2-fold regulated by androgen treatment in control-transfected condition (B).
Figure 2. Real time RT-PCR validation of microarray results.
LNCaP cells were transfected with siRNAs targeting SRF or non-specific control siRNAs (c) as described. SRF-dependency and androgen responsiveness of genes identified by the microarray approach were evaluated by real-time RT-PCR using the primer pairs listed in Table S1. Target gene mRNA levels were normalized with the values obtained from glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression and are expressed as relative expression values, taking the value obtained from one of the vehicle-treated control transfected conditions as 1. Columns, means of values obtained from three independent biological replicates; bars, SEM values.
Figure 3. SRF-dependent androgen-responsive gene signature is subject to an indirect mechanism of androgen action.
LNCaP cells were transfected with siRNAs targeting AR or control siRNAs and real-time RT-PCR was done as described (c) (A). LNCaP cells were seeded in medium supplemented with charcoal stripped serum (CSS). Two days later, medium was changed and cells were treated with 0, 0.1 or 1 nM R1881, with 10μM of the antiandrogen Casodex (bicalutamide) or a combination of R1881 and Casodex (B), or with 0, 1, 10 or 100 nM DHT (C). Forty-eight hours later, cells were harvested and real-time RT-PCR was performed (B-C). LNCaP cells were seeded in medium supplemented with CSS. Two days later, medium was changed and cells were treated with 1 nM R1881 (+) or vehicle (−) for 4, 8, 16, 24 and 48 hours. Real-time RT-PCR was performed as described (D). Columns, means of values obtained from three independent biological replicates; bars, SEM values.
SRF-dependent androgen-responsive gene functions are relevant to cancer biology
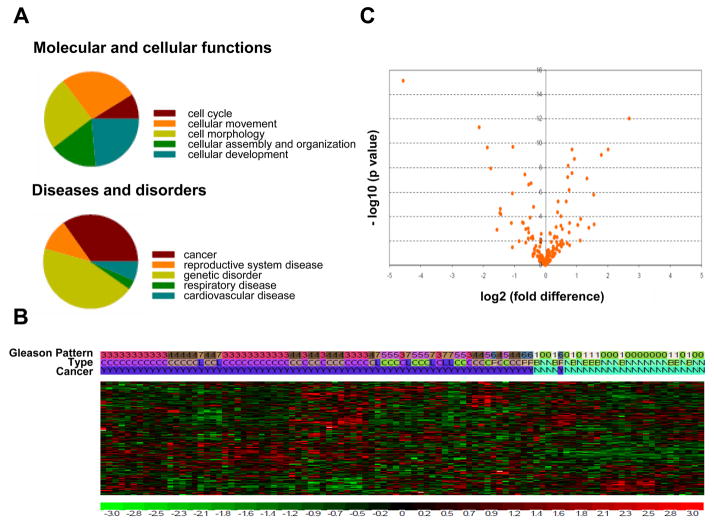
While some of the genes belonging to the newly identified 158 gene signature had been reported to be androgen-regulated (14,18,35), the majority of these genes have not been described previously as targets for androgen action in PCa cells. Interestingly, genes belonging to the SRF- and AR-dependent gene profile fulfill roles in processes as diverse as cell division (e.g. CDC25A), lipid synthesis (e.g. DGAT2), chemokine activity (e.g. CCL8), immediate early response (e.g. IER5) and extracellular matrix binding (e.g. CYR61), all of which are relevant to cancer cell biology. Moreover, Ingenuity Pathway Analysis (IPA) primarily assigned functions in cell cycle, cell morphology and cellular movement, development, assembly and organization to this gene signature. IPA indicated a significant association between this 158 gene expression profile and cancer as well as genetic disorders. Reproductive, respiratory and cardiovascular diseases were also associated with this gene signature (Fig. 4A). These observations suggest that the novel mechanism of androgen action may play a critical role in PCa.
Figure 4. SRF- and androgen-dependent gene signature is relevant to PCa.
Ingenuity Pathway Analysis identifies the molecular and cellular functions (left panel) and diseases and disorders (right panel) which most significantly associate with the 158 SRF-dependent and androgen-responsive gene signature (A). Unsupervised clustering using the 158 gene/178 probe sets signature separates normal from cancerous prostate samples. Gleason pattern nomenclature 0, 1, 6 and 7 correspond to normal epithelium, BPH, PIN and LN metastases samples, respectively. Tissue type designation C, L, B, N and P refers to localized cancer, LN metastases, BPH, normal epithelium and PIN, respectively. Cancer samples are marked as ‘Y’ and non-cancer samples are marked as ‘N’ (B). Differential expression analysis for the 178 probe sets between cancer (PIN, primary and metastatic prostate cancer) and non-cancer (BPH and normal prostate) samples was conducted using parametric t test. A volcano plot was created to visualize the results using – log 10 (p value) as y-axis and log2 fold change as x-axis. Probe sets which are differentially expressed (p<0.05) are listed in Table S4 (C).
SRF-dependent androgen-responsive gene signature is sufficient to separate benign and malignant prostate tissues
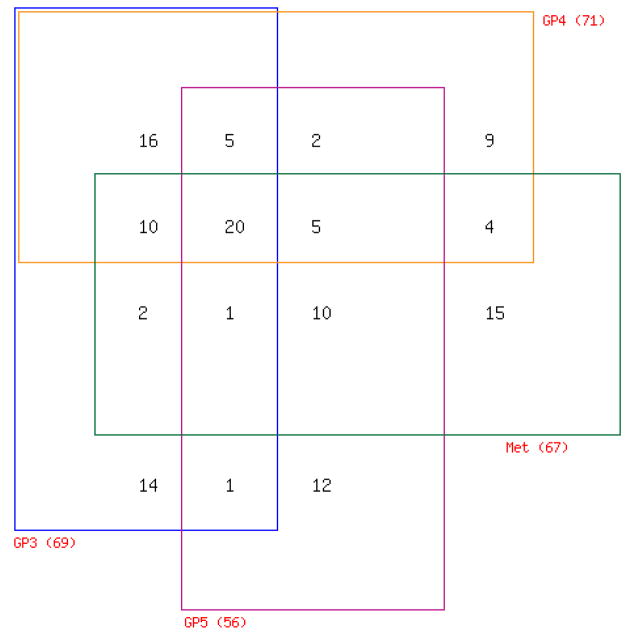
To validate the relevance of the SRF-dependent androgen-responsive gene signature for the clinical situation, the expression of the 178 probe sets was analyzed against mRNA expression datasets derived from human prostate specimens that were generated before (36). Prostate tissues included in this profiling study consist of normal epithelium (n=17), benign prostatic hyperplasia (BPH) (n=10), prostatic intraepithelial neoplasia (PIN) (n=4), PCas of Gleason Pattern (GP) 3 (n=31), GP4 (n=20), GP5 (n=10) and lymph node (LN) metastasis (n=7). An increase in GPs corresponds to a poorer prognosis, as does the presence of LN metastases at the time of prostatectomy (37,38). RNA was obtained from prostate tissue that was isolated by laser capture microdissection from radical prostatectomy specimens from patients with no preoperative treatment as described (36) and was analyzed using U133 Plus 2.0 microarrays. For initial analysis, normal epithelium and BPH samples were designated as non-cancer cases; whereas PIN lesions, localized GP3, GP4, GP5 and LN cases were considered cancer cases. Unsupervised clustering of these 99 samples by the 178 probe set signature alone generated 2 major clusters, which strikingly separated “normal” versus “malignant” prostate tissues. One case of PIN, considered a premalignant lesion to PCa, was the only exception and clustered with “normal” prostate tissue (Fig. 4B). A t-test comparison revealed that this separation is due to differential expression of 78 probe sets between “normal” and “cancerous” prostate samples (Fig. 4C, Table S4). Subsequent analyses did not include BPH samples. Pair-wise comparison between normal epithelial prostate samples and PIN, GP3, GP4, GP5 and LN specimens respectively, showed differential expression of 28, 69, 71, 56 and 67 probes sets. Noteworthy, altered expression of some probe sets was unique to a particular tissue type; whereas changes in expression of others were common to 2 or more tissue types (Fig. 5, Table S5). As shown in Figure 5, a core of 20 probe sets was found to be consistently deregulated with altered expression in GP3, GP4, GP5 and LN tissues compared to normal prostate epithelium. For validation purposes, the expression patterns of these 20 probe sets were examined in prostate tissue mRNA profiles available through the Oncomine database. This search indicated that the 20 core probe sets are consistently differentially expressed between normal and malignant prostate samples in 12 independent profiling studies, which had used different tissue procurement methods, RNA extraction procedures and microarray platforms (Table S6). In contrast, similar analyses for random sets of 20 direct AR target genes indicate that 35–40% of gene expression is inconsistently altered between malignant and benign prostate samples in different datasets (data not shown). These findings confirm the significance of the SRF- and AR-dependent core gene signature in PCa and corroborate the validity of our microarray experiment. Literature review provided additional evidence to support differential expression of target genes at the RNA and protein level (e.g. 39–43). Moreover, comparisons between paired normal prostate epithelium and localized PCas (n=9) and between matched localized cancer and LN metastases (n=4) yielded 52 and 23 differentially expressed probe sets (paired t-test, p<0.05), respectively (Tables S7 and S8).
Figure 5. Differentially expressed SRF-dependent androgen-responsive probe sets in normal epithelium, localized cancer and metastatic cancer.
Pairwise comparisons were made between PIN, GP3, GP4, GP5 and LN metastases and normal epithelium, respectively, using the limma package implemented in R to identify differentially expressed probe sets. Expression patterns of probe sets that are common or unique in GP3, GP4, GP5 and LN metastatic tissues are summarized in a 4 way Venn diagram. A detailed description of probe set tissue distribution is listed in Table S5.
SRF-dependent androgen-responsive gene signature correlates with aggressive disease
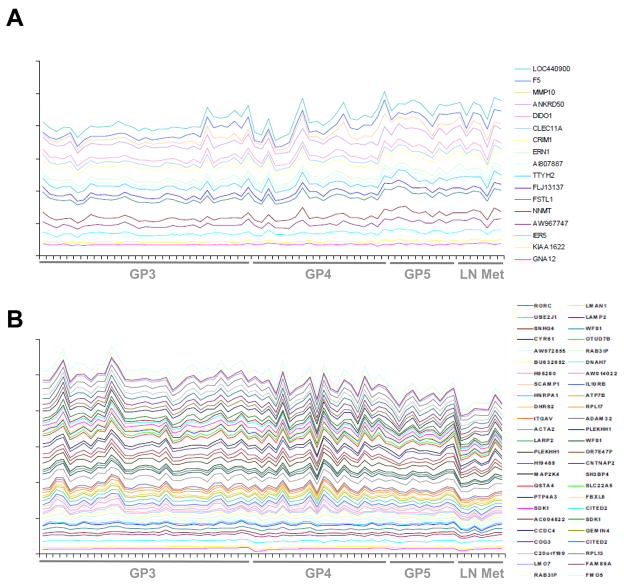
The relevance of the SRF-dependent androgen-responsive gene signature to PCa progression was assessed by exploring its correlation with GP number and metastatic status. Application of a linear regression model demonstrated that expression of 67 transcripts is positively (n=17) or negatively (n=50) associated with aggressive disease (p<0.05) (Fig. 6, Table S9). These results suggest that this gene signature may also be indicative of clinical outcome after initial surgical treatment. Consequently, the correlation between expression of the 178 probe sets and PSA failure was explored. PSA failure was defined as detectable levels of the PCa serum marker PSA (>0.4 ng/dl) after prostatectomy, which heralds the onset of CRPC and failure of primary treatment. For 56 of 61 patients with localized PCa (GP3, GP4 or GP5) at the time of surgery, follow-up data and PSA measurements were available (Table S10). Despite the limited number of patients who experienced PSA failure (n=15) and the relatively short time of follow-up (mean=45 months), expression of 15 probe sets was significantly associated with PSA failure (Table S11) (p<0.05). The same analysis was done for 10 sets of 178 randomly selected AR-target gene probes, which were taken from a pool of 1,228 probe sets that correspond to 452 AR-target genes (Table S12). 8 to 17 AR-target gene probe sets associated with PSA failure with statistical significance (mean = 11.6, standard deviation = 3.1). One-sided t statistics analysis demonstrated that these numbers are significantly lower than 15 (p = 0.003), which indicates that the fraction of SRF- and AR- dependent genes associated with PSA failure is larger than the fraction of direct AR-target genes (data not shown).
Figure 6. Expression of SRF-dependent androgen-responsive probe sets correlates with aggressive disease.
A linear regression model assesses the correlation of probe set expression with GP number and presence of LN metastatic disease. Probe sets that correlate significantly (p<0.05) are plotted in a stacked line graph. The Y axis represents a log 2 scale. Probe set expression is plotted as relative expression levels. Top panel: probe sets that correlate positively with tumor aggressiveness (A). Bottom panel: probe sets that correlate negatively with aggressiveness of disease (B). Probe sets that associate with aggressive disease are listed in Table S9.
In order to validate these findings, the expression of the 178 SRF- and AR- dependent probe sets was evaluated in a second, independent PCa gene expression profiling dataset ((44), available at http://cbio.mskcc.org/prostate-portal/). Here, mRNA expression was derived from 131 localized primary prostate tumors (27 of which experienced biochemical recurrence), 19 metastatic prostate tumors and 29 normal prostate specimens (Table S13). Tissues were macrodissected, mRNA profiling was done without amplification and Affymetrix Human Exon 1.0 ST Arrays were used. The 178 probe sets correspond to 142 annotated and unique genes, 139 of which mapped to the Exon Array dataset by gene symbol. Survival analysis was performed for the 139 genes using Cox proportional hazard model where PSA recurrence was the end point. Eighteen genes were identified that were associated significantly with biochemical failure (Table S14). The overlap with Table S11 consists of 1 gene, DGAT2, which is positively associated with biochemical recurrence. As limited overlap for outcome markers between microarray based profiling studies is not uncommon (e.g. (45)), gene set analysis was conducted for the 139 gene set as a whole, which showed that it is significantly associated with biochemical failure (p = 0.02) (Table 1). Next, gene set analysis was conducted for both the 139 SRF- and AR- dependent genes and 10 randomly selected sets of 142 AR-dependent genes (142 = same number of unique and annotated genes) using LS/KS permutation statistics. These two tests find gene sets that have more genes correlated with survival times (here time to PSA failure) than expected by chance through 100,000 permutations. Table 1 shows that the SRF- and AR-dependent gene set ranks number 1 in both tests with permutation p value 0.02 and 0.08, respectively. These results indicate that the SRF- and AR-dependent gene set is associated more significantly with PSA recurrence than similarly-sized sets of AR-target genes and that the association with biochemical recurrence is less likely a chance finding. Gene set analysis of totally random gene sets of similar size also failed to demonstrate a significant association with biochemical failure (Table S15). Evaluation of gene set enrichment between PCa and normal prostate tissues indicated that all 10 random AR-target gene sets are significantly enriched in normal tissues (FDR<0.25, negative enrichment scores), which is consistent with previous observations (46). In contrast, the SRF- and AR- dependent gene set is enriched in cancer tissue (FDR 0.12, positive enrichment score) (data not shown). Moreover, evaluation of 10 totally random gene sets found one set to be enriched in normal prostate tissues whereas the remaining 9 gene sets were not significantly enriched in either benign or malignant prostate (data not shown).
Table 1.
Gene set enrichment analysis evaluates association of gene expression with biochemical failure using LS/KS permutation statistics.
| Gene set | Number of genes | LS permutation p-value | KS permutation p-value |
|---|---|---|---|
| AR-SRF focus set | 139 | 0.02882 | 0.07976 |
| AR random set 8 | 142 | 0.06031 | 0.54397 |
| AR random set 6 | 142 | 0.07680 | 0.76182 |
| AR random set 1 | 142 | 0.13604 | 0.74264 |
| AR random set 2 | 142 | 0.14234 | 0.72544 |
| AR random set 10 | 142 | 0.18899 | 0.90306 |
| AR random set 5 | 142 | 0.21640 | 0.94449 |
| AR random set 9 | 142 | 0.35668 | 0.95764 |
| AR random set 3 | 142 | 0.55384 | 0.96665 |
| AR random set 4 | 142 | 0.62122 | 0.76469 |
| AR random set 7 | 142 | 0.67887 | 0.85191 |
Discussion
Here, we report the isolation of, to our knowledge, the first discrete androgen-dependent signaling pathway that is relevant to the clinical situation in PCa. Despite the dependence of PCa cells on AR signaling (3–12), previous attempts to identify genes that are critical for disease progression and metastasis and are under androgen control have been largely unsuccessful. The difficulties in validating the androgen-responsive transcriptomes derived from in vitro model systems using expression profiles from clinical samples may in part be due to the use of different platforms and variations in RNA procurement and processing protocols. More likely, the analysis was hampered by the inherent assumptions that all androgen regulation of AR target genes occurs via a similar transcriptional mechanism and that every gene found to be subject to androgen regulation in model systems also plays a significant role in the clinical setting. Efforts to sort gene expression profiling data based on the extent, direction and kinetics of androgen-regulation have isolated distinct clusters of genes (16,18,19), but do not allow for correlation between cell based models and clinical specimens.
We reasoned that co-dependency of the AR on secondary TFs, which are increasingly recognized to convey selectivity to the regulation of subsets of AR target genes (21–25), can be exploited to delineate distinct mechanisms of androgen action in PCa and that thoughtful selection of a candidate TF with potentially important roles in PCa cell biology can identify gene signatures that contribute to disease progression. To test this hypothesis, a microarray-based approach was undertaken to identify the spectrum of genes that rely on SRF, a versatile TF which functions in the immediate early response and the organization of the cytoskeleton (29,30), to achieve full androgen responsiveness.
This approach revealed a gene profile that corresponds to less than 6 percent of the androgen-dependent transcriptome in the PCa cell line LNCaP. Strikingly, even without prior knowledge of levels of expression for these genes or further manipulation of the data, this gene signature was able to successfully separate benign from malignant prostate. Moreover, the gene expression profile correlated with aggressive disease and poor post-operative outcome. Validation studies using an independent PCa profiling study with suitable tissue and RNA procurement methods, complete clinical annotation and reliable follow-up data (44) confirmed the association of the SRF- and AR-dependent gene signature with biochemical recurrence. Moreover, fewer genes that are both androgen-regulated and harbor genomic AR binding sites (Table S12) correlate with PSA failure. In stark contrast with the SRF- and AR-dependent gene signature, random sets of AR target genes are significantly enriched in normal rather than malignant prostate tissues.
The overall design of the experiment, particularly the concentration of androgen used and the timing of the different steps, aimed to avoid skewing the results towards effects that are merely attributable to changes in cell proliferation and survival. In keeping with this premise, the resulting 158 gene signature displayed a striking enrichment for genes that function in cell adhesion, cell-cell communication and cell-cell interaction. IPA pathway analysis associated this profile with development and function of the cardiovascular, connective tissue and skeletal and smooth muscle systems (data not shown). Independent Gene Ontology analysis of the probe sets which are expressed differentially between benign and malignant prostate confirmed involvement in musculature and central nervous system (data not shown), both systems where SRF as well as AR have been shown to fulfill indispensable roles (47–48). At the cellular and molecular level, IPA as well as Gene Ontology analysis point towards a disproportional contribution (~ 30%) of genes with actin cytoskeleton-related functions to the aberrantly expressed SRF-and androgen-dependent gene signature. In addition to the differentially expressed genes that encode components and regulators of the cytoskeleton (e.g. ACTA2, CNN2), many are associated with cellular events that rely heavily on the cell’s actin cytoskeleton such as chemotaxis (e.g. CYR61), cell division (e.g. CDC25A), formation of cellular protrusions (e.g. WWTR1) and endosomal and vacuolar transport (e.g. VPS26B). (Re)organization of the cytoskeleton is vital for cancer cells to invade surrounding tissue, migrate and generate metastatic lesions, and may underlie the correlation of the SRF-dependent mechanism of androgen action with more aggressive disease. It should be noted that the 158 gene signature also contains genes involved in lipid synthesis, transcription and protein synthesis, which have been shown to be important in PCa, as well as several genes involved in ion homeostasis. Ion channels have been implicated in cancer cell invasive behavior and may be viable pharmacological targets (49).
Apart from its clinical significance, this novel mechanism of androgen action represents a different means of SRF activation at the molecular level. According to its classical model of action, SRF is bound constitutively to CArG boxes in target genes, where its activity is regulated by signaling cascades and/or by interaction with one or more of its cofactors (50). Induction of SRF activity by extracellular stimuli typically results in rapid changes in the levels of its target genes. In contrast, androgen-responsiveness of CArG-box containing genes for which SRF binding has been confirmed by ChIP (e.g. COG3, SDK1) (data not shown) requires 8 to 16 hours of androgen treatment. The need for even longer androgen exposure for some genes (e.g. CCL8) most likely reflects indirect regulation by SRF. Ongoing work in our laboratory indicates that androgens affect the activity but not expression levels, cellular localization or CArG box-binding potential of SRF. Moreover, SRF is not differentially expressed between benign and malignant prostate specimens (data not shown). These observations suggest AR- and ARE-dependent modulation of regulators of SRF activity.
Taken together, the data presented here provide a proof-of-principle for the existence of discrete modes of AR action with clinical relevance. Thus, novel ADT approaches may be developed that are geared selectively toward the molecular events by which AR controls PCa progression, metastasis and the lethal phenotype.
Supplementary Material
Acknowledgments
Financial support: Supported by NIH Grants CA121277, CA91956, CA15083, CA125747, DK65236, T.J. Martell Foundation, Mayo Clinic Prostate Cancer SPORE (HVH) and Department of Defense Prostate Cancer Research Program (HVH).
Footnotes
Potential conflicts of interest: none
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Klein EA, Ciezki J, Kupelian PA, Mahadevan A. Outcomes for intermediate risk prostate cancer: are there advantages for surgery, external radiation, or brachytherapy? Urol Oncol. 2009;27:67–71. doi: 10.1016/j.urolonc.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Miyamoto H, Messing EM, Chang C. Androgen deprivation therapy for prostate cancer: current status and future prospects. Prostate. 2004;61:332–53. doi: 10.1002/pros.20115. [DOI] [PubMed] [Google Scholar]
- 4.Litvinov IV, De Marzo AM, Isaacs JT. Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab. 2003;88:2972–82. doi: 10.1210/jc.2002-022038. [DOI] [PubMed] [Google Scholar]
- 5.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488–90. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 6.Mohler JL. Castration-recurrent prostate cancer is not androgen-independent. Adv Exp Med Biol. 2008;617:223–34. doi: 10.1007/978-0-387-69080-3_21. [DOI] [PubMed] [Google Scholar]
- 7.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–8. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attard G, Reid AH, Yap TA, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26:4563–71. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 9.Attard G, Reid AH, Olmos D, de Bono JS. Antitumor activity with CYP17 blockade indicates that castration-resistant prostate cancer frequently remains hormone driven. Cancer Res. 2009;69:4937–40. doi: 10.1158/0008-5472.CAN-08-4531. [DOI] [PubMed] [Google Scholar]
- 10.Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attard G, Cooper CS, de Bono JS. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell. 2009;16:458–62. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Clegg NJ, Scher HI. Anti-androgens and androgen-depleting therapies in prostate cancer: new agents for an established target. Lancet Oncol. 2009;10:981–91. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 14.DePrimo SE, Diehn M, Nelson JB, et al. Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 2002;3:RESEARCH0032. doi: 10.1186/gb-2002-3-7-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson PS, Clegg N, Arnold H, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99:11890–5. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segawa T, Nau ME, Xu LL, et al. Androgen-induced expression of endoplasmic reticulum (ER) stress response genes in prostate cancer cells. Oncogene. 2002;21:8749–58. doi: 10.1038/sj.onc.1205992. [DOI] [PubMed] [Google Scholar]
- 17.Febbo PG, Lowenberg M, Thorner AR, Brown M, Loda M, Golub TR. Androgen mediated regulation and functional implications of fkbp51 expression in prostate cancer. J Urol. 2005;173:1772–7. doi: 10.1097/01.ju.0000155845.44729.ba. [DOI] [PubMed] [Google Scholar]
- 18.Velasco AM, Gillis KA, Li Y, et al. Identification and validation of novel androgen- regulated genes in prostate cancer. Endocrinology. 2004;145:3913–24. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- 19.Ngan S, Stronach EA, Photiou A, Waxman J, Ali S, Buluwela S. Microarray coupled to quantitative RT-PCR analysis of androgen-regulated genes in human LNCaP prostate cancer cells. Oncogene. 2009;28:2051–63. doi: 10.1038/onc.2009.68. [DOI] [PubMed] [Google Scholar]
- 20.Dehm SM, Tindall DJ. Molecular regulation of androgen action in prostate cancer. J Cell Biochem. 2006;99:333–44. doi: 10.1002/jcb.20794. [DOI] [PubMed] [Google Scholar]
- 21.Massie CE, Adryan B, Barbosa-Morais NL, et al. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 2007;8:871–8. doi: 10.1038/sj.embor.7401046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–17. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia L, Berman BP, Jariwala U, et al. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE. 2008;3:e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verhoeven G, Swinnen JV. Indirect mechanisms and cascades of androgen action. Mol Cell Endocrinol. 1999;151:205–12. doi: 10.1016/s0303-7207(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 27.Heemers HV, Verhoeven G, Swinnen JV. Androgen activation of the sterol regulatory element-binding protein pathway: Current insights. Mol Endocrinol. 2006;20:2265–77. doi: 10.1210/me.2005-0479. [DOI] [PubMed] [Google Scholar]
- 28.Heemers HV, Regan KM, Dehm SM, Tindall DJ. Androgen induction of the androgen receptor coactivator four and a half LIM domain protein-2: evidence for a role for serum response factor in prostate cancer. Cancer Res. 2007;67:10592–9. doi: 10.1158/0008-5472.CAN-07-1917. [DOI] [PubMed] [Google Scholar]
- 29.Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–7. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Chen G, Streb JW, et al. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–99. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–68. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franco CA, Mericskay M, Parlakian A, et al. Serum response factor is required for sprouting angiogenesis and vascular integrity. Dev Cell. 2008;15:448–61. doi: 10.1016/j.devcel.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Korenchuk S, Lehr JE, MClean L, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001:163–8. [PubMed] [Google Scholar]
- 35.Gupta V, Bhasin S, Guo W, et al. Effects of dihydrotestosterone on differentiation and proliferation of human mesenchymal stem cells and preadipocytes. Mol Cell Endocrinol. 2008;296:32–40. doi: 10.1016/j.mce.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kube DM, Savci-Heijink CD, Lamblin AF, et al. Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol Biol. 2007;21:8–25. doi: 10.1186/1471-2199-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epstein JI. An update of the Gleason grading system. J Urol. 2010;183:433–40. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 38.Swanson GP, Thompson IM, Basler J. Current status of lymph node-positive prostate cancer: Incidence and predictors of outcome. Cancer. 2006;107:439–50. doi: 10.1002/cncr.22034. [DOI] [PubMed] [Google Scholar]
- 39.Darby S, Sahadevan K, Khan MM, Robson CN, Leung HY, Gnanapragasam VJ. Loss of Sef (similar expression to FGF) expression is associated with high grade and metastatic prostate cancer. Oncogene. 2006;25:4122–7. doi: 10.1038/sj.onc.1209428. [DOI] [PubMed] [Google Scholar]
- 40.Zisman-Rozen S, Fink D, Ben-Izhak O, et al. Downregulation of Sef, an inhibitor of receptor tyrosine kinase signaling, is common to a variety of human carcinomas. Oncogene. 2007;26:6093–8. doi: 10.1038/sj.onc.1210424. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M, Sumizawa T, Mutoh M, et al. Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 2000;60:1312–1316. [PubMed] [Google Scholar]
- 42.Riddick AC, Shukla CJ, Pennington CJ, et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer. 2005;92:2171–80. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu YT, Han HY, Leung SC, et al. CDC25A functions as a novel Ar corepressor in prostate cancer cells. J Mol Biol. 2009;385:446–56. doi: 10.1016/j.jmb.2008.10.070. [DOI] [PubMed] [Google Scholar]
- 44.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106:2824–8. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 47.Zuloaga DG, Puts DA, Jordan CL, Breedlove SM. The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Horm Behav. 2008;53:613–26. doi: 10.1016/j.yhbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zofková I. Hormonal aspects of the muscle-bone unit. Physiol Res. 2008;57 (Suppl 1):S159–169. doi: 10.33549/physiolres.931501. [DOI] [PubMed] [Google Scholar]
- 49.Fraser SP, Pardo LA. Ion channels: functional expression and therapeutic potential in cancer. Colloquium on Ion Channels and Cancer. EMBO Rep. 2008;9:512–5. doi: 10.1038/embor.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.