Abstract
Experiments in vivo and in cell culture demonstrated that oestradiol induces dopamine β-hydroxylase (DBH) gene transcription. Here we examined oestrogen-responsive elements of the rat DBH gene promoter to characterize the mechanisms of oestradiol-induced DBH transcription. Various mutations and deletions of DBH promoter reporter constructs were tested for responsiveness to 17 β-oestradiol (E2). Mutation of the half palindromic oestrogen response element (ERE) at position −759 reduced the response to E2 in PC12 cells co-transfected with ERα indicating a functional role for this motif. In cells co-transfected with ERβ, mutations at the −759 site were unresponsive to E2. To characterize the additional E2 responsive elements, mediated by ERα, the DBH promoter was truncated to the proximal 249 or 200 nucleotides upstream of the transcription start site. Despite either truncation, 10 nM E2 still elicited about a two-fold induction of DBH promoter activity. Mutation of a possible ERE like sequence at −59 had no effect. The lack of a functional ERE in the proximal region of the rat DBH promoter despite E2-mediated DBH promoter activity, suggests regulation by a non-classical mechanism, such as a membrane-initiated signaling pathway. Moreover, the induction of DBH promoter activity and the rise in DBH mRNA levels were observed within hours. To determine whether membrane-initiated E2 signaling is involved in rat DBH gene transcription, a membrane impermeable E2 conjugate, E2BSA, was used. Incubation with E2BSA induced luciferase promoter activity and elicited a significant rise in DBH mRNA levels in the ERα transfected cells. The findings indicate two different mechanisms whereby DBH transcription is regulated by E2 in the presence of ERα. The results implicate both genomic and membrane-initiated mechanisms, mediated by ERα, in E2-induced DBH gene transcription.
Introduction
Oestrogens play a role in sex-specific differences observed in many disorders. For example, men have a greater risk of cardiovascular disease and hypertension than premenopausal women, with equal incidence between men and postmenopausal women, whereas women have a greater risk of depression, eating disorders and nicotine induced lung cancer (1–4). The role of oestrogens in the regulation of these sex-specific differences is likely by acting on numerous targets. The dopamine β-hydroxylase (DBH) gene has been implicated as one such target.
The DBH gene encodes the catecholamine biosynthetic enzyme that catalyzes the hydroxylation of dopamine to form norepinephrine (NE). NE is an important hormone and neurotransmitter, controlling cardiovascular functions, such as blood pressure, as well as other functions including memory, learning, attention, mood, pain, and the stress response (5–9). Sex differences are found in the morphology of locus coeruleus (LC), the center of the majority of the brain’s noradrenergic neurons in adult rats and humans (10). In female rats, the volume of the LC is greater and contains more DBH immunoreactive neurons than in male rats (11, 12). Plasma DBH protein activity, which correlates with DBH protein levels (13), has been shown to be greater in female rats throughout the lifetime, as compared to males (14).
We previously demonstrated that injections of oestradiol benzoate (EB) to ovariectomized rats elevate DBH mRNA levels in the LC (15) as well as in the adrenal medulla (16). Furthermore, treatment of PC12 cells, exogenously expressing either oestrogen receptor (ER) α or β, with 17 β-oestradiol (E2) induces DBH promoter activity, indicating that E2 regulation of DBH occurs at the transcriptional level (15, 17).
Oestrogens can regulate gene transcription by binding to ERs, which function as ligand-activated transcription factors. Oestrogen-induced modulation of transcription through ERs occurs via several different mechanisms (reviewed in (18, 19)). In the classical mechanism, oestrogen bound to ERs induces dimerization and subsequent binding to oestrogen response elements (EREs) of target gene promoters. ERs can also regulate transcription by an ERE-independent mechanism, whereby ERs tether to transcription factors via protein-protein interactions for binding to target gene promoters via cognate cis-regulatory elements. ERs can also be activated by a ligand-independent mechanism via phosphorylation by growth factor-activated protein kinase cascades, leading to binding to EREs of target gene promoters. Whereas the effects of these genomic mechanisms are often observed on the time scale of hours to days, oestrogens can also act by a more rapid non-genomic mechanism initiated by ERs localized at the plasma membrane [reviewed in (20, 21)]. These membrane ERs activate signaling cascades leading to second messenger production and subsequent transcription factor phosphorylation, and therefore modulate gene expression. The genomic and non-genomic mechanisms may thus converge to potentiate transcriptional regulation by oestrogens (19, 22).
Although we have previously shown that E2 induces DBH transcription both in vivo and in cell culture (15, 17), the underlying mechanisms are not yet known. In this study, we aimed to identify the oestrogen-responsive elements of the rat DBH gene promoter and to characterize the mechanisms of E2-induced DBH transcription. The results indicate the importance of both classical and non-classical mechanisms in ERα-mediated induction of DBH gene transcription in response to E2.
Materials and Methods
PC12 cell culture and maintenance
PC12 cells (23) were grown in 100 mm tissue culture dishes (Falcon, Lincoln Park, NJ) with Dulbecco's Modified Eagle's Medium (DMEM, Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA), 5% heat inactivated horse serum (Gemini Bio-Products, Woodland, CA) and 0.5% penicillin-streptomycin (Gibco BRL, Grand Island, NY) at 37°C in a humidified incubator at 7% CO2 with the media changed every other day as previously described (15, 24).
To reduce oestrogens or oestrogenic compounds in the media, cells were pretreated for at least one day at approximately 50% density in phenol red free DMEM (Gibco BRL, Grand Island, NY) supplemented with 10% fetal bovine serum, 5% heat inactivated horse serum, and 0.5% penicillin-streptomycin in 6-well tissue culture plates (Falcon, Lincoln Park, NJ). This media was replaced with stripped media, containing phenol red free DMEM supplemented with 10% charcoal stripped fetal bovine serum (Atlanta Biologicals, Norcross, GA), 5% dialyzed donor horse serum (Gibco BRL, Grand Island, NY), and 0.5% penicillin-streptomycin one day prior to transfection.
DBH promoter constructs and site direct mutagenesis
The rDBH(−1624/+21)/Luc plasmid, containing the first 1624 bp of the rat DBH promoter relative to the transcriptional start site (McMahon and Sabban 1992), fused to the firefly luciferase (Luc) gene in the pGL3-basic vector (Promega, Madison, WI), was constructed by digesting the rDBH(−2236/+21)/Luc plasmid (Serova et al. 2002) with KpnI, followed by religation. The rDBH(−249/+21)/Luc construct was made as previously described (25).
The mutant (mut) reporter constructs −759ERE/Mut1 and −759ERE/Mut2 were generated using the wild-type plasmid rDBH(−1624/+21)/Luc as the template for PCR using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The deletion (del), mutation, and truncation reporter constructs −56ERE/Del, −56ERE/LeftMut, −56ERE/RightMut, rDBH(−200/+21)/Luc, and rDBH(−100/+21)/Luc were generated using the wild-type plasmid rDBH(−249/+21)/Luc as the template. The forward and reverse oligonucleotides used to generate the different constructs are shown in Table 1. All plasmids were isolated using the EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA) and dissolved in TE buffer. All deletions, mutations, and truncations were confirmed by sequencing (Davis Sequencing, Davis, CA).
Table 1.
Primers for deletion, mutation, and truncation of rDBH/Luc reporter constructs
| rDBH/Luc Construct | Forward (top) and reverse (bottom) primers (listed 5’ to 3’) |
|---|---|
| −759 ERE/Mut1 | GCTGAGCAATAGGAAGGTCTCAACTTCGGACTGGTGG CCACCAGTCCGAAGTTGAGACCTTCCTATTGCTCAGC |
| −759 ERE/Mut2 | GCTGAGCAATAGGAAGTACAAAACTTCGGACTGGTGG CCACCAGTCCGAAGTTTTGTACTTCCTATTGCTCAGC |
| −56 ERE/Del | CCAGACAAATGTGATTACAACCCCACCGAAC GTTCGGTGGGGTTGTAATCACATTTGTCTGG |
| −56 ERE/LeftMut | CCACCAGACAAATGTGATTACTTATGGCCTGGCCCAACCCCACC GGTGGGGTTGGGCCAGGCCATAAGTAATCACATTTGTCTGGTGG |
| −56 ERE/RightMut | GACAAATGTGATTAGGTACAGCCTTCTACAACCCCACCGAACAGACATAA TTATGTCTGTTCGGTGGGGTTGTAGAAGGCTGTACCTAATCACATTTGTC |
| rDBH(−200/+21)/Luc | CGAGGTCGACGGTATCATGGCCATCTGCTTC GAAGCAGAATGGCCATGATCACGTCGACCTCG |
| rDBH(−100/+21)/Luc | GAGGTCGACGGTATTGGGAGAGCCAC GTGGCTCTCCCAATACCGTCGACCTC |
Transient transfection of PC12 Cells
Transient co-transfection of PC12 cells with rDBH/Luc reporters and expression vectors for mouse ERα (pcDNA3/mERα) or ERβ (pSG5/mERβ), kindly provided by Dr. E. R. Levin (University of California, Irvine, CA), was performed as previously described (15, 17). In some experiments, cells were additionally co-transfected with β-galactosidase expression vector (pSV β-gal; Promega, Madison, WI) to normalize the transfection efficiencies.
For transfection, 1.5 µg of luciferase reporter plasmid DNA, 1.5 µg expression vector plasmid DNA (ERα or ERβ) and 0.75 µg pSV β-gal DNA were diluted to 150 µl in phenol red-free DMEM, and mixed with SuperFect Transfection Reagent (Qiagen, Valencia, CA) in the ratio of 1:2 (7.5 µl SuperFect for 3.75 µg total DNA), according to the manufacturer’s protocol. After 10 min the transfection complexes were diluted with 600 µl of stripped media and the total volume was gently added to the cells and incubated at 37°C in a humidified incubator at 5% CO2. After 3 h the complexes were replaced with fresh stripped media and the cells were incubated for an additional 24 h.
Preparation of stock E2BSA
β-estradiol-6-(O-carboxy-methyl) oxime-bovine serum albumin (E2BSA) (Sigma, St. Louis, MO) was carefully prepared just prior to use to remove any free, contaminating oestradiol, as previously described (26, 27). Briefly, a 10 µM E2BSA stock solution was prepared in 50 mM Tris-HCl, pH 8.5 and filtered through a Microcon YM-3 (membrane molecular weight cut-off of 3,000) centrifugal filter device, according to conditions described previously (27).
Pharmacological treatments and luciferase assay
After transfection for 24 h, the cells were treated with either 17 β-oestradiol (E2; Sigma, Saint Louis, MO) dissolved in ethanol, not exceeding 0.01% of the final concentration, or purified E2BSA. Controls for E2 and E2BSA treatments were the vehicle or BSA in the vehicle, respectively. The cells were then further incubated under the same conditions for various times from 1–48 h (see figure legends). After incubation, the cells were harvested in phosphate buffered saline and collected by centrifugation. The lysates were prepared by re-suspending the cell pellet in Passive Lysis Buffer (Promega, Madison, WI). For luciferase reporter assays, an aliquot of each lysate was mixed with five volumes of luciferase assay reagent (Promega, Madison, WI) and assayed for firefly luciferase activity within the linear range by immediately measuring in a luminometer (TD-20/20 Turner, Sunnyvale, CA). β-galactosidase activity was assayed using the β-Galactosidase Enzyme Assay System by Promega (Madison, WI) according to the manufacturer’s protocol. Otherwise, the concentration of protein in the lysates was determined with Bio-Rad Protein Assay Reagent (Bio-Rad, Hercules, CA) using Synergy HT plate reader (BioTek, Winooski, VT). Luciferase activity was normalized to the β-galactosidase activity or protein levels in each lysate. Each experimental group contained 5–6 replicate culture wells and each experiment was repeated at least twice.
Isolation of RNA from PC12 cells, northern blot analysis, and quantitative RT-PCR
For RNA isolation, the cells were homogenized in RNA-Stat-60 (Tel-Test, Inc., Friendswood, TX) according to the manufacturer’s protocol. Northern blot analysis was performed as previously described (16). Briefly, RNA was fractionated on 1.2% agarose gels and transferred to Gene-Screen Plus membranes (New England Nuclear, Boston, MA). Hybridization was performed with a [32P]α-dCTP rat DBH cDNA probe and DNA for 18S rRNA as a control in ULTRA-Hyb solution (Ambion, Austin, TX) at 42°C. The blots were washed and exposed to BioMax film (Kodak, Rochester, NY) within the linear signal range. The blots were subsequently re-probed using a probe for 18S rRNA. Autoradiograms were analyzed by Image-Pro-Analysis software (Media Cybernetics). DBH mRNA levels were normalized to 18S rRNA levels.
For quantitative RT-PCR, total RNA (800 ng), quantitated by spectrophotometric analysis, was reverse transcribed in 5 µl total volume using AMV reverse transcriptase (Roche, Indianapolis, IN). The 5 µl RT mixture contained 1× RT buffer (Roche, Indianapolis, IN), 1 mM dNTP mix, 5 units of RNAse inhibitor (Roche, Indianapolis, IN), 1 µM specific reverse transcription primer (5’-AGGCTGCAAGGCTTCTGTGATGGC-3’) and 5 units of AMV reverse transcriptase (Roche, Indianapolis, IN).
PCR reactions, with a total volume of 20 µl, were set up with a final concentration of 1× LightCycler DNA Master SYBR Green I, 0.5 µM of each the forward (5’-CCACGCCATGCAGTTCTTCACCA-3’) and reverse (5’-AGGCTGCAAGGCTTCT-GTGATGGC-3’) primers, 3 mM MgCl2, and 2 µl of the standard DBH cDNA or cDNA with unknown concentration. A standard curve plotted using serial dilutions from 2 ng to 0.2 pg of DBH cDNA was used for quantification by the Fit Points method. The specificity of the amplified target sequences was confirmed with melting curve analysis by comparing its melting temperature with the melting temperatures of the standards as a positive control. The values for DBH mRNA were normalized to levels of total RNA.
Statistical analysis
Data are presented as mean ± SEM of representative experiments. Statistical significance was evaluated by Student’s t-test for two experimental groups or by one way ANOVA followed by Fisher’s post-hoc comparisons for more than two experimental groups. A value of p ≤ 0.05 was considered significant.
Results
Mapping oestrogen responsive elements in the rat DBH promoter
We have previously shown that treatment of PC12 cells with E2 induces reporter activity under the control of the proximal 2236 bases of the rat DBH promoter [rDBH(−2236/+21)/Luc] (15) Analysis of the cis-regulatory elements in this promoter region identified perfect half palindromic EREs (GGTCA or TGACC) at positions −1968, −759 and −739, relative to the transcriptional start site [MatInspector and Transcription Element Search Software (TESS)]. Truncation of this construct to contain the proximal 1624 bases of the rat DBH promoter [rDBH(−1624/+21)/Luc] demonstrated a similar ability to induce luciferase activity after 2.5 or 10 nM E2 treatment in cells co-transfected with ERα (Fig. 1B) or ERβ (data not shown).
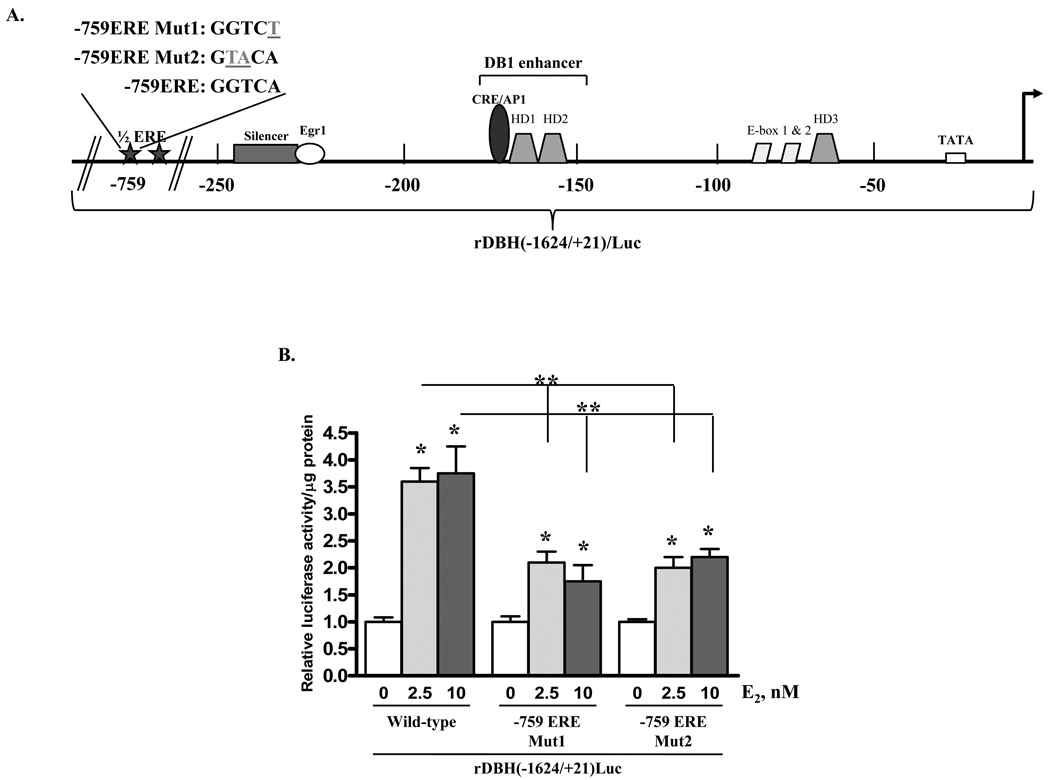
Figure 1.
17 β-oestradiol induces rat DBH promoter-driven luciferase activity partially through the ERE at position −759 in PC12 cells transfected with ERα. (A) Schematic of functional cis-regulatory elements of the rat DBH promoter. (B) PC12 cells were co-transfected with ERα expression vector and either wild-type rDBH(−1624/+21)/Luc or rDBH(−1624/+21)/Luc mutated at the −759 ERE (−759ERE/Mut1 or −759ERE/Mut2), as shown in (A). Cells were treated with 0, 2.5, or 10 nM E2 for 24 h and assayed for luciferase activity. Levels of luciferase activity were normalized to protein levels in each sample and are expressed relative to the respective untreated control taken as 1. * p≤0.05 compared to untreated control; ** p≤0.05 compared to wild-type rDBH(−1624/+21)/Luc.
To determine whether the half ERE at −759 is functional, the GGTCA sequence at −759 of rDBH(−1624/+21)/Luc was mutated to either GGTCT or GTACA (Fig. 1A). In PC12 cells co-transfected with ERα expression vector, treatment with 2.5 and 10 nM E2 were able to induce luciferase activity under the control of both mutated DBH promoter constructs, but the induction was significantly reduced compared with the wild-type DBH promoter construct (Fig. 1B). This indicates that the site at −759 is functional and plays a role in the response to E2 in the presence of ERα. In cells co-transfected with ERβ, mutants of the −759 site in the rDBH(−1624/+21)/Luc vector, in contrast to the wild type promoter, were unresponsive to treatment with 2.5 or 10 nM E2 (data not shown), indicating that this site is sufficient for the response to E2 with ERβ. However, at least one additional oestrogen responsive motif exists downstream of the −759 position of the rat DBH promoter for the response with ERα. Therefore, we focused on identifying the additional ERα-dependent oestrogen responsive motifs of the rat DBH promoter.
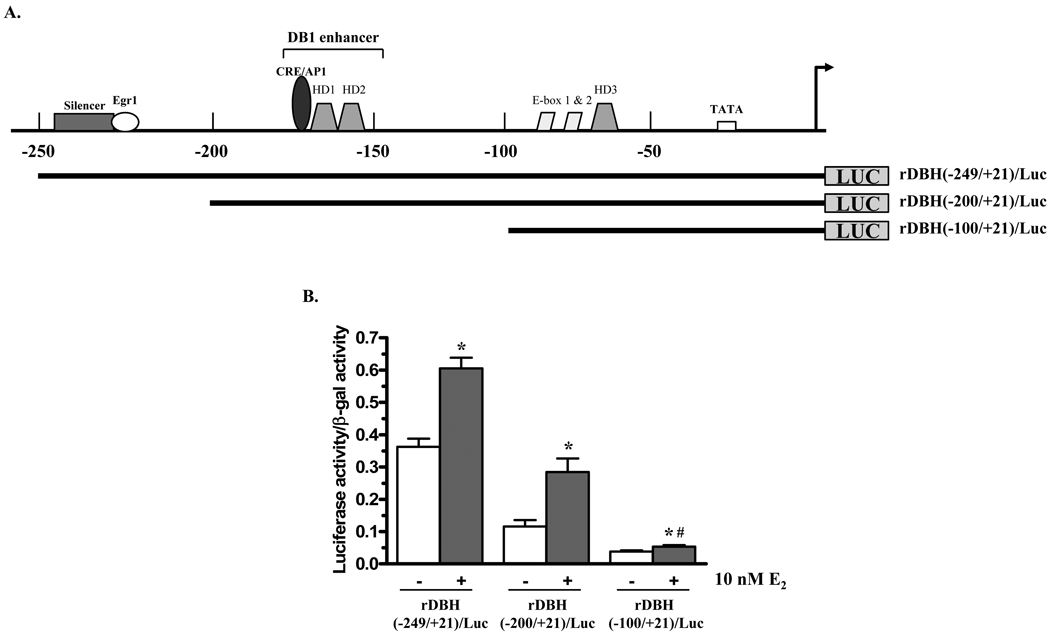
The rDBH(−1624/+21)/Luc plasmid was truncated to determine the additional ERα-dependent oestrogen responsive motif(s) of the rat DBH promoter (Fig. 2A). The constructs with the proximal 249 or 200 nucleotides upstream of the transcription start site display about two-fold elevation in reporter activity in response to 10 nM E2. With rDBH(−100/+21)/Luc reporter construct the response to E2, while still significant, was greatly reduced (Fig. 2B).
Figure 2.
Effect of truncation of the rat DBH promoter on response to E2. (A) Schematic of truncations of DBH promoter. (B) PC12 cells were co-transfected with ERα and β-galactosidase (β-gal) expression vectors and either rDBH(−249/+21)/Luc, rDBH(−200/+21)/Luc, or rDBH(−100/+21)/Luc. Cells were treated with 0 or 10 nM E2 for 24 h and assayed for luciferase activity. Luciferase activity is normalized to β-gal activity and expressed relative to untreated controls. * p≤0.05 compared to respective untreated control; # p≤0.05 compared to rDBH(−200/+21)/Luc treated with E2.
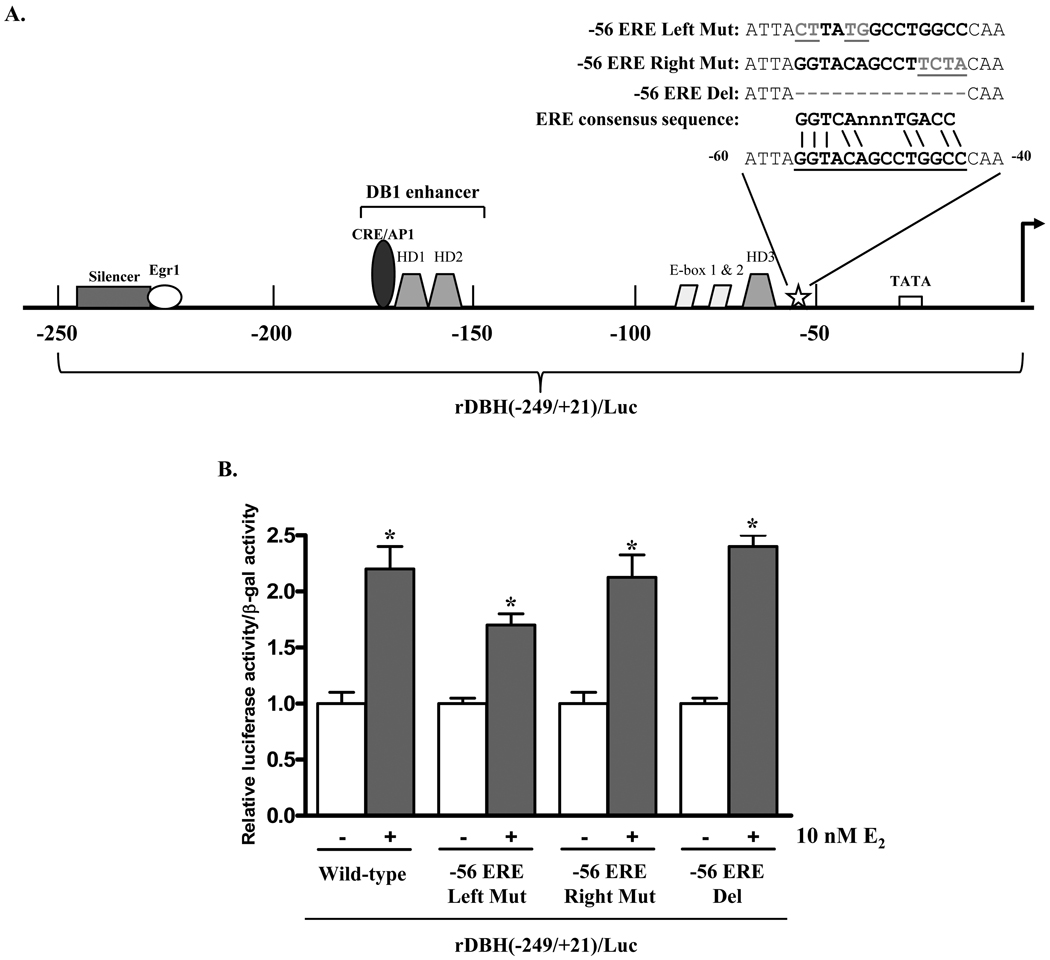
Further analysis of the rat DBH promoter identified a motif with some homology to an ERE (28) at position −56 to −43 (Fig. 3A). To determine the functionality of this element, site-directed mutagenesis was used to mutate or delete this region of the rDBH(−249/+21)/Luc plasmid. Upon treatment with 10 nM E2, luciferase activity was induced by about two-fold with all three mutants or with the wild type plasmid (Fig. 3B). Therefore, the −56 to −43 site is not a functional ERE.
Figure 3.
The putative ERE at −56 to −43 is not a functional ERE. (A) Schematic of mutations of DBH promoter. (B) PC12 cells were co-transfected with ERα and β-gal expression vectors and either wild-type rDBH(−249/+21)/Luc or mutant rDBH(−249/+21)/Luc, mutated at the −56 ERE (−56ERE/Del, −56ERE/Right Mut or −56ERE/Left Mut). Cells were treated with 0 or 10 nM E2 for 24 h and assayed for luciferase activity. Luciferase activity was normalized to β-gal activity and is expressed relative to the respective untreated control taken as 1. * p≤0.05 compared to untreated control.
Time course of the changes with oestradiol
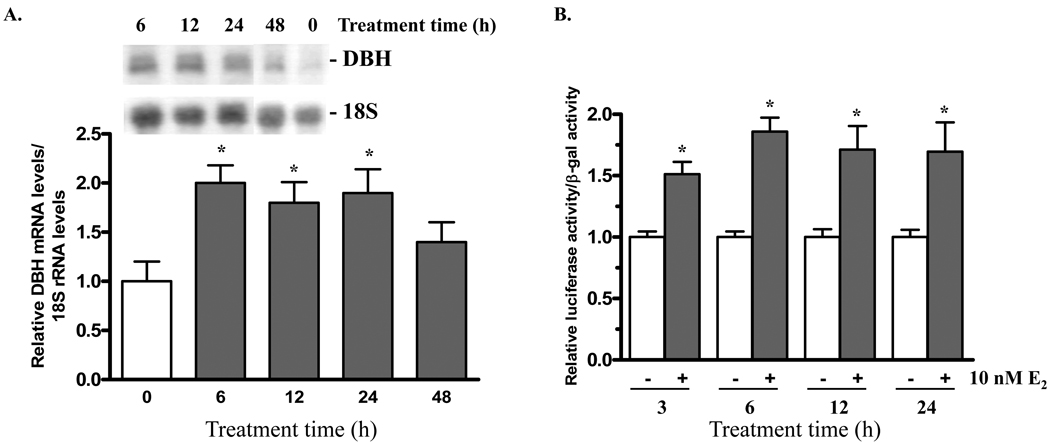
The time course of the response of DBH mRNA and DBH promoter activity with the short rDBH(−200/+21)/Luc plasmid to E2 was examined in PC12 cells transfected with ERα expression vector and treated with 10 nM E2. DBH mRNA levels are elevated approximately two-fold after 6, 12, and 24 h of treatment with 10 nM E2 (Fig. 4A). By 48 h the levels were no longer increased.
Figure 4.
17 β-oestradiol induces increased rat DBH promoter activity and elevated mRNA levels within hours. (A) PC12 cells were transfected with ERα expression vector and treated with 10 nM E2 for 0, 6, 12, 24, or 48 h. Total RNA was isolated and analyzed by northern blot. DBH mRNA levels are normalized to 18S rRNA levels and expressed relative to the 0 h group and taken as 1. n = 4–5 replicates per group * p≤0.05 compared to 0 h control. (B) PC12 cells were co-transfected with ERα and β-gal expression vectors and rDBH(−200/+21)/Luc. Cells were treated with 0 or 10 nM E2 for 3, 6, 12 or 24 h. Luciferase activity was normalized to β-gal activity and is expressed relative to the respective untreated control taken as 1. * p≤0.05 compared to untreated control.
The promoter activity was determined at various times after addition of E2 to PC12 cells co-transfected with rDBH(−200/+21)/Luc and ERα. A rise in luciferase activity was evident by 3 h and remained significant during 24 h (Fig. 4B).
Involvement of a membrane-initiated oestradiol signaling pathway
The lack of a functional ERE in the proximal region of the rat DBH promoter despite E2-mediated DBH promoter activity, and the relatively rapid induction of promoter activity suggests regulation by a non-classical mechanism, such as a membrane-initiated signaling pathway.
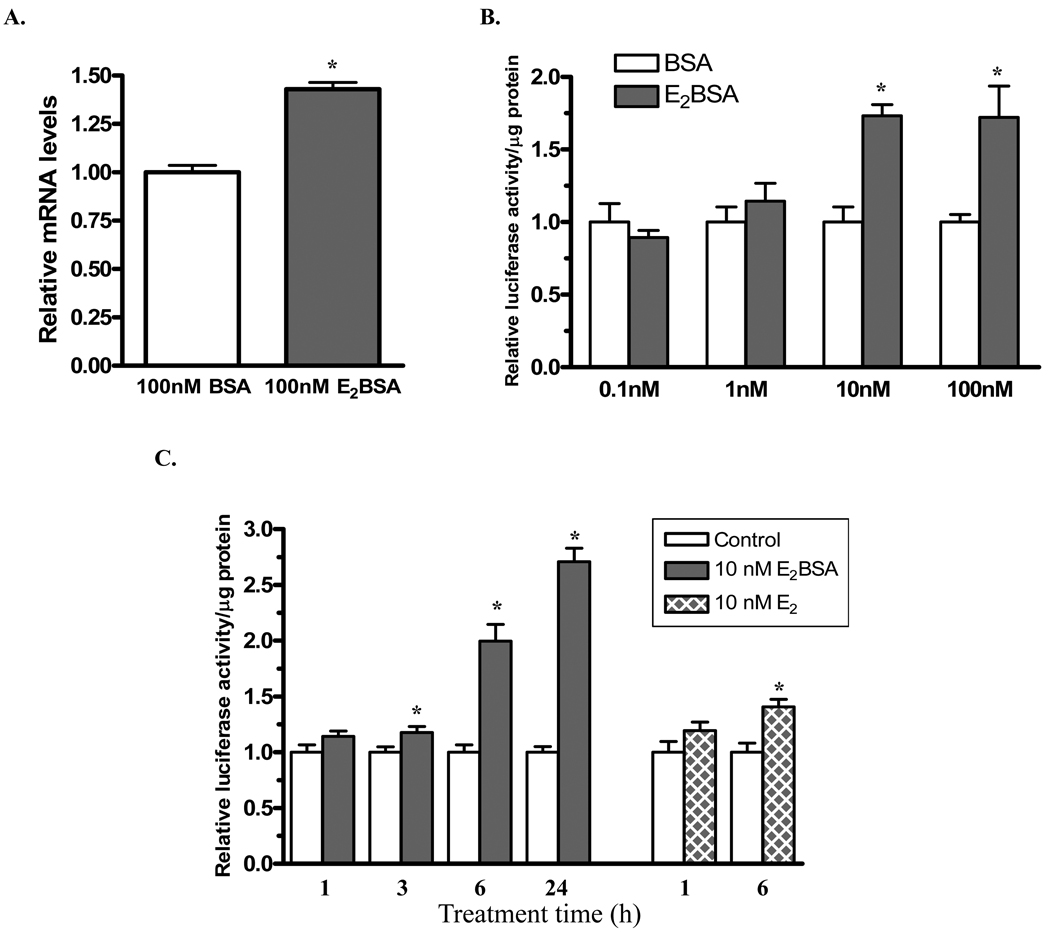
To determine whether membrane-initiated E2 signaling is involved in rat DBH gene transcription, a membrane impermeable E2 conjugate, E2BSA, was used. As in our previous study (27), even 100 nM E2BSA did not induce a luciferase reporter construct driven by a promoter containing three canonical EREs in tandem, showing that there was no dissociated free E2 (data not shown). PC12 cells were transfected with ERα expression vector then treated with 100 nM E2BSA or BSA for 24 h. DBH mRNA levels were elevated compared to BSA-treated controls (Fig. 5A) demonstrating that E2, acting by an ERα-mediated membrane-initiated signaling pathway can induce endogenous DBH mRNA levels.
Figure 5.
A membrane impermeable form of estradiol, E2BSA, induces rat DBH promoter activity and increases rat DBH mRNA levels. (A) PC12 cells were transfected with ERα expression vector and treated with 100 nM E2BSA or BSA for 24 h. Total RNA was isolated and analyzed by quantitative RT-PCR. DBH mRNA levels are normalized to total RNA and expressed relative to the BSA-treated control taken as 1. n=6 *p≤0.05 compared to BSA-treated control.
(B) PC12 cells were co-transfected with ERα expression vector and rDBH(−249/+21)/Luc, treated with 0.1, 1, 10, or 100 nM E2BSA or BSA for 24 h and assayed for luciferase activity. Luciferase activity is normalized to total protein and expressed relative to the respective BSA-treated control taken as 1. *p≤0.05 compared to BSA-treated control. (C) PC12 cells were co-transfected with ERα expression vector and rDBH(−249/+21)/Luc, treated with 10 nM E2BSA or BSA (control), or 10 nM E2 or vehicle (control), for 1, 3, 6, or 24 h and assayed for luciferase activity. Data are normalized to total protein and expressed relative to the respective control and taken as 1. *p≤0.05 compared to respective control.
Next the effects of various concentrations of E2BSA, from 0.1 to 100 nM, were examined for their effect on DBH promoter activity. Both 10 and 100 nM E2BSA elevated luciferase promoter activity in PC12 cells co-transfected with ERα expression vector and rDBH(−249/+21)/Luc (Fig. 5B). These results demonstrate that DBH gene transcription is regulated by a membrane-initiated E2 signaling pathway via an element within the proximal 249 bases of the rat DBH promoter in PC12 cells exogenously expressing ERα. The time course of the 10 nM E2BSA triggered induction of DBH promoter activity shows a small, but significant elevation after 3 h with induction of nearly 2 and 3 fold after 6 and 24 h respectively (Fig. 5C).
Discussion
The findings of this study indicate two different mechanisms whereby DBH transcription is regulated by oestradiol in the presence of ERα. Mutational analysis of the rat DBH promoter reveals that E2-induced DBH transcription is mediated by at least one functional ERE half-site at −759 as well as additional element(s) within the proximal 249 bp of the rat DBH promoter. The latter is mediated by membrane impermeable E2-initiated signalling. Together, these results implicate both genomic and membrane-initiated mechanisms, mediated by ERα, in the modulation of E2-induced DBH gene transcription.
The findings pinpoint the importance of the half palindromic ERE at −759 in the DBH promoter. Although half EREs generally confer weak oestrogen responsiveness, there are examples for which transcription is induced by two or more ERE-half palindromic elements, for example corticotrophin releasing hormone, platelet activating receptor transcript 2 and prothymosin α [reviewed in (28)]. Chromatin immunoprecipitation in genome wide analysis revealed diverse ERα binding sites, with about 25% of them identified as ERE half sites (29). While constructs with a mutated half palindromic ERE at −759 in the DBH promoter still responded to E2 in cells transfected with ERα, they were unresponsive in cells transfected with expression vector for ERβ. These results suggest differences in the response of the DBH promoter to oestrogens in the presence of ERα or ERβ. We have previously shown that treatment with E2 induces DBH promoter activity and this induction is greater and more sensitive to E2 with ERα than ERβ (17). In this regard, our recent experiments showed that injections of rats with an ERα agonist (PPT), but not an ERβ agonist, effectively elevated DBH mRNA levels in the LC and in the rostral medial and caudal nucleus of the solitary tract (NTS) (30). This is despite the fact that both ERα and ERβ are expressed in these noradrenergic neurons (31).
Although the −759 half palindromic ERE of the rat DBH promoter is functional, our results indicate that the GGTCA site at −1968 is unimportant for the response to oestrogen since a similar response to E2 was observed with the construct containing 2236 or 1624 bp of the promoter (15). Further upstream GGTCA sequences in the rat, such as those at −2268, −2304, −2538 and −2670, as well as any additional upstream sites, were not examined. The promoter region containing the −759 half ERE in the rat is not homologous to any region of the human DBH promoter. There are, however, numerous sites in the human DBH promoter with the same GGTCA half palindromic ERE sequence. For example, within the proximal 5 kb of the human DBH promoter, these sites are located at −413, −3192, −3800, −4406, and −4924, with additional sites located further upstream. Similarly, this sequence is also found in the proximal 5 kb of the mouse DBH promoter (−1113, −1820, −2381, −2654, −3308, −3840, −4217, −4525, and −4633). Which, if any, of these GGTCA half palindromic ERE sequences are functional remains to be determined.
In addition to the functional role of the −759 half ERE in the rat, the results of this study show that truncation to the proximal −249 bp of the rat DBH promoter is still responsive to E2 or E2BSA in the presence of ERα. There is a great deal of homology between rat, mouse and human DBH genes in the approximately 300 bp upstream of the transcription start site. Functional elements in this region of the rat DBH promoter include CRE/AP1, Egr1, homeodomain, and E box motifs, many of which are highly conserved in the human DBH promoter [reviewed in (7)]. Although the element(s) responsible for mediating E2-induced DBH gene transcription via membrane initiated signaling in the rat is currently unknown, the high degree of homology between the proximal region of the rat and human DBH promoter support the likelihood of similar mechanisms.
The findings reveal the importance of membrane initiated signalling in the oestrogen triggered regulation of DBH transcription with ERα. It is likely that ERα is acting as the membrane E2 receptor, as exogenously expressed ERα has been reported to localize to the plasma membrane of PC12 cells (32, 33). While these findings are so far restricted to PC12 cells and need to be confirmed in normal noradrenergic and adrenergic cells, they emphasize the importance of both classical genomic and membrane initiated mechanisms. The physiological importance of membrane initiated oestrogen signalling is increasingly apparent, for example in preventing bone loss or by providing cardioprotection in mice (34, 35). The in vivo effects on catecholaminergic systems remain to be determined. However, the actions at the membrane are likely integrated with classical effects. In neuroblastoma cells, membrane initiated actions of oestrogen, by E2BSA, were found to potentiate subsequent transcriptional activity at a classical ERE motif (22). Since our findings display that DBH possesses both types of responses with ERα, this may enable DBH to manifest a potentiated response to oestrogens in ERα expressing cells, and may explain our finding that an ERα agonist is especially effective in inducing DBH mRNA levels in rat LC and NTS (30).
Oestrogens however have paradoxical effects on catecholaminergic systems. While they enhance basal DBH transcription they reduce or attenuate the response of DBH gene expression to stress (16, 30). Selective ERα agonists were especially effective in attenuating the response of DBH in the LC and NTS to even repeated immobilization stress (30). The finding of membrane initiated and classical responses in regulation of DBH may help explain this paradox. Long term exposure to estrogens, especially via membrane bound ERα may modulate the signalling pathways responsible for the elevation of DBH gene expression upon exposure to stress, and may be responsible for the attenuated autonomic response to stress in women between puberty and menopause [reviewed in (36)].
Our findings are consistent with a membrane-initiated E2 signaling pathway via ERα in PC12 cells which regulates transcription of the tyrosine hydroxylase (TH) gene, which encodes the first and major rate-limiting enzyme in catecholamine biosynthesis (27). E2 or E2BSA elicited rapid phosphorylation of CREB and of extracellular signal regulated kinase (ERK), especially ERK1. Specific inhibitors indicated that both protein kinase A and MEK signaling pathways are required for the response of the TH gene to membrane impermeant E2.
DBH transcription and gene expression are also regulated by activation of protein kinase A activity (37–40). An adjacent CRE/AP1 motif slightly upstream from two homeobox motifs is present in the rat DBH promoter and involved in the response of the DBH promoter to cAMP and to phorbol esters (41). There is transcriptional synergism between the homeodomain proteins Arix/Phox2a and protein kinase A pathway in regulation of DBH transcription (42). A similar CRE/AP1 motif in the human DBH gene binds to CREB (43) and its phosphorylation is implicated in the activation of DBH transcription in response to ethanol (44). However, in the rat DBH promoter, AP1 proteins and to a lesser extent CREB are involved in mediating the transcriptional response to cAMP (45). It remains to be determined if the protein kinase A and/or MEK pathways mediate the membrane initiated response of DBH to E2 in cells expressing ERα. However, the activation of the MEK pathway may not be involved in the induction of DBH by membrane initiated E2 signaling since activation of ERK is actually associated with cytokine suppression of DBH promoter activity, at least in neuroblastoma cells (46) and ERK1/2 is a negative regulator of the homeodomain protein Arix/Phox2a involved in DBH transcriptional regulation (47).
What might be the physiological significance of regulation of DBH transcription by oestradiol? While DBH is not generally considered the rate limiting enzyme in catecholamine biosynthesis under certain circumstances DBH can be rate limiting [rev in (48)]. A recent review (49) summarizes the evidence supporting the hypothesis that extracellular DA in the cerebral cortex originates not only from dopaminergic terminals but also from LC derived noradrenergic ones, and that DA can act not only as a precursor for NE but also as a co-transmitter. Thus an increase in DBH by oestrogen could alter the NE/DA ratio in the medial prefrontal cortex (PFC). This could be very significant as the medial PFC is involved in the regulation of cognitive and emotional processing and the LC noradrenergic projections to this region have modulatory effects on working memory and attention (50).
The regulation of DBH gene expression by oestrogen may be responsible for the greater number of DBH immunoreactive neurons in female than in male rats (11, 12). This could be involved in sex specific differences in vigilance and alertness and in memory.
Overall, the findings of this study provide new mechanistic insight into the physiological and possibly patho-physiological roles of oestrogens on the regulation of DBH transcription and, on a larger scale, regulation of catecholaminergic systems.
Acknowledgements
We are grateful to funding from NIH grant NS28869 and Senior Development Award 0130102 N (to LIS) from the American Heart Association and to Dr. Ellis R. Levin (University of California, Irvine, CA) for the ER expression vectors.
References
- 1.Reckelhoff JF. Gender differences in the regulation of blood pressure. Hypertension. 2001;37(5):1199–1208. doi: 10.1161/01.hyp.37.5.1199. [DOI] [PubMed] [Google Scholar]
- 2.Seeman MV. Psychopathology in women and men: focus on female hormones. Am J Psychiatry. 1997;154(12):1641–1647. doi: 10.1176/ajp.154.12.1641. [DOI] [PubMed] [Google Scholar]
- 3.Stabile LP, Siegfried JM. Estrogen receptor pathways in lung cancer. Current oncology reports. 2004;6(4):259–267. doi: 10.1007/s11912-004-0033-2. [DOI] [PubMed] [Google Scholar]
- 4.Gillies GE, McArthur S. Estrogen actions in the brain and the basis for differential action in men and women: a case for sex-specific medicines. Pharmacol Rev. 62(2):155–198. doi: 10.1124/pr.109.002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Bockstaele EJ, Reyes BA, Valentino RJ. The locus coeruleus: A key nucleus where stress and opioids intersect to mediate vulnerability to opiate abuse. Brain Res. 2010;1314:162–174. doi: 10.1016/j.brainres.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein DS. Stress, Catecholamines and Cardiovascular Disease. Oxford: Oxford University Press; 1995. [Google Scholar]
- 7.Kvetnansky R, Sabban EL, Palkovits M. Catecholaminergic Systems in Stress: Structural and Molecular Genetic Approaches. Physiol Rev. 2009;89:535–606. doi: 10.1152/physrev.00042.2006. [DOI] [PubMed] [Google Scholar]
- 8.Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu Rev Neurosci. 2005;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- 9.Sara SJ. The locus coeruleus and noradrenergic modulation of cognition. Nat Rev Neurosci. 2009;10(3):211–223. doi: 10.1038/nrn2573. [DOI] [PubMed] [Google Scholar]
- 10.Busch C, Braak H, Bohl J, Ohm T. The human nucleus coeruleus in aginag - A sterological analysis of females and males. Neurobiol Aging. 1995;19:1565. [Google Scholar]
- 11.Guillamon A, de Blas MR, Segovia S. Effects of sex steroids on the development of the locus coeruleus in the rat. Brain Res. 1988;468(2):306–310. doi: 10.1016/0165-3806(88)90143-5. [DOI] [PubMed] [Google Scholar]
- 12.Luque JM, de Blas MR, Segovia S, Guillamon A. Sexual dimorphism of the dopamine-beta-hydroxylase-immunoreactive neurons in the rat locus ceruleus. Brain Res Dev Brain Res. 1992;67(2):211–215. doi: 10.1016/0165-3806(92)90221-h. [DOI] [PubMed] [Google Scholar]
- 13.Dunnette J, Weinshilboum R. Human serum dopamine beta-hydroxylase: correlation of enzymatic activity with immunoreactive protein in genetically defined samples. Am J Hum Genet. 1976;28(2):155–166. [PMC free article] [PubMed] [Google Scholar]
- 14.Koudelova J, Mourek J. Influence of age, sex and hypoxia on plasma dopamine-beta-hydroxylase activity in the rat. Physiol Bohemoslov. 1990;39(5):409–416. [PubMed] [Google Scholar]
- 15.Serova L, Rivkin M, Nakashima A, Sabban EL. Estradiol stimulates gene expression of norepinephrine biosynthetic enzymes in rat locus coeruleus. Neuroendocrinology. 2002;75(3):193–200. doi: 10.1159/000048237. [DOI] [PubMed] [Google Scholar]
- 16.Serova LI, Maharjan S, Sabban EL. Estrogen modifies stress response of catecholamine biosynthetic enzyme genes and cardiovascular system in ovariectomized female rats. Neuroscience. 2005;132(2):249–259. doi: 10.1016/j.neuroscience.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 17.Sabban EL, Maharjan S, Nostramo R, Serova LI. Divergent effects of estradiol on gene expression of catecholamine biosynthetic enzymes. Physiol Behav. 2010;99(2):163–168. doi: 10.1016/j.physbeh.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81(4):1535–1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- 19.Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19(4):833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 20.Levin ER. Plasma membrane estrogen receptors. Trends Endocrinol Metab. 2009;20(10):477–482. doi: 10.1016/j.tem.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriarty K, Kim KH, Bender JR. Minireview: estrogen receptor-mediated rapid signaling. Endocrinology. 2006;147(12):5557–5563. doi: 10.1210/en.2006-0729. [DOI] [PubMed] [Google Scholar]
- 22.Vasudevan N, Pfaff DW. Non-genomic actions of estrogens and their interaction with genomic actions in the brain. Front Neuroendocrinol. 2008;29(2):238–257. doi: 10.1016/j.yfrne.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73(7):2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maharjan S, Serova L, Sabban EL. Transcriptional regulation of tyrosine hydroxylase by estrogen: opposite effects with estrogen receptors alpha and beta and interactions with cyclic AMP. J Neurochem. 2005;93(6):1502–1514. doi: 10.1111/j.1471-4159.2005.03142.x. [DOI] [PubMed] [Google Scholar]
- 25.Cheng SY, Serova LI, Glazkova D, Sabban EL. Regulation of rat dopamine beta-hydroxylase gene transcription by early growth response gene 1 (Egr1) Brain Res. 2008;1193:1–11. doi: 10.1016/j.brainres.2007.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi Y, Koslowski M, Bodenner DL. Binding of estrogen receptor with estrogen conjugated to bovine serum albumin (BSA) Nuclear receptor. 2004;2(1):5. doi: 10.1186/1478-1336-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maharjan S, Serova LI, Sabban EL. Membrane-initiated estradiol signaling increases tyrosine hydroxylase promoter activity with ERalpha in PC12 cells. J Neurochem. 2010;112(1):42–55. doi: 10.1111/j.1471-4159.2009.06430.x. [DOI] [PubMed] [Google Scholar]
- 28.Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. Anatomy of the estrogen response element. Trends Endocrinol Metab. 2004;15(2):73–78. doi: 10.1016/j.tem.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3(6):e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serova LI, Harris HA, Maharjan S, Sabban EL. Modulation of responses to stress by estradiol benzoate and selective estrogen receptor agonists. J Endocrinol. 2010;205(3):253–262. doi: 10.1677/JOE-10-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997;388(4):507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Watson CS, Alyea RA, Hawkins BE, Thomas ML, Cunningham KA, Jakubas AA. Estradiol effects on the dopamine transporter - protein levels, subcellular location, and function. Journal of molecular signaling. 2006;1:5. doi: 10.1186/1750-2187-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alyea RA, Laurence SE, Kim SH, Katzenellenbogen BS, Katzenellenbogen JA, Watson CS. The roles of membrane estrogen receptor subtypes in modulating dopamine transporters in PC-12 cells. J Neurochem. 2008;106(4):1525–1533. doi: 10.1111/j.1471-4159.2008.05491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kousteni S, Chen JR, Bellido T, Han L, Ali AA, O'Brien CA, Plotkin L, Fu Q, Mancino AT, Wen Y, Vertino AM, Powers CC, Stewart SA, Ebert R, Parfitt AM, Weinstein RS, Jilka RL, Manolagas SC. Reversal of bone loss in mice by nongenotropic signaling of sex steroids. Science. 2002;298(5594):843–846. doi: 10.1126/science.1074935. [DOI] [PubMed] [Google Scholar]
- 35.Chambliss KL, Wu Q, Oltmann S, Konaniah ES, Umetani M, Korach KS, Thomas GD, Mineo C, Yuhanna IS, Kim SH, Madak-Erdogan Z, Maggi A, Dineen SP, Roland CL, Hui DY, Brekken RA, Katzenellenbogen JA, Katzenellenbogen BS, Shaul PW. Non-nuclear estrogen receptor alpha signaling promotes cardiovascular protection but not uterine or breast cancer growth in mice. J Clin Invest. 120(7):2319–2330. doi: 10.1172/JCI38291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kajantie E, Phillips DI. The effects of sex and hormonal status on the physiological response to acute psychosocial stress. Psychoneuroendocrinology. 2006;31(2):151–178. doi: 10.1016/j.psyneuen.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Serova LI, Gueorguiev V, Cheng SY, Sabban EL. Adrenocorticotropic hormone elevates gene expression for catecholamine biosynthesis in rat superior cervical ganglia and locus coeruleus by an adrenal independent mechanism. Neuroscience. 2008;153(4):1380–1389. doi: 10.1016/j.neuroscience.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamouroux A, Houhou L, Biguet NF, Serck-Hanssen G, Guibert B, Icard-Liepkalns C, Mallet J. Analysis of the human dopamine beta-hydroxylase promoter: transcriptional induction by cyclic AMP. J Neurochem. 1993;60(1):364–367. doi: 10.1111/j.1471-4159.1993.tb05861.x. [DOI] [PubMed] [Google Scholar]
- 39.McMahon A, Sabban EL. Regulation of expression of dopamine beta-hydroxylase in PC12 cells by glucocorticoids and cyclic AMP analogues. J Neurochem. 1992;59(6):2040–2047. doi: 10.1111/j.1471-4159.1992.tb10092.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim KS, Ishiguro H, Tinti C, Wagner J, Joh TH. The cAMP-dependent protein kinase regulates transcription of the dopamine beta-hydroxylase gene. J Neurosci. 1994;14(11 Pt 2):7200–7207. doi: 10.1523/JNEUROSCI.14-11-07200.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaskus J, Greco D, Asnani LP, Lewis EJ. A bifunctional genetic regulatory element of the rat dopamine beta-hydroxylase gene influences cell type specificity and second messenger-mediated transcription. J Biol Chem. 1992;267(26):18821–18830. [PubMed] [Google Scholar]
- 42.Swanson DJ, Adachi M, Lewis EJ. The homeodomain protein Arix promotes protein kinase A-dependent activation of the dopamine beta-hydroxylase promoter through multiple elements and interaction with the coactivator cAMP-response element-binding protein-binding protein. J Biol Chem. 2000;275(4):2911–2923. doi: 10.1074/jbc.275.4.2911. [DOI] [PubMed] [Google Scholar]
- 43.Seo H, Yang C, Kim HS, Kim KS. Multiple protein factors interact with the cis-regulatory elements of the proximal promoter in a cell-specific manner and regulate transcription of the dopamine beta-hydroxylase gene. J Neurosci. 1996;16(13):4102–4112. doi: 10.1523/JNEUROSCI.16-13-04102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hassan S, Duong B, Kim KS, Miles MF. Pharmacogenomic analysis of mechanisms mediating ethanol regulation of dopamine beta-hydroxylase. J Biol Chem. 2003;278(40):38860–38869. doi: 10.1074/jbc.M305040200. [DOI] [PubMed] [Google Scholar]
- 45.Swanson DJ, Zellmer E, Lewis EJ. AP1 proteins mediate the cAMP response of the dopamine beta-hydroxylase gene. J Biol Chem. 1998;273(37):24065–24074. doi: 10.1074/jbc.273.37.24065. [DOI] [PubMed] [Google Scholar]
- 46.Dziennis S, Habecker BA. Cytokine suppression of dopamine-beta-hydroxylase by extracellular signal-regulated kinase-dependent and -independent pathways. J Biol Chem. 2003;278(18):15897–15904. doi: 10.1074/jbc.M212480200. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh MM, Lupas G, Rychlik J, Dziennis S, Habecker BA, Lewis EJ. ERK1/2 is a negative regulator of homeodomain protein Arix/Phox2a. J Neurochem. 2005;94(6):1719–1727. doi: 10.1111/j.1471-4159.2005.03333.x. [DOI] [PubMed] [Google Scholar]
- 48.Cubells JF, Zabetian CP. Human genetics of plasma dopamine beta-hydroxylase activity: applications to research in psychiatry and neurology. Psychopharmacology (Berl) 2004;174(4):463–476. doi: 10.1007/s00213-004-1840-8. [DOI] [PubMed] [Google Scholar]
- 49.Devoto P, Flore G. On the Origin of Cortical Dopamine: Is it a Co-Transmitter in Noradrenergic Neurons? Curr Neuropharmacol. 2006;4(2):115–125. doi: 10.2174/157015906776359559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramos BP, Arnsten AF. Adrenergic pharmacology and cognition: focus on the prefrontal cortex. Pharmacol Ther. 2007;113(3):523–536. doi: 10.1016/j.pharmthera.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]