Abstract
Small molecule fluorometric boron dipyrromethene probes were developed to bind hepatitis C virus-encoded NS5A protein and aid subcellular distribution studies. These molecules did not co-locate with NS5A, therefore alternative ‘silent’ azide reporters were used to obtain a more relevant picture of their distribution. Following pre-incubation with replicon cells, click chemistry was used to append a fluorophore to the azide that confirmed the co-localisation of the small molecule with the NS5A protein, thus providing greater insight into the antiviral mode of action of this chemotype.
Electronic supplementary material
The online version of this article (doi:10.1007/s12154-010-0047-1) contains supplementary material, which is available to authorized users.
Keywords: Hepatitis C virus, Click chemistry, Labelling, Small molecules
Introduction
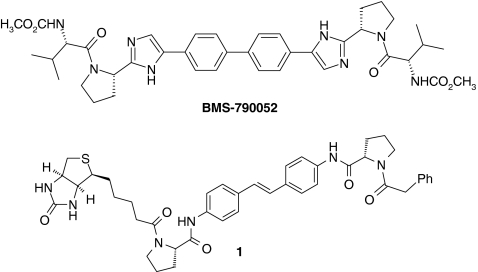
Hepatitis C virus (HCV) infection is a global public health problem and it has been estimated that 3% of the world’s population are infected with HCV (http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index1.html). Gao et al. [1] have previously described a chemical proteomics strategy to identify NS5A, an HCV-encoded zinc-binding phosphoprotein, as the target of the potent inhibitor BMS-790052, a clinical candidate for the treatment of HCV. In this work, the structurally-related biotinylated stilbene derivative 1 was incubated with genotype 1b replicon cells and NS5A was affinity-isolated using streptavidin–agarose beads following cell lysis, to aid identification of NS5A as the relevant biological target of the inhibitor (Fig. 1) [1].
Fig. 1.
HCV replication inhibitor BMS-790052 and biotinylated probe 1
In tissue culture cells replicating HCV RNA, NS5A is sequestered in replication complexes, which are essentially factories for HCV genome synthesis [2, 3]. Replication complexes are thought to form from invaginations in the membrane of the endoplasmic reticulum (ER) to create vesicles that house HCV-encoded proteins, specific host factors required by the virus for RNA synthesis, and replicating HCV RNA [2–4]. Consequently, NS5A is localised to the ER in cells replicating HCV RNA [2].
We decided to conjugate the boron dipyrromethene (BODIPY®) fluorophore to the stilbene template in 1 to assess the subcellular distribution of this chemotype in the 1b replicon cells and to confirm the co-localisation with NS5A protein following immunofluorescence staining to further support target identification [5–7]. The BODIPY® fluorophore was chosen due to the high extinction coefficients and quantum efficiencies associated with these derivatives. Additionally, a number of BODIPY® succinimidyl esters are commercially available [8] thus aiding the synthetic tractability of conjugation to the stilbene scaffold. Moreover, a range of absorption/emission wavelengths are available and we decided the BODIPY® 558/568 derivative was appropriate for confocal microscopy visualisation (absorption 558 nm/emission 568 nm) [9].
A short and long (polyethylene glycol, PEG) linker to the fluorophore was used to explore the effect of tether length on subcellular distribution. BODIPY® dyes 3 and 4 were prepared following amidation of the known amine 2 (Scheme 1) [1, 10].
Scheme 1.
Preparation of BODIPY® dyes 3 and 4. (i) BODIPY® 558/568 succinimidyl ester, NEt3, CH2Cl2; (ii) BocNH(CH2CH2O)4CH2CH2CO2H, azabenzotriazolyl tetramethyluromium hexafluorophosphate, (HATU), Hünigs base, DMF; (iii) trifluoroacetic acid, CH2Cl2, water; (iv) BODIPY® succinimidyl ester, NEt3, CH2Cl2
Both compounds were confirmed as inhibitors of HCV replication (Table 1). Genotype 1b replicon cells were then treated with 3 or 4, fixed and visualised, and then immunostained for NS5A protein (see Experimental section). Interestingly, neither compound appeared to co-localise with the NS5A protein (Fig. 2).
Table 1.
Replicon 1b potencies
| Compound | 3 | 4 | 5 | 6 |
|---|---|---|---|---|
| Replicon 1b EC50 (μM) | 0.80 | 1.0 | 0.013 | 0.11 |
| cLogP | 9.5 | 8.8 | 6.3 | 5.7 |
Fig. 2.
Treatment of cells with BODIPY® dyes 3 (a–c) and 4 (d–i). a, d, g BODIPY® inhibitors; b, e, h NS5A; c, f, i merged images. g, h, i Enlarged images of the areas in the white rectangle
It appeared that the presence of the large and lipophilic boron dipyrromethene moiety could significantly perturb the distribution characteristics of the NS5A replication inhibitors [11] (see Table 1 for calculated lipophilicity, cLogP). We therefore sought a method to append a fluorophore to a molecule that more closely resembled the parent, in structure and physicochemical properties, after exposure of cells to the inhibitor (e.g., cLogP of BMS-790052 is 4.7). Copper-catalysed Huisgen [3+2] cycloaddition chemistry between an alkyne and an azide [12, 13] is ideally suited to this task due to its bio-orthogonality and specificity, thereby eliminating potential background reactions [14]. Moreover, recent work using copper-free cycloadditions has broadened the scope of click chemistry for cellular imaging and target identification studies as it avoids the use of toxic copper that can compromise the viability of living cells [15, 16].
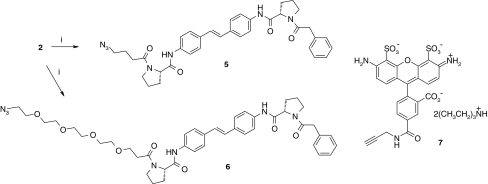
We decided to append an azide tag to the stilbene template for subsequent click chemistry labelling, and adopted a similar strategy to explore the effect of tether length; therefore, 5 and 6 were prepared (Scheme 2). The lipophilicity of the resulting probes was significantly reduced (Table 1).
Scheme 2.
Preparation of azido-NS5A inhibitors 5, 6 and structure of alkyne-Alexa Fluor® 488 7. (i) Acid, azabenzotriazolyl tetramethyluromium hexafluorophosphate, (HATU), Hünigs base, THF
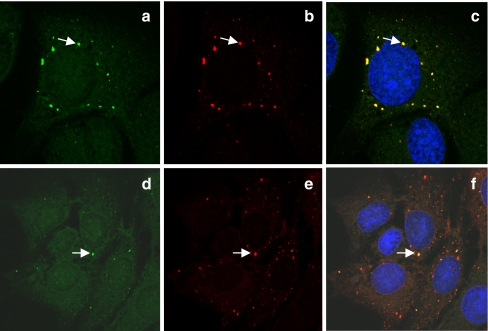
Compounds 5 and 6 were more potent inhibitors of replication than BODIPY-labelled molecules 3 and 4 (Table 1) presumably due to the smaller azide more readily satisfying the structure–activity relationships of the NS5A protein versus the larger BODIPY® dye. Pleasingly, when cells were incubated with probes 5 and 6, then fixed and labelled using the Click-iT™ alkyne-tethered Alexa Fluor® 488 7 (http://products.invitrogen.com/ivgn/product/A10267) (see Experimental section), the molecules are clearly seen to co-localise with the NS5A protein, thus vindicating our original strategy (Fig. 3). There appeared to be no difference between the short and long-tethered azide inhibitors in the labelling efficiency or localisation.
Fig. 3.
Imaging of azido inhibitors 5 (a–c) and 6 (d–e) following click reaction with fluorophore 7. a, d Alexa Fluor® 488; b, e NS5A; c, f merged images
As a control experiment, we treated 1b replicon cells with DMSO and then performed the click reaction with 7 in the absence of azido inhibitor to assess the intrinsic distribution profile of the alkyne dye. Figure 4 shows that 7 partially co-locates with NS5A, unlike the reaction with the azido inhibitors where there is full co-localisation (particularly clear in Fig. 3c). These results are important to understand the natural propensity for certain dyes to locate to specific subcellular regions [11]. The partial co-localisation observed between 7 and NS5A, in the absence of the azide-tagged NS5A-targeting molecules, presumably reflects non-specific association with components of methanol-fixed cells. This observation may be related to the fixation conditions, the particular cell line used, or the physical–chemical properties of the alkyne-tagged Alexa fluorophore. However, despite this, co-localisation with NS5A appeared absolute in the presence of the azide-tagged NS5A-targeting molecules. Further work is warranted to explain these unusual observations.
Fig. 4.
Imaging of DMSO control: a Alexa Fluor® 488, b NS5A, c merged image
In conclusion, we have created azide-containing HCV replication inhibitors that can be imaged in 1b replicon cells following click attachment of a fluorometric dye. These probes were shown to co-localise with the viral protein NS5A, thus supporting the mode of action of these molecules. These results demonstrate that careful design of ‘clickable’ small molecule probes can enable efficient subcellular distributions to be delineated. In this work we were also able to show that a covalent linkage between the small molecule inhibitor and the target protein is not necessary for imaging purposes. Further work in our group includes the exploration of copper-free alkyne-azide cycloaddition chemistry to enable imaging in live cells. Also, the effects of NS5A-targeting small molecules on the intracellular distribution of NS5A will be the subject of a subsequent publication.
Experimental
Cell culture
Replicon cells (Huh-7 human hepatoma-derived cells supporting constitutive replication of con1 strain HCV genotype 1b subgenomic RNA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum, 500 μg/ml G418, and 1× each of penicillin/streptomycin, sodium pyruvate, non-essential amino acids, and l-glutamine.
Compound addition
Replicon cells (1 × 105) were seeded onto 13-mm glass coverslips placed in each well of a 24-well cell culture plate. Compounds were added to replicon cells to give a final concentration of 500 nM (click probes) or 5 μM (BODIPY®-tagged compounds) in 1% DMSO, control cells received DMSO only. Cells were then incubated for 17 h at 37°C.
Cell fixation
Cells were washed in PBS and fixed in 100% methanol at −20°C for 25 min. Following subsequent washing with PBS, cells were either subjected to the click reaction or, if treated with BODIPY®-tagged compounds, processed directly for indirect immunofluorescence.
Click reaction
Cells were treated with 100 μM N-methyl-maleimide for 15 min prior to the addition of the click labeling solution (300 μM CuSO4, 1.2 mM TBTA-OH, 5 mM aminoguanidine, 5 mM ascorbate, 5 mM Alexa Fluor® 488-alkyne) prepared in PBS. Cells were incubated in the dark for 1.5 h, then washed with PBS, and stained for NS5A by indirect immunofluorescence.
Indirect immunofluorescence
Cells were probed for the presence of HCV-encoded NS5A protein using a sheep polyclonal antiserum specific for NS5A. Use of the NS5A antiserum has been reported previously [2, 17, 18]. It exhibits very little non-specific background and, in methanol-fixed HCV replicon-containing cells, reliably detects authentic distribution of NS5A in replication complexes co-localized with the ER membrane [2]. Recognition of the NS5A antigen is independent of NS5A conformation, i.e. the antiserum will still bind to NS5A protein following denaturing polyacrylamide gel electrophoresis and transfer to nitrocellulose membrane (Western blot). Bound primary antibody was detected through the use of donkey anti-sheep secondary antibodies appropriately conjugated with Alexa Fluor® dyes. Confocal images were captured using a Leica TCS SP5 inverted confocal microscope and associated software.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 48 kb)
References
- 1.Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun J-H, O’Boyle DR, II, Lemm JA, Wang C, Knipe JO, Chien C, Colonno RJ, Grasela DM, Meanwell NA, Hamann LG. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Targett-Adams P, Boulant S, McLauchlan J. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinkert D, Bartenschlager R, Lohmann V. J Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnsson N, Johnsson K. ACS Chem Biol. 2007;2:31–38. doi: 10.1021/cb6003977. [DOI] [PubMed] [Google Scholar]
- 6.Rao J, Dragulescu-Andrasi A, Yao H. Curr Opin Biotechnol. 2007;18:17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Sadaghiani AM, Verhelst SH, Bogyo M. Curr Opin Chem Biol. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Molecular Probes (web edition) The handbook—a guide to fluorescent probes and labeling technologies, Chapter 1.4
- 9.Cole L, Davies D, Hyde GJ, Ashford AE. J Microsc. 2000;197:239. doi: 10.1046/j.1365-2818.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- 10.Bachand C, Belema M, Deon DH, Good AC, Goodrich J, James CA, Lavoie R, Lopez OD, Martel A, Meanwell NA, Nguyen VN, Romine JL, Ruediger EH, Snyder LB, St. Laurent DR, Yang F, Langley DR, Wang G, Hamann LG (2008) WO2008021927
- 11.Cunningham CW, Mukhopadhyay A, Lushington GH, Blagg BSJ, Prisinzano TE, Krise JP. Mol Pharm. 2010;7:1301–1310. doi: 10.1021/mp100089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 14.Yang P-Y, Liu K, Ngai MH, Lear MJ, Wenk MR, Yao SQ. J Am Chem Soc. 2010;132:656–666. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- 15.Fokin VV. ACS Chem Biol. 2007;2:775–778. doi: 10.1021/cb700254v. [DOI] [PubMed] [Google Scholar]
- 16.Sletten EM, Bertozzi CR. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macdonald A, Crowder K, Street A, McCormick C, Saksela K, Harris M. J Biol Chem. 2003;278:17775–17784. doi: 10.1074/jbc.M210900200. [DOI] [PubMed] [Google Scholar]
- 18.Targett-Adams P, McLauchlan J. J Gen Virol. 2005;86:3075–2080. doi: 10.1099/vir.0.81334-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(DOC 48 kb)