Abstract
Genome-wide association studies (GWAS) have identified three genomic regions, at 15q24-25.1, 5p15.33 and 6p21.33, which associate with risk of lung cancer. Large meta-analyses of GWA data have failed to find additional associations of genome-wide significance. In this study, we sought to confirm 7 variants with suggestive association to lung cancer (P<10−5) in a recently published meta-analysis. In a GWA dataset of 1,447 lung cancer cases and 36,256 controls in Iceland, three correlated variants on 15q15.2 (rs504417, rs11853991 and rs748404) showed a significant association with lung cancer whereas rs4254535 on 2p14, rs1530057 on 3p24.1, rs6438347 on 3q13.31 and rs1926203 on 10q23.31 did not. The most significant variant, rs748404, was genotyped in additional 1,299 lung cancer cases and 4,102 controls from the Netherlands, Spain and the USA and the results combined with published GWAS data. In this analysis, the T allele of rs748404 reached genome-wide significance (OR=1.15, P=1.1×10−9). Another variant at the same locus, rs12050604, showed association with lung cancer (OR=1.09, 3.6×10−6) and remained significant after adjustment for rs748404 and vice versa. rs748404 is located 140 kb centromeric of the TP53BP1 gene that has been implicated in lung cancer risk. Two fully correlated, non-synonymous coding variants in TP53BP1, rs2602141 (Q1136K) and rs560191 (E353D), showed association with lung cancer in our sample set; however, this association did not remain significant after adjustment for rs748404. Our data show that one or more lung cancer risk variants of genome-wide significance and distinct from the coding variants in TP53BP1 are located at 15q15.2.
Keywords: Lung cancer, genome-wide association studies, GWAS, 15q15.2, TP53BP1
Introduction
Lung cancer is the most common form of cancer and causes more deaths worldwide than any other cancer (1). The single most important factor influencing the risk of lung cancer is smoking. However, studies on lung cancer risk in family members of lung cancer probands have shown that genetic factors also contribute to the risk of developing the disease (2, 3). Furthermore, although smoking is the primary cause of lung cancer, only about 15% of lifelong smokers develop the disease, pointing to a role for genetic susceptibility (4, 5).
In the last two years, genome-wide association studies (GWAS) have yielded variants at three loci that associate with lung cancer in populations of European descent. First, sequence variants within a cluster of nicotinic acetylcholine receptor genes on chromosome 15q25 were simultaneously shown to associate with the number of cigarettes smoked per day (6), nicotine dependence (6, 7) and smoking-related diseases such as lung cancer (6, 8, 9) and peripheral arterial disease (6). The other two loci are at 5p15.33 in a region that includes the TERT and CLPTM1L genes (10-12) and 6p21.33 within the MHC locus (8, 11). Neither locus is associated with smoking behavior.
Recently, Broderick et al. reported a meta-analysis of lung cancer based on 7,560 cases and 8,205 controls, genotyped for 21,620 SNPs (13). Apart from variants at the 3 previously reported loci, no variants showed a significant association with lung cancer after correcting for the number of tests (P<1×10−7). Seven SNPs showed suggestive associations with P values less than 10−5. The objective of this study was to test these variants in several independent lung cancer case control sample sets in order to determine if they could be confirmed as lung cancer associated variants of genome-wide significance.
Materials and Methods
Study populations (summarized in Table 1)
Table 1.
Case control groups used in the study
| Study Group | cases (n) | Average age at diagnosis (range) |
% males (cases) |
controls (n) |
Study type |
|---|---|---|---|---|---|
| Iceland alla | 1,487 | 67 (16-97) | 53 | 36,256 | Population based |
| Iceland genotypedb | 848 | 68 (20-95) | 50 | 36,256 | Population based |
| The Netherlands, Nijmegen | 552 | 62 (25-93) | 81 | 1,832 | Population based |
| Spain, Zaragoza | 548 | 65 (18-92) | 87 | 1,432 | Hospital-based |
| USA, Denver | 196 | 66 (29-85 ) | 71 | 838 | Hospital-based |
|
| |||||
| Total | 2,783 | 40,358 | |||
Total number of Icelandic subjects; genotyped directly or by familial imputation.
Directly genotyped Icelandic subjects only
Iceland
The Icelandic lung cancer study population was previously described (6). In brief, information on all lung cancer cases diagnosed from 1955 to 2008 was obtained from the Icelandic Cancer Registry. Samples from 848 cases were genotyped on the Human Hap300 or HumanCNV370-duo Bead Arrays. In addition to using data from directly genotyped cases, we used a method where genotypes of relatives are used to provide information on lung cancer cases not genotyped (familial imputation) (14). Using genotypes from relatives of 2,172 un-genotyped lung cancer cases, it was possible to infer genotypes that are equivalent to 639 lung cancer cases. The 36,256 controls used in this study consisted of individuals from other ongoing genome-wide association studies at deCODE that have been genotyped with the Human Hap300 or HumanCNV370-duo Bead Arrays. No individual disease group accounts for more than 6% of the total control group. The group of 15,310 smokers were a part of a study on smoking behavior and nicotine dependence that is described in detail in a recent publication (15). Study protocols were approved by the National Bioethics Committee of Iceland and all subjects gave written informed consent.
Nijmegen, the Netherlands
The Dutch study population was previously described (12). The 552 patients with lung cancer were identified through the population-based cancer registry of the Comprehensive Cancer Center IKO, Nijmegen, the Netherlands, and recruited through several independent studies (12). All participants gave informed consent for DNA-related research and linkage with disease registries. The 1,832 cancer-free controls (46% males) were selected from participants of the “Nijmegen Biomedical Study” (NBS) (16). All controls are of self-reported European descent. The study protocols of the NBS were approved by the Institutional Review Board of the RUNMC and all study subjects signed a written informed consent form.
Zaragoza, Spain
The Spanish study population was previously described (12). The 548 lung cancer cases were recruited from the Oncology Department of Zaragoza Hospital in Zaragoza, from June 2006 to June 2009. The 1,432 Spanish controls attended the University Hospital in Zaragoza for diseases other than cancer between November 2001 and May 2007. All patients and controls are of self-reported European descent. Study protocols were approved by the Institutional Review Board of Zaragoza University Hospital. All subjects gave written informed consent.
Denver, Colorado
DNA samples from blood and clinical data were provided by the University of Colorado Cancer Center under COMIRB protocol 08-0380. Blood samples were collected from 1,217 patients diagnosed with different diseases enrolled in any of 20 clinical research trials carried out at Colorado SPORE protocols between 1993 and 2008. Of these 1,217 patients, 246 were lung cancer cases and 971 had never had lung cancer at the time of sample shipment. Lung cancer cases were identified either from data matches with the Colorado Central Cancer Registry or by having malignant lung tissue collected via enrollment in a surgical protocol. Work in this study was limited to cases and controls of self-reported European ancestry (196 cases and 838 controls).
Data from published GWAS studies
In addition to the sample sets described above, we used association results from previously published studies. The study by Broderick et al., from which we selected the 7 SNPs to follow-up, uses sample sets from UK, IARC and Texas and does not overlap with the populations directly genotyped for this study (Iceland, the Netherlands, Spain and USA-Denver) (13). The study by Landi et al., contains data from 13,300 cases and 19,666 controls and this dataset partly overlaps with the Icelandic sample set (17). In order to use only non-overlapping data when performing meta-analysis for rs12050604, we combined the data presented by Landi et al. only with data from the Netherlands, Spain and USA-Denver in the combined analysis.
Genotyping
The Icelandic cases and controls and the Dutch controls were assayed using the Human Hap300 and HumanCNV370-duo Bead Arrays (Illumina) (6). All other samples, i.e. the Dutch cases and the cases and controls from Spain and the USA were genotyped using the Centaurus (Nanogen) platform (18). The quality of each Centaurus SNP assay was evaluated by genotyping each assay in the CEU HapMap samples and comparing the results with the HapMap publicly released data. Assays with >1.5% mismatch rate were not used and a linkage disequilibrium (LD) test was used for markers known to be in LD. Approximately 10% of the Icelandic cases that were genotyped on the Illumina platform were also genotyped using the Centaurus assays and the observed mismatch rate was less than 0.5%.
Statistical analysis
A likelihood procedure implemented in the NEMO software was used for the association analyses (19). We tested the association of an allele to cancer using a standard likelihood ratio statistic that, if the subjects were unrelated, would have asymptotically a χ2 distribution with one degree of freedom under the null hypothesis. Allelic frequencies rather than carrier frequencies are presented for the markers in the main text. Allele-specific ORs and associated P values were calculated assuming a multiplicative model for the two chromosomes of an individual. To adjust for possible population stratification and the relatedness amongst the Icelandic study subjects, we divided the χ2 test statistics from the whole-genome lung cancer scan using the method of genomic control (20), i.e. the 304,073 χ2 test statistics were divided by their means, which were 1.06 for the Icelandic subjects that were chip-genotyped and 1.10 for the combined dataset of genotyped and un-genotyped subjects whose genotypes were derived by familial imputation. The sample sets from the Netherlands, Spain and the USA used in this study were recruited on a population basis and assumed to be unrelated. Results from multiple case-control groups were combined using a Mantel-Haenszel model in which the groups were allowed to have different population frequencies for alleles, haplotypes and genotypes but were assumed to have common relative risks (21). This method was used both for combining data within study groups genotyped by us and to combine those results with published results (allelic ORs and P values) from Broderick et al. (13) and Landi et al. (17). All reported P values are two-sided.
Results
We assessed the 7 SNPs reported by Broderick et al. to have a suggestive association with lung cancer (P< 10−5) in our independent GWAS dataset corresponding to 1,447 lung cancer cases and 36,256 controls from Iceland. Only the 3 SNPs on 15q15.2 (rs504417, rs11853991 and rs748404) showed an effect in the same direction as previously reported with an association with lung cancer that remained significant for all three SNPs when taking the 7 tests into account (P<0.05/7 = 0.007, Table 2). The SNPs are highly correlated (r2>0.8) and in agreement with the results of Broderick et al., the strongest association was to the T allele of rs748404 (OR=1.20, P=5.2×10−4). The other 4 SNPs, located on chromosomes 2p14, 3p24.1, 3q13.31 and 10q23.31, did not show significant association with lung cancer (all P≥0.1, Table 2).
Table 2.
Association results in the Icelandic population for 7 markers previously reported to have suggestive association with lung cancer.
| SNP(allele) | CHR | cases (N) | cases (F) | cont. (N) | cont. (F) | OR | 95%CI | P | OR* |
|---|---|---|---|---|---|---|---|---|---|
| rs4254535(T) | 2p14 | 1,452 | 0.672 | 36203 | 0.68 | 0.97 | 0.89-1.05 | 0.45 | 0.89 |
| rs1530057(A) | 3p24.1 | 1,429 | 0.049 | 36224 | 0.048 | 1.01 | 0.83-1.23 | 0.93 | 1.26 |
| rs6438347(G) | 3q13.31 | 1,454 | 0.209 | 36259 | 0.223 | 0.92 | 0.83-1.02 | 0.10 | 0.88 |
| rs1926203(T) | 10q23.31 | 1,466 | 0.671 | 36273 | 0.674 | 0.99 | 0.90-1.08 | 0.76 | 1.12 |
| rs504417(G) | 15q15.2 | 1,447 | 0.275 | 36228 | 0.301 | 0.88 | 0.80-0.96 | 0.0043 | 0.89 |
| rs11853991(G) | 15q15.2 | 1,451 | 0.274 | 36246 | 0.301 | 0.88 | 0.80-0.96 | 0.0036 | 0.89 |
| rs748404(T) | 15q15.2 | 1,447 | 0.825 | 36256 | 0.797 | 1.20 | 1.08-1.33 | 0.00052 | 1.15 |
Shown are the SNP name, allele tested, chromosome location, the number of individuals (N) and the allelic frequency (F) of the variant in cases and controls, allelic odds-ratio (OR), 95% CI and P values based on the multiplicative model and the OR reported by Broderick et al. (OR*). All P values are two-sided.
In order to further explore the association between rs748404-T and lung cancer, we genotyped the SNP in sample sets from the Netherlands, Spain and USA (Table 1). In combined analysis of the Icelandic and follow-up sample sets, the association became more significant (P=2.5×10−4) and the effect was identical to the effect reported by Broderick et al. (OR=1.15) (Table 3). When all the data in our study (2,739 cases and 40,485 controls) were combined with the summary results of Broderick et al., using the Mantel-Haenszel model, the association of rs748404-T with lung cancer reached the commonly used threshold for genome-wide significance (P=5×10−8) with a combined OR of 1.15 and a combined P value of 1.1×10−9 (Table 3). We imputed all HapMap CEU (phase II) SNPs in a 450 kb region centered on rs748404 into the chip-genotyped Icelandic samples (808 cases and 36,256 controls) and repeated the association analysis. Of the 201 SNPs imputed in the region, no SNP showed stronger association with lung cancer than rs748404. rs748404-T was not associated with gender (P=0.64) or age at diagnosis (P=0.69) among lung cancer cases. Furthermore, we found no association between rs748404 and smoking quantity as measured by cigarettes smoked per day in a large group of Icelandic smokers (N=15,310, P=0.2), indicating that the variant is not likely to be acting through smoking quantity. We analyzed the effect of rs748404 in different histological types of lung cancer versus controls and observed a significant association between the T allele and the risk of adenocarcinoma (N=854, OR=1.17, P=0.0095) and squamous cell carcinoma (N=672, OR=1.16, P=0.030) (Supplementary Table 1). The number of cases with other histological types of lung cancer was too small (N<350) to draw definitive conclusions.
Table 3.
Association between rs748404-T and lung cancer in 7 case control sample sets
| Population | OR | CI95 | P | cases (N) | cases (F) |
cont. (N) |
cont.(F) | Phetc | I2d |
|---|---|---|---|---|---|---|---|---|---|
| ICELAND a | 1.20 | 1.08-1.33 | 0.00052 | 1,447 | 0.825 | 36,256 | 0.797 | ||
| USA | 1.19 | 0.89-1.59 | 0.246 | 186 | 0.796 | 838 | 0.766 | ||
| NETHERLANDS | 1.07 | 0.90-1.27 | 0.456 | 528 | 0.777 | 1,832 | 0.766 | ||
| SPAIN | 1.10 | 0.93-1.29 | 0.27 | 548 | 0.759 | 1,432 | 0.742 | ||
| OVERALL b | 1.15 | 1.07-1.24 | 0.00025 | 2,709 | 0.79 | 40,358 | 0.768 | 0.63 | 0 |
| From Broderick et al. | 1.15 | 1.09-1.20 | 1.08×10−6 | 7,560 | 8,205 | ||||
|
| |||||||||
| All combined | 1.15 | 1.10-1.20 | 1.1×10−9 | 10,269 | 48,563 | 0.78 | 0 | ||
Shown are the allelic odds-ratio (OR), 95% CI and P values based on the multiplicative model, the number of individuals (N) and the allelic frequency (F) of the variant in cases and controls. All P values are two-sided.
The Icelandic results are obtained by combining data from individuals genotyped directly or by familial imputation. Results for the Icelandic population were adjusted by the method of genomic control.
For the combined study populations, the reported control frequency was the average, unweighted control frequency of the individual populations, while the OR and the P value were estimated using the Mantel-Haenszel model.
Phet denotes the tests of heterogeneity performed by comparing the null hypothesis of the effect being the same in all populations to the alternative hypothesis of each population having a different effect using a likelihood ratio test.
I2 takes values between 0% and 100% and describes the proportion of the total variation in estimates that is due to heterogeneity.
Landi et al. reported the results of a meta-analysis of over 13,000 cases and 19,000 controls which included samples from the Icelandic sample set used here (719 cases and 6,030 controls), and the UK sample set used by Broderick et al. (1,978 cases and 1,438 controls) (17). In this meta-analysis, five SNPs at 15q15.2 had suggestive association with lung cancer (P values ranging from 3.5×10−6 to 4.2×10−5). The association between these 5 SNPs and lung cancer in the Icelandic chip-genotyped individuals, a part of which was included in the dataset used by Landi et al., is shown in Supplementary table 2. The 5 SNPs are correlated with rs748404 with r2 ranging from 0.13 to 0.69 (Supplementary table 3). Of the 5 SNPs, the two fully-correlated variants rs2277532 and rs12050604 (r2=1 in CEU) showed the strongest association with lung cancer in the report by Landi et al. (OR=1.11, P=3.5×10−6) (17). Those two SNPs are also the least correlated to rs748404 (r2≤0.14), potentially suggesting a second signal at this locus. We genotyped rs12050604 in the samples from USA, the Netherlands and Spain and combined these data with the published summary results from Landi et al., giving us a total of 14,480 cases and 22,824 controls (Supplementary table 4). In this large dataset, the C allele of rs12050604 had an OR of 1.09 and a P value of 3.6×10−6, not reaching genome-wide significance.
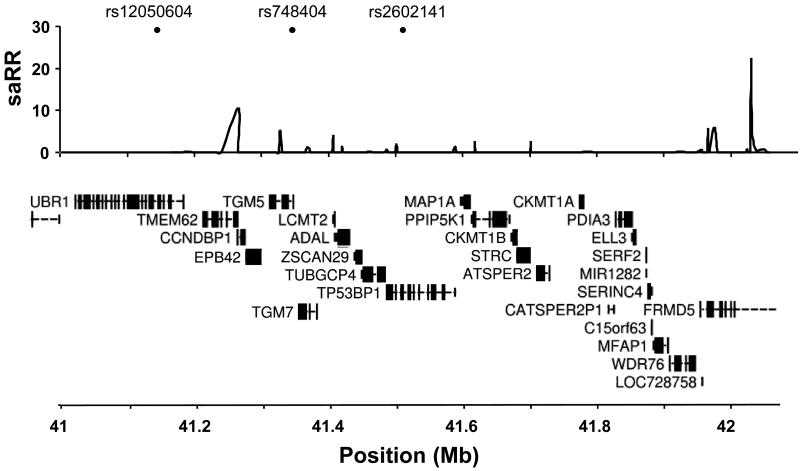
The variant rs748404 is located between two transglutaminases genes, TGM5 and TGM7, and approximately 200 kb telomeric to rs12050604 which is in an intron of the E3 ubiquitin-protein ligase UBR1 gene (Figure 1). The two variants are separated by a recombination hotspot and the r2 between them is only 0.126. When we adjust the results for each SNP in a logistic regression, using the other SNP as a covariate and only including individuals who we genotyped for both markers (Iceland, the Netherlands, Spain and USA), results for rs748404 remain significant after adjusting for rs12050604 (OR=1.09, P=0.037) and rs12050604 remains significant after adjusting for rs748404 (OR 1.12, P=0.0028) (Supplementary table 5). Overall, these results indicate that neither rs748404 nor rs12050604 can, by themselves, fully account for the association observed between sequence variants in this region and lung cancer. This observation suggests that either a unique variant, capturing the effects of both rs748404 and rs12050604, remains to be discovered or that the region contains more than one variant that predisposes to lung cancer.
Figure 1.
A schematic view of the lung cancer associated region on chromosome 15q15.2. Top panel: Location of the three main SNPs tested in the study, rs12050604, rs748404 and rs2602141. Center panel: The estimated recombination rates (saRR) in cM / Mb from the HapMap (v22) Phase II data (25). Bottom panel: Location of known genes in the region (from the UCSC browser).
The variant that shows a genome-wide significant association with lung cancer, rs748404, is located at the beginning of a 750 kb LD block containing several genes (Figure 1). Of those, the most notable with respect to lung cancer is TP53BP1, a DNA damage checkpoint protein that is located 140 kb telomeric to rs748404. TB53BP1 contains three common SNPs (MAF>5% in HapMap CEU) that encode missense mutations, rs2602141 (Q1136K), rs560191 (E353D) and rs689647 (S412G). rs2602141 and rs560191 are fully correlated (r2=1) and the common alleles of these SNPs have previously been reported to be associated with risk of lung cancer (22, 23). In our Icelandic dataset of 1,417 cases and 36,256 controls, the T allele of rs2602141 was associated with lung cancer (OR=1.11, P=0.011). We genotyped rs2602141 in our sample sets from the Netherlands, Spain and USA and combined the data with the Icelandic data, as well as with data from previous candidate gene studies by Rudd et al. (22) and Truong et al. (23). This analysis shows that the missense variant rs2602141-T (and consequently, the fully correlated missense variant rs560191-G) is significantly associated with risk of lung cancer (OR=1.11, P=8.60×10−9) (Supplementary table 6). The variant rs748404 is moderately correlated with rs2602141 and rs560191 (r2=0.578). By adjusting the results for the coding SNP rs2602141 with rs748404 and vice versa, using only individuals we genotyped for both markers (a total of 2,239 cases and 40,418 controls), we found that results for rs748404 remain significant after adjustment for rs2602141 (OR=1.12, P =0.027) whereas results for rs2602141 do not remain significant after adjustment for rs748404 (OR=1.02, P =0.74) (Supplementary Table 7). Thus, there is no evidence for an association signal from the coding variants, rs2602141 and rs560191, that is not captured by rs748404. The third missense mutation in TP53BP1, rs689647-T, did not show significant association with lung cancer in a group of 819 lung cancer cases and 10,724 controls from the Icelandic population (OR=0.94, P=0.53).
Discussion
In this study, we have confirmed that a region on 15q15.2 is the fourth lung cancer risk region discovered through GWA studies in Europeans. By combining several independent datasets, the association of rs748404 reaches genome-wide significance; however, our results suggest that rs748404 does not fully explain the association observed at the locus and that either there may be a yet-unidentified variant or that there may be more than one variant at this locus that associates with lung cancer risk. In this regard, the locus has similarities to the TERT locus on 5p15.33 that has been shown to contain several independent variants that associate with lung cancer (10-12). Furthermore, we observed that although the missense mutation in TP53BP1, rs2602141, is significantly associated with lung cancer, this association no longer remains when adjusted for rs748404.
We noted that rs748404 was not among the top 200 SNPs (all P values < 8×10−5) reported by Landi et al. in the meta-analysis of over 13,000 lung cancer cases and 19,000 controls although a SNP with an OR=1.15 and MAF~0.77 should have a probability of ~80% to be included on this list. However, the list of top 200 SNPs in the meta-analysis includes 2 SNPs that have an r2 of 0.69 with rs748404 and P values of 2×10−5, supporting the conclusion that rs748404 or correlated SNPs are associated with lung cancer.
We are aware that we do not currently know all the variants that exist at the 15q15.2 locus. Comprehensive genomic sequencing will eventually allow a more complete dissection of the association between variants in the region and lung cancer risk. Currently, whole genome sequencing efforts, such as the 1000 Genomes (1000G) project (24), are leading to the discovery of a large number of variants that will be imputed into large datasets for association analysis. However, it may not be possible to impute all these variants with certainty, requiring direct genotyping on large datasets to test if they associate with defined phenotypes.
Mapping of lung cancer risk regions is an important step towards understanding of the pathogenesis of lung cancer. Lung cancer risk variants could fall into at least three categories, i.e. variants that affect risk of lung cancer regardless of smoking status, variants that increase vulnerability to the harmful effects of smoking in smokers and variants that affect smoking behavior. For this last category, in addition to the variants at 15q25, large meta-analyses on smoking behavior have yielded sequence variants at 8p11 and 19q13 that show genome-wide significant association with smoking quantity and nominally significant association with lung cancer (6, 15). Large scale collaborative efforts will be required to uncover new variants that associate with lung cancer and refine the association signals already discovered.
Supplementary Material
Acknowledgements
We thank the individuals that participated in the study and whose contribution made this work possible. We also thank the nurses at deCODE’s participant recruitment center and the personnel at deCODE’s core facilities. We acknowledge the Icelandic Cancer Registry for assistance in the ascertainment of the Icelandic cancer patients.
Financial support: This work was supported by European Union (EU) FP7-MC-IAPP Grant agreement no. 218071 (CancerGene), the EU grant GENADDICT: LSHM-CT-2004-005166 and SPORE in Lung Cancer grant (P50 CA058187) and EDRN grant CA85070 from the National Cancer Institute.
References
- 1.Devesa SS, Bray F, Vizcaino AP, Parkin DM. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–299. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Hemminki K. Familial and second lung cancers: a nation-wide epidemiologic study from Sweden. Lung Cancer. 2003;39:255–263. doi: 10.1016/s0169-5002(02)00535-4. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson S, Thorsteinsdottir U, Gudbjartsson DF, et al. Familial risk of lung carcinoma in the Icelandic population. Jama. 2004;292:2977–2983. doi: 10.1001/jama.292.24.2977. [DOI] [PubMed] [Google Scholar]
- 4.Brennan P, Crispo A, Zaridze D, et al. High cumulative risk of lung cancer death among smokers and nonsmokers in Central and Eastern Europe. Am J Epidemiol. 2006;164:1233–1241. doi: 10.1093/aje/kwj340. [DOI] [PubMed] [Google Scholar]
- 5.Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ. 2000;321:323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 9.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wetzels JF, Kiemeney LA, Swinkels DW, Willems HL, den Heijer M. Age- and gender-specific reference values of estimated GFR in Caucasians: the Nijmegen Biomedical Study. Kidney Int. 2007;72:632–637. doi: 10.1038/sj.ki.5002374. [DOI] [PubMed] [Google Scholar]
- 17.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutyavin IV, Milesi D, Belousov Y, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 20.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 21.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 22.Rudd MF, Webb EL, Matakidou A, et al. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006;16:693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong T, Sauter W, McKay JD, et al. International Lung Cancer Consortium: coordinated association study of 10 potential lung cancer susceptibility variants. Carcinogenesis. 2010;31:625–633. doi: 10.1093/carcin/bgq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. www.1000genomes.org.
- 25.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.