Abstract
RNA-based protein synthesis produces L-amino acid-containing proteins and peptides. D-amino acid-containing peptides (DAACPs) can be generated from L-amino acid peptides via post-translational modification. In the nervous system, the conformational change of a single L-amino acid in a peptide to its D-form results in altered bioactivity, with some DAACPs having orders-of-magnitude enhanced efficacy. However, this modification is often overlooked when characterizing endogenous peptides. Here, using matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF)/TOF mass spectrometry, neuropeptides that have the second residue isomerized to the D-isoform are distinguished from their L-epimers via differences in the relative amounts of specific fragment ions during tandem MS. By choosing the appropriate fragment ions, and in some cases with the use of metal adducts, epimer discrimination is optimized. Specifically, the cardioexcitatory peptide Asn-DTrp-Phe-amide (NdWFa) was assayed directly from neurons isolated from the sea slug Aplysia californica; the fraction of the peptide with the second residue (W) in the D- versus L-form was 90 ± 10%. We demonstrate that this approach is well suited for confirming DAACPs directly from cells and tissue, advancing our understanding of the L to D modification and the role it plays in cell-to-cell signaling.
INTRODUCTION
Neuropeptides act as cell-to-cell signaling molecules and perform important roles in the central nervous system in animals. They are generated via the processing of larger precursor proteins into biologically active peptides and their function is often dependent on chemical modifications.1,2 One of the more unusual post-translational modifications (PTMs) in neuropeptides is the isomerization of an L-amino acid residue into its D-form.3
D-amino acid-containing peptides (DAACPs) share the same sequence with their all-L isomers, but in many cases, the D-amino acids confer distinct and at times, dramatically enhanced receptor binding affinity.4,5 Consequently, the modification is sometimes a requisite for the induction of physiological effects. For example, NdWFa, a cardioexcitatory peptide first isolated from Aplysia kurodai, was found to be highly active in the modified form, but NWFa only induces minimal effects.6 As another example, achatin-I (Gly-DPhe-Ala-Asp), isolated from the African giant land snail Achatina fulica, demonstrated potent neuroexcitatory effects, while the all-L-form isomer achatin-II (Gly-Phe-Ala-Asp) lacked significant activity.7 How is this enhancement possible? The isomerization event changes secondary structure,8–10 and in longer peptides, even disrupts tertiary structures.11 Moreover, because binding affinity depends on shape, and is also dependent on electronic interactions and hydrogen bonding between the peptide and the receptor, changes in shape can dramatically shift the bioactivity of these cell-cell signaling molecules.
Over the last three decades, multiple DAACPs have been reported in mollusks, such as the neuropeptides NdWFa, achatin-I and its homolog in octopus, ocp-1 and ocp-4,12 fulicin (FdNEFVa) and fulyal (YdAEFLa), with the latter two peptides from the fulicin gene-related peptide precursor.7,13,14 In total, ~30 DAACPs (including hormones and antimicrobial peptides) have been found in a variety of species.6,12,15–34 In several cases, the PTM has been created by a peptidylaminoacyl-L/D-isomerase. Several isomerases have been reported, including enzymes from the funnel web spider Agelenopsis aperta,35 the frog species Bombina variegate and Bombina orientalis,36 and the primitive mammal, the platypus.37 These enzymes do not share the same catalytic mechanisms, and more interestingly, the preferred site of isomerization is distinct. The enzyme(s) responsible for the formation of DAACP in several molluskan species (e.g., NdWFa, FGRPs, achatin) are not known, but they share a strikingly conserved activity in the formation of these specific DAACPs; the second position from the N-terminal is the amino acid that is isomerized.
A separate class of naturally occurring D-amino acids in peptides is spontaneous racemization, for example, a D-Ser residue in the beta-amyloid protein [D-Ser26]Aβ1–40 is generated in an age-dependent process. This non-toxic form is processed by a protease to form the more problematic peptide [D-Ser26]Aβ25–35/40, which may be related to the transition of the soluble Aβ to the pathological fibrillar form.38
How many neuropeptides contain a D-amino acid residue but have yet to be discovered due to the lack of a sensitive assay? Questions about how DAACPs function cannot be answered unless there is an efficient method to characterize them. Unfortunately, conventional sequencing techniques such as Edman degradation and mass spectrometry (MS) do not distinguish D- from L-amino acids, raising the possibility that previously “characterized” peptides may have been tested for bioactivity on the non-physiological, all-L-amino acid-containing forms. Therefore, in order to achieve complete characterization of a novel peptide, techniques that allow diastereomeric characterization of the amino acids in peptides are required, especially in cases where the L-form has not been shown to have robust activity.
How are unidentified DAACPs characterized? While MS is the most common approach for characterizing an unknown peptide,39,40 because this PTM has no associated mass change, improved approaches are needed. One option is enzymatic screening, which uses specific enzymes to selectively degrade peptides that contain only L-amino acids.41 An enzyme-aided screening method, when combined with MS, can be used to identify peptides containing a D-amino acid near the N-terminal, even from a complex mixture. The advantage of this type of screening approach is that standards are not required; note however, that other peptide modifications can cause false positives. Confirming that a peptide contains a D-residue entails additional experimental steps, and often requires L- and D-amino acid-containing peptide standards to accomplish. One of the more interesting MS-based techniques used to distinguish diastereomers relies on the stability of complexes formed between an added metal species and the diastereomers, known as the kinetic method.42 The semi-stable complexes formed with the peptide isomers have different thermochemical properties that lead to different branched fragmentation profiles, thereby enabling discrimination.
These approaches can be extended by application of direct tandem MS (MS/MS) methods to investigate the protonated peptides. By measuring the differential fragmentation patterns, the diastereomers can be discriminated. Relatively soft ionization methods such as electrospray ionization (ESI) can allow enough solution-phase structure to carry into the gas phase to allow such discrimination.43 Recently, Adams et al.44 reported using ESI-Fourier transform MS with electron capture dissociation to distinguish and quantify DAACPs and proteins with a limit of detection of 1%; later this method was extended to MALDI MS by Sachon el al.45 In the latter work, two short peptides were studied and discrimination made by measuring the abundance of corresponding fragment ions generated from the different epimers; for DAACPs with a substitution of the 2nd residue, N-terminal acetylation was introduced to enable the distinction. These methods serve as downstream approaches to confirm the DAACP candidates, but to date have used only synthetic peptides.
For this study, we adapted the MALDI MS method described above,45 combining it with the direct MALDI measurement of complex mixtures of peptides in individual and small numbers of neurons.46,47 Cells were deposited directly onto a MALDI target and the DAACPs distinguished and quantified using collision-induced dissociation (CID) MS/MS. We validated this approach with known molluskan DAACPs. Using a working curve derived from standards, we characterized the fractions of NWFa and NdWFa within single cells—obtaining amounts consistent with prior reports—and demonstrated the ability to discriminate other putative Aplysia DAACPs. For those peptides where the two isomers fragmented similarly for the measured fragments, the use of various additives was shown to enhance chiral discrimination. Thus, this approach appears to be general, at least for all of the peptides tested here.
EXPERIMENTAL SECTION
Materials
Synthetic peptides: NWFa, YAEFLa, GFAD, GFFD, FMRFa, and the corresponding five analogs with D-amino acids in the second position (Table 1), were synthesized by the Protein Sciences Facility, University of Illinois at Urbana-Champaign. Chemicals and reagents: Water was purified from a Milli-Q system (Millipore, Bedford, MA,); the antibiotics, 2,5-dihydroxybenzoic acid (DHB), NaCl, KCl, CaCl2, MgCl2, CoCl2, MgSO4, HEPES, and acetonitrile (ACN) containing 0.01% trifluoroacetic acid (TFA), were obtained from Sigma-Aldrich (St. Louis, MO).
Table 1.
The five pairs of synthetic peptides, their chirality-reporting fragment ions and the corresponding chiral recognition factors.
| (M+H)+M/Z (mono) | Peptide | Sequence | Chirality-reporting fragments | Ratio of chirality-reporting fragments (measured by peak intensity) | Rchiral=RD/RL |
|---|---|---|---|---|---|
| 465.2 | N(l/d)WFa | Asn-Trp-Phe-NH2 | b3 and b2 | 1.28 ± 0.03 | 2.97 |
| Asn-DTrp-Phe-NH2 | 3.78 ± 0.06 | ||||
| 641.3 | Y(l/d)AEFLa | Tyr-Ala-Glu-Phe-Leu-NH2 | b4 and y2 | 0.043 ± 0.002 | 2.51 |
| Tyr-DAla-Glu-Phe-Leu-NH2 | 0.111 ± 0.002 | ||||
| 485.2 | G(l/d)FFD | Gly-Phe-Phe-Asp | Y2 and MH+H2O | 0.13 ± 0.02 | 1.78 |
| Gly-DPhe-Phe-Asp | 0.23 ± 0.02 | ||||
| 599.3 | F(l/d)MRFa | Phe-Met-Arg-Phe-NH2 | b3 (or y3-NH3) and y3 (or c3) | 7.65 ± 0.18 | 1.14 |
| Phe-DMet-Arg-Phe-NH2 | 8.7 ± 0.2 | ||||
| 657.3 | F(d/l)MRFa Cobalt adduct | [M−H++Co]+ | b3+Co and 615 | 0.30 ± 0.1 | 2.80 |
| 0.73 ± 0.09 | |||||
| b3+Co and 612 | 0.10 ± 0.03 | 2.88 | |||
| 0.33 ± 0.05 | |||||
| b3+Co and a3+Co | 0.26 ± 0.01 | 1.62 | |||
| 0.40 ± 0.03 | |||||
| b3+Co and c3 (or y3) | 0.101 ± 0.008 | 1.60 | |||
| 0.16 ± 0.02 | |||||
| 409.2 | G(l/d)FAD | Gly-Phe-Ala-Asp | b3 and y2 | 1.00 ± 0.02 | 1.20 |
| Gly- DPhe-Ala-Asp | 1.21 ± 0.01 | ||||
| 431.2 | G(d/l)FAD Sodium adduct | [M+Na]+ | c3+Na and y2-H2O | 7.6 ± 5.3 | 3.44 |
| 26 ± 12 | |||||
| c3+Na and a3+Na | 2.02 ± 0.08 | 1.29 | |||
| 2.6 ± 0.1 | |||||
| c3+Na and b3+Na | 1.9 ± 0.2 | 1.28 | |||
| 2.4 ± 0.3 |
Single letter codes are used for the amino acids, with a terminal “a” indicating an amidated C-terminus, and a small “l” or “d” denoting that the following residue is in the L- or the D-form.) Rchiral is distinct for each pairs of chirality reporters, RL and RD calculated by dividing the peak area of the former by the latter chirality reporters in the table. Enhanced discrimination power of this method on F(l/d)MRFa and G(l/d)FAD are realized via metal-complexed cations (cobalt adducts and sodium adducts respectively) instead of protonated peptides as precursors for fragmentation. A different set of product ions are found with metal adducts, which have different Rchiral. With such adducts, a higher discrimination power is measured but with higher standard deviation.
Animals
Aplysia californica (120–170 g) were purchased from the University of Miami/NIH National Resource for Aplysia and kept in an aquarium containing aerated and filtered artificial seawater (Instant Ocean, Aquarium Systems Inc., Mentor, OH) at around 14 °C until used. Prior to dissection, animals were anesthetized by injection of isotonic MgCl2 (30 to 50% of body weight) into the body cavity.
Neuron Isolation
The abdominal ganglia were dissected and incubated in artificial sea water (ASW: 460 mM NaCl, 10 mM KCl, 10 mM CaCl2, 22 mM MgCl2, 6 mM MgSO4, and 10 mM HEPES, pH 7.8) supplemented with antibiotics (penicillin G, gentamycin and streptomycin) and containing 1% (w/v) protease (Type IX: Bacterial; Sigma-Aldrich) at 32 °C for 45 min to loosen the connective tissue sheath. Following a 1–3 h rinse in ASW with antibiotics to remove the bulk of the protease, the ganglia were stretched onto a silicone elastomer (Sylgard, Dow Corning, Midland, MI) layer in a petri dish containing 3–4 mL of the ASW-antibiotic medium using 0.15 mm diameter tungsten needles (WPI, Sarasota, FL) and connective tissue was surgically removed to expose neurons.
Clusters of peptidergic neurons from individual A. californica containing N(d/l)WFa were identified according to their position, size, and pigmentation.48,49 Isolated neuron clusters were briefly rinsed in Milli-Q water to remove excess salts and ASW before depositing onto the MALDI target. Excess liquid was aspirated from the target and spot-to-spot cell transfers were performed for sampling of individual neurons as described previously.50 Between 0.2–0.4 μL of 10 mg/mL DHB matrix was applied by the dried droplet method over the lysed cells. Ten animals were tested (biological replicates), and at least three technical replicates were made from each biological sample.
MALDI TOF/TOF MS Analysis
MS measurements were performed using an UltrafleXtreme MALDI time-of-flight (TOF)/TOF mass spectrometer (Bruker Daltonics, Billerica, MA) in positive-ion reflector mode. MS/MS measurements were performed in “LIFT” mode with CID turned on. Collision was performed with argon gas, creating a source pressure of 6 × 10−6 mbar, and the collisional energy was typically set to 27 keV, including a potential raise in the “LIFT” chamber. External peptide standards were used for calibration of the MS and MS/MS in the low mass range.
For the synthetic peptides, 0.7 μL of 1 mg/mL of each was dissolved in water, deposited on the target, and mixed 1:1 (v/v) with matrix: 10 mg/mL of DHB in 50% ACN with 0.01% TFA. In some experiments, 0.5 μL of 1 mM CoCl2 was co-spotted with sample and matrix in order to form metal-peptide complexes. For each synthetic peptide sample, six replicate spots were characterized. For each replicate, the spectrum was an average of 5,000 laser pulses. The same instrument settings were used for the synthetic peptide samples that included mixtures of the D- and L-standards. For calibration curve plotting, the D-epimer mole fraction was incremented in 20% steps, and data collection was the same as with the pure isomers. For the single-cell measurements, conditions were the same, except that laser intensity was adjusted to observe peaks with a signal-to-noise ratio higher than 100 but not saturated; this value was kept consistent across the cell samples from each animal (with n = 10).
The MS/MS spectra were analyzed using the FlexAnalysis and Biotools software packages (Bruker Daltonics). Statistics was performed with intensity/area values exported from FlexAnalysis. Statistical calculations, including mean and standard deviation for bar charts and calibration curves, were performed using Microsoft Excel 2010. To compare fragmentation patterns between two epimers, intensity values for each fragment ion were normalized to the sum of all major fragment ions produced from the particular sample spot (except for results from metal adducts, which were normalized to the most intense peak as the metal adduct example contained large numbers of low intensity peaks). The observed fragment ion peaks were mass matched to predicted fragment ions using Protein Prospector (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct).
RESULTS and DISCUSSION
Our goal was to determine if the neuropeptides known to contain D-amino acids have enough differences in their fragmentation patterns to enable the peptide fraction in each form to be characterized. While ESI MS is normally considered to be more reproducible, we use MALDI TOF/TOF MS here because of its compatibility with direct cell and tissue measurements, thereby enabling smaller-volume neuropeptide characterization.39,51,52
The D-amino acid-containing neuropeptides NdWFa, YdAEFLa, and GdFAD previously reported from animal tissues, and another two short peptides, GdFFD and FdMRFa, and their five respective L-epimers were used for MS/MS measurements. These peptides yield fragment ions, with b- and y- being the major types; the product ions resulting from further water losses can be detected as well (see Table 1, with additional supporting data presented in the Supporting Information). The peak intensity and area value of each major fragment ion were both measured and the results did not differ significantly (data not shown); therefore, the peak intensity measurements were used for analyses.
A reasonable reproducibility of the peak intensity measurements allows comparison between epimers; however, we found that the intensities of product ions can vary significantly across six replicate spots. In order to improve run-to-run consistency, instrumental parameters, including laser fluence, remained unchanged throughout the samples tested. In addition, all fragment ion intensities were normalized, either to the sum of product ions generated from the respective replicate, or to the most intense peak (the latter was done in the case of the metal adducts, where there were large numbers of low intensity product ions). With this normalization procedure, a small standard deviation was observed (less than 1% relative standard deviation for major fragment ions), allowing clear discrimination between epimers.
For the five peptide pairs tested, pure D/L peptides demonstrated significant stereochemical effects that allowed discrimination of their fragment ion fingerprints (Figure S1, with F(d/l)MRFa shown in Figure S2). Not surprisingly, this technique demonstrated differing abilities in characterizing some pairs over others, reflected in the Rchiral values of each pair, where Rchiral represents the ratio of RD and RL, with each being the ratio of a pair of chirality-reporting fragment ions for the D- and L-epimer, respectively. While one could use the entire spectra to distinguish the two forms, here we determined the specific fragment ion(s) that were most useful for this discrimination. The relative quantification of the differentially generated fragments, as shown in the bar charts (Figure S1), led to the identification of indicator ions critical for discrimination. For example, the ratios of fragments b3 to b2 of NWFa and NdWFa (referred to as RD and RL, respectively) were used as chirality reporters to distinguish the D- versus L-form and determine the fraction of the D-peptide.
Interestingly, the fragment ions that showed the most pronounced changes in relative abundance between the D- and L-forms were not always the flanking residues of the D-amino acid, as also observed by others.11,45 For the peptides we examined, the a4, b4, z4 and y2 ions varied the most between YAEFLa and YdAEFLa (Figure S1C), as can be visualized with the representative spectra in Figure S3; and y2 and MH-H2O showed the most significant changes between the two epimers for GFFD and GdFFD (Figure S1D). This suggests that the substitution of a single D-amino acid has an impact on the overall conformation of the peptide, rather than only on the neighboring residues, resulting in complex changes in fragmentation patterns.
Using direct fragmentation we successfully discriminated all five pairs of peptide epimers but found that the chirality-reporting product ion abundances for GFAD and GdFAD, and FMRFa and FdMRFa, were fairly similar. Therefore, to maximize the sensitivity of these assays we explored the fragmentation differences in metal-complexed cations of these epimers. The sodium adducts present in the peptide standards have shown promise for enhancing chiral discrimination when protonated peptides only show minimal differences.53 As seen in Figure S4, the Na+-cationized peptides generated a different set of fragment ions from the protonated peptides and several pairs of product ions were used for chiral discrimination, some of which led to an enhanced chiral recognition factor (Rchiral=RD/RL). This effect may be due to different binding sites or the orientation of sodium ions on the peptide.53 The variation between runs is significant when compared to the fragmentation of protonated peptides, partly due to the low intensities of the product ions. It may be that this is a result of a low abundance of salt adducts, or a lower fragmentation efficiency of these adducts due to the charge stabilization effect.
Another option is to use added metal species to enhance the discrimination power. As adapted from the kinetic method,42,54 transition metal ions can be added to the sample for metal-peptide complex formation. Here, cobalt chloride was used to discriminate FMRFa and FdMRFa. These two peptides are difficult to discriminate when assayed directly, but showed obvious differences with cobalt adduct fragmentation (Figure S2).
When one assays a tissue sample, partially processed and final peptide products will be present. Thus, it is expected that the peptide will be present in both the L- and D-forms, and so measuring the relative abundance of each helps us to understand the importance of DAACPs as signaling molecules. Among the four pairs of peptides, protonated NWFa and NdWFa had the most distinct fragmentation profiles, with the b2 and b3 ions demonstrating a large Rchiral of 3.18, making this pair a good model to be used for isomeric content measurement.
According to the following calculations, the RM measured from two chiral indicator ions of the mixture is correlated with the mole fraction of the D-epimer.44 For an L- and D-form mixture of a peptide, α being the mole fraction of the D-form:44,55
| (Eq. 1) |
with this leading to:
| (Eq. 2) |
Therefore, ln RM is linearly proportional to α. In this equation, ΔG is the free energy change in the fragmentation process, Δ (ΔG) is the difference of ΔG between two fragmentation pathways, R is the branching ratio of two fragmentation pathways, which approximately equals the abundance ratio of the two fragment ions, RM represents the measured abundance ratio of these two chirality-reporting ions obtained from the isomeric mixture, and Rchiral equals the ratio of RL and RD.
Based on Equation 2, a calibration curve (Figure S5) was created using a mixture of the D- and L-peptide standards, thus allowing the isomeric content to be calculated (ln RM=0.1495 +1.1181*α, with the squared correlation coefficient R2 = 0.9757); standards were co-spotted with an endogenous peptide mixture extracted from the buccal ganglion of A. californica that did not contain the target analytes. Adding the endogenous peptide mixtures without target analytes allows the complexity expected in later biological samples to be mimicked without perturbing the measured isomeric ratio. The high data consistency and linearity shown in this curve demonstrates that the approach can be used for accurate isomeric determinations. The deviations from linearity may result from an unequal affinity of the two epimers to salt adducts.
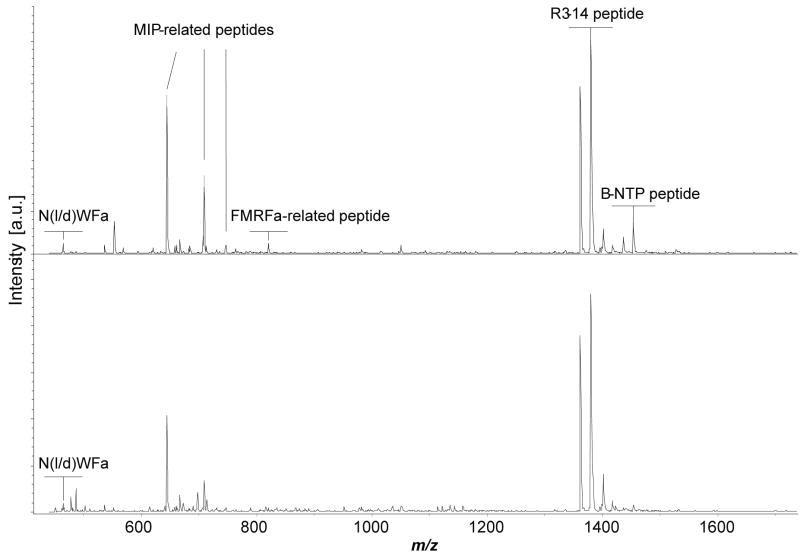
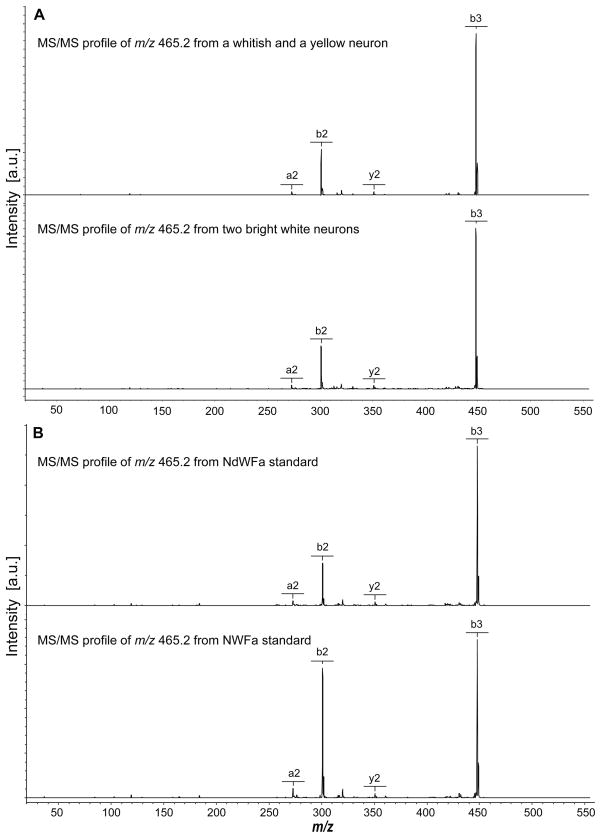
We applied this method to determine the conformation of NdWFa/NWFa in a direct assay from individually isolated neurons. Although Morishita et al.6 reported the NWFa peptide in the R3-R13 neurons, we did not detect it in those cells; instead, a mass corresponding to NWFa was detected in a small cluster of whitish, medium-sized neurons located on the lateral edge of the upper right rostral quadrant of the abdominal ganglion, as well as in several white neurons at the medial edge of the quadrant (the mass fingerprint of these cells is shown in Figure 1). Because the target peptides are of relative low-abundance, the cell salt rinsing step was critical to reducing interference of salts from the ASW and enhanced MS detection. DHB was found to be more suitable for these measurements because of its higher salt tolerance (as found in neurons) compared with α-cyano-4-hydroxycinnamic acid, and the improved peak resolution relative to concentrated DHB matrices. MS/MS analysis was performed using these peptidergic neurons (Figure 2).
Figure 1.
Two representative mass spectra demonstrating the peptide profile of the isolated neurons from A. californica. These neurons contain several peptides from the MIP-related peptide precursor and R3-14 peptide precursor.
Figure 2.
MS/MS profiles of endogenous N(l/d)WFa compared with synthetic NWFa and NdWFa standards. A. Two typical MS/MS mass spectrums generated from m/z 465.2, acquired from relevant neurons isolated from the abdominal ganglia of A. californica. B. Fragmentation patterns of the NdWFa and NWFa standards.
Ten animals were used for the isomeric content measurements. The average percentage of NdWFa measured is 89.5 ± 9.6%, indicating that the content of the DAACP epimer is surprisingly high in these relevant neurons. This is in agreement with previous capillary electrophoresis measurements48 and may indicate that the equilibrium is toward converting most of the peptide into the D-amino acid-containing form. Furthermore, the high levels of the D-form suggest that this is the bioactive peptide, as suggested by the observation that it takes three orders of magnitude higher concentration of NWFa than NdWFa to induce the same heart beating amplitude.6 Of course, when observing peptide contents from cells, one observes both the final products and processing intermediates, and so the small amount of the L-form peptide may indicate that the conversion occurs on a faster time scale than the release of these peptides. Regardless of the reasons, our data demonstrates that most of the NWFa exists as the NdWFa peptide.
Overall, this study shows that DAACPs with the 2nd-residue substitution were distinguished from their all-L variants using direct MS/MS methods; the isomeric content of a DAACP was quantified within a complex mixture in samples as small as several neurons. With the use of synthetic standards, chirality-reporting fragment ions are identifiable by comparing the abundance of respective major fragments from the epimers having the 2nd-position isomerization using MALDI MS. The chiral recognition factor varies with specific peptide sequences, but most of the biologically relevant peptides tested are discriminated, sometimes with the use of metal-complexed cations. In biological samples, the ratio RM calculated from these reporters can be used to determine the natural isomeric content based on calibration curves obtained with standards. If prior knowledge about the target peptide is available, single cells or tissue samples can be assayed and the fraction in each form characterized. This option provides an advantage because the direct cell measurements involve samples where the target peptide is more concentrated and the ratios provide information on peptide forms within a biological context.
Although no general features associated with DAACPs have been determined that allow their “blind” identification, this study demonstrates a fast and sensitive downstream approach to confirm candidates that potentially contain a D-amino acid. Using MALDI MS rather than other ionization techniques allows multiple experiments to be conducted within the same sample spots, so that peptides can be surveyed in a high-throughput fashion and then selected samples can be tested using the chiral measurements. This method, together with high-throughput enzymatic screening of complex peptide samples,41 provides an important addition to the approaches needed to characterize novel DAACPs.
Supplementary Material
Acknowledgments
We thank Dr. Peter M. Yau and Dr. Brian Imai, from the Protein Sciences Facility at the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, with peptide synthesis. The project described was supported by Award No. CHE-0400768 from the National Science Foundation (NSF) and by Award No. 5RO1NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS). The content is solely the responsibility of the authors and does not necessarily represent the official views of NSF or NINDS.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Predel R, Brandt W, Kellner R, Rapus J, Nachman RJ, Gäde G. Eur J Biochem. 1999;263:552. doi: 10.1046/j.1432-1327.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- 2.Gualillo O, Lago F, Casanueva FF, Dieguez C. Mol Cell Endocrinol. 2006;256:1–8. doi: 10.1016/j.mce.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Bai L, Sheeley S, Sweedler JV. Bioanal Rev. 2009;1:7–24. doi: 10.1007/s12566-009-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broccardo M, Erspamer V, Falconierierspamer G, Improta G, Linari G, Melchiorri P, Montecucchi PC. Br J Pharmacol. 1981;73:625–631. doi: 10.1111/j.1476-5381.1981.tb16797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreil G, Barra D, Simmaco M, Erspamer V, Erspamer GF, Negri L, Severini C, Corsi R, Melchiorri P. Eur J Pharmacol. 1989;162:123–128. doi: 10.1016/0014-2999(89)90611-0. [DOI] [PubMed] [Google Scholar]
- 6.Morishita F, Nakanishi Y, Kaku S, Furukawa Y, Ohta S, Hirata T, Ohtani M, Fujisawa Y, Muneoka Y, Matsushima O. Biochem Biophys Res Commun. 1997;240:354–358. doi: 10.1006/bbrc.1997.7659. [DOI] [PubMed] [Google Scholar]
- 7.Kamatani Y, Minakata H, Kenny PTM, Iwashita T, Watanabe K, Funase K, Sun XP, Yongsiri A, Kim KH, Novalesli P, Novales ET, Kanapi CG, Takeuchi H, Nomoto K. Biochem Biophys Res Commun. 1989;160:1015–1020. doi: 10.1016/s0006-291x(89)80103-2. [DOI] [PubMed] [Google Scholar]
- 8.Lee DL, Powers JPS, Pflegerl K, Vasil ML, Hancock REW, Hodges RS. J Pept Res. 2004;63:69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause E, Beyermann M, Dathe M, Rothemund S, Bienert M. Anal Chem. 1995;67:252–258. doi: 10.1021/ac00098a003. [DOI] [PubMed] [Google Scholar]
- 10.Mangoni ML, Papo N, Saugar JM, Barra D, Shai YC, Simmaco M, Rivas L. Biochem. 2006;45:4266–4276. doi: 10.1021/bi052150y. [DOI] [PubMed] [Google Scholar]
- 11.Adams CM, Kjeldsen F, Patriksson A, van der Spoel D, Graslund A, Papadopoulos E, Zubarev RA. Int J Mass spectrom. 2006;253:263–273. [Google Scholar]
- 12.Iwakoshi E, Hisada M, Minakata H. Peptides. 2000;21:623–630. doi: 10.1016/s0196-9781(00)00201-1. [DOI] [PubMed] [Google Scholar]
- 13.Ohta N, Kubota I, Takao T, Shimonishi Y, Yasudakamatani Y, Minakata H, Nomoto K, Muneoka Y, Kobayashi M. Biochem Biophys Res Commun. 1991;178:486–493. doi: 10.1016/0006-291x(91)90133-r. [DOI] [PubMed] [Google Scholar]
- 14.YasudaKamatani Y, Kobayashi M, Yasuda A, Fujita T, Minakata H, Nomoto K, Nakamura M, Sakiyama F. Peptides. 1997;18:347–354. doi: 10.1016/s0196-9781(96)00343-9. [DOI] [PubMed] [Google Scholar]
- 15.Barra D, Mignogna G, Simmaco M, Pucci P, Severini C, Falconierierspamer G, Negri L, Erspamer V. Peptides. 1994;15:199–202. doi: 10.1016/0196-9781(94)90002-7. [DOI] [PubMed] [Google Scholar]
- 16.Mor A, Delfour A, Sagan S, Amiche M, Pradelles P, Rossier J, Nicolas P. FEBS Lett. 1989;255:269–274. doi: 10.1016/0014-5793(89)81104-4. [DOI] [PubMed] [Google Scholar]
- 17.Erspamer V, Melchiorri P, Falconierierspamer G, Negri L, Corsi R, Severini C, Barra D, Simmaco M, Kreil G. Proc Natl Acad Sci U S A. 1989;86:5188–5192. doi: 10.1073/pnas.86.13.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mignogna G, Simmaco M, Kreil G, Barra D. EMBO J. 1993;12:4829–4832. doi: 10.1002/j.1460-2075.1993.tb06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez EC, Olivera BM, Gray WR, Cruz LJ. J Biol Chem. 1996;271:28002–28005. doi: 10.1074/jbc.271.45.28002. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez EC, Craig AG, Watkins M, Hillyard DR, Gray WR, Gulyas J, Rivier JE, Cruz LJ, Olivera BM. Biochem. 1997;36:989–994. doi: 10.1021/bi962840p. [DOI] [PubMed] [Google Scholar]
- 21.Jacobsen RB, Jimenez EC, De la Cruz RGC, Gray WR, Cruz LJ, Olivera BM. J Pept Res. 1999;54:93–99. doi: 10.1034/j.1399-3011.1999.00093.x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen R, Jimenez EC, Grilley M, Watkins M, Hillyard D, Cruz LJ, Olivera BM. J Pept Res. 1998;51:173–179. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- 23.Buczek O, Yoshikami D, Watkins M, Bulaj G, Jimenez EC, Olivera BM. FEBS J. 2005;272:4178–4188. doi: 10.1111/j.1742-4658.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- 24.Buczek O, Yoshikami D, Bulaj G, Jimenez EC, Olivera BM. J Biol Chem. 2005;280:4247–4253. doi: 10.1074/jbc.M405835200. [DOI] [PubMed] [Google Scholar]
- 25.Buczek O, Jimenez EC, Yoshikami D, Imperial JS, Watkins M, Morrison A, Olivera BM. Toxicon. 2008;51:218–229. doi: 10.1016/j.toxicon.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Dutertre S, Lumsden NG, Alewood PF, Lewis RJ. FEBS Lett. 2006;580:3860–3866. doi: 10.1016/j.febslet.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Han YH, Huang FJ, Jiang H, Liu L, Wang Q, Wang YF, Shao XX, Chi CW, Du WH, Wang CG. FEBS J. 2008;275:1976–1987. doi: 10.1111/j.1742-4658.2008.06352.x. [DOI] [PubMed] [Google Scholar]
- 28.Pisarewicz K, Mora D, Pflueger FC, Fields GB, Mari F. J Am Chem Soc. 2005;127:6207–6215. doi: 10.1021/ja050088m. [DOI] [PubMed] [Google Scholar]
- 29.Fujisawa Y, Ikeda T, Nomoto K, Yasudakamatani Y, Minakata H, Kenny PTM, Kubota I, Muneoka Y. Compara Biochem Physiol C-Pharmacol Toxicol & Endocrinol. 1992;102:91–95. doi: 10.1016/0742-8413(92)90049-d. [DOI] [PubMed] [Google Scholar]
- 30.Soyez D, Vanherp F, Rossier J, Lecaer JP, Tensen CP, Lafont R. J Biol Chem. 1994;269:18295–18298. [PubMed] [Google Scholar]
- 31.Yasuda A, Yasuda Y, Fujita T, Naya Y. Gen Comp Endocrinol. 1994;95:387–398. doi: 10.1006/gcen.1994.1138. [DOI] [PubMed] [Google Scholar]
- 32.Heck SD, Kelbaugh PR, Kelly ME, Thadeio PF, Saccomano NA, Stroh JG, Volkmann RA. J Am Chem Soc. 1994;116:10426–10436. [Google Scholar]
- 33.Torres AM, Menz I, Alewood PF, Bansal P, Lahnstein J, Gallagher CH, Kuchel PW. FEBS Lett. 2002;524:172–176. doi: 10.1016/s0014-5793(02)03050-8. [DOI] [PubMed] [Google Scholar]
- 34.Torres AM, Tsampazi C, Geraghty DP, Bansal PS, Alewood PF, Kuchel PW. Biochem J. 2005;391:215–220. doi: 10.1042/BJ20050900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shikata Y, Watanabe T, Teramoto T, Inoue A, Kawakami Y, Nishizawa Y, Katayama K, Kuwada M. J Biol Chem. 1995;270:16719–16723. doi: 10.1074/jbc.270.28.16719. [DOI] [PubMed] [Google Scholar]
- 36.Jilek A, Mollay C, Tippelt C, Grassi J, Mignogna G, Mullegger J, Sander V, Fehrer C, Barra D, Kreil G. Proc Natl Acad Sci U S A. 2005;102:4235–4239. doi: 10.1073/pnas.0500789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres AM, Tsampazi M, Tsampazi C, Kennett EC, Belov K, Geraghty DP, Bansal PS, Alewood PF, Kuchel PW. FEBS Lett. 2006;580:1587–1591. doi: 10.1016/j.febslet.2006.01.089. [DOI] [PubMed] [Google Scholar]
- 38.Kubo T, Nishimura S, Kumagae Y, Kaneko I. J Neurosci Res. 2002;70:474–483. doi: 10.1002/jnr.10391. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Sweedler JV. Ann Rev Anal Chem. 2008;1:451–483. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 40.Svensson M, Skold K, Svenningsson P, Andren PE. J Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 41.Ewing MA, Wang J, Sheeley SA, Sweedler JV. Anal Chem. 2008;80:2874–2880. doi: 10.1021/ac7025173. [DOI] [PubMed] [Google Scholar]
- 42.Tao WA, Zhang DX, Nikolaev EN, Cooks RG. J Am Chem Soc. 2000;122:10598–10609. [Google Scholar]
- 43.Loo JA. Int J Mass spectrom. 2000;200:175–186. [Google Scholar]
- 44.Adams CM, Zubarev RA. Anal Chem. 2005;77:4571–4580. doi: 10.1021/ac0503963. [DOI] [PubMed] [Google Scholar]
- 45.Sachon E, Clodic G, Galanth C, Amiche M, Ollivaux C, Soyez D, Bolbach G. Anal Chem. 2009;81:4389–4396. doi: 10.1021/ac9002886. [DOI] [PubMed] [Google Scholar]
- 46.Hummon AB, Sweedler JV, Corbin RW. Trac-Trends Anal Chem. 2003;22:515–521. [Google Scholar]
- 47.Li LJ, Garden RW, Romanova EV, Sweedler JV. Anal Chem. 1999;71:5451–5458. doi: 10.1021/ac9907181. [DOI] [PubMed] [Google Scholar]
- 48.Sheeley SA, Miao H, Ewing MA, Rubakhin SS, Sweedler JV. Analyst. 2005;130:1198–1203. doi: 10.1039/b504717j. [DOI] [PubMed] [Google Scholar]
- 49.Morishita F, Nakanishi Y, Sasaki K, Kanemaru K, Furukawa Y, Matsushima O. Cell Tissue Res. 2003;312:95–111. doi: 10.1007/s00441-003-0707-3. [DOI] [PubMed] [Google Scholar]
- 50.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. J Mass Spectrom. 1996;31:1126–1130. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 51.Rubakhin SS, Jurchen JC, Monroe EB, Sweedler JV. Drug Discov Today. 2005;10:823–837. doi: 10.1016/S1359-6446(05)03458-6. [DOI] [PubMed] [Google Scholar]
- 52.Floyd PD, Li LJ, Rubakhin SS, Sweedler JV, Horn CC, Kupfermann I, Alexeeva VY, Ellis TA, Dembrow NC, Weiss KR, Vilim FS. J Neurosci. 1999;19:7732–7741. doi: 10.1523/JNEUROSCI.19-18-07732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsunematsu H, Ikeda H, Hanazono H, Inagaki M, Isobe R, Higuchi R, Goto Y, Yamamoto M. J Mass Spectrom. 2003;38:188–195. doi: 10.1002/jms.428. [DOI] [PubMed] [Google Scholar]
- 54.Serafin SV, Maranan R, Zhang KL, Morton TH. Anal Chem. 2005;77:5480–5487. doi: 10.1021/ac050490j. [DOI] [PubMed] [Google Scholar]
- 55.Cooks RG, Wong PSH. Acc Chem Res. 1998;31:379–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.