Abstract
Background & Aims
We previously reported that the NS2 protein of hepatitis C virus (HCV) inhibits the expression of reporter genes driven by a variety of cellular and viral promoters. The aim of the study was to determine whether the broad transcriptional repression is caused by endoplasmic reticulum (ER) stress.
Methods
Phosphorylation of the translation initiation factor eIF2α and HCV replication were detected by Western and Northern blot, respectively. De novo protein synthesis was measured by metabolic labeling. Activation of ER stress responsive genes was determined by promoter reporter assay, as well as mRNA and protein measurement by real time PCR and Western blot.
Results
Transient or inducible NS2 protein expression increased eIF2α phosphorylation and reduced de novo protein synthesis. It up-regulated promoter activities and transcript levels of ER stress inducible genes including GRP78, ATF6, and GADD153, as well as GRP78 protein level. The same effect was observed when NS2 was synthesized as part of the core-E1-E2-p7-NS2 polypeptide. NS2 protein also inhibited reporter gene expression from the HCV internal ribosome entry site and consequently reduced HCV replication. The full-length HCV replicon activated GRP78, ATF6, and GADD153 promoters more efficiently than the subgenomic replicon lacking the coding sequence for both the structural proteins and NS2. Abrogation of HCV infection/replication, by an inhibitor of the NS3 protease, relieved ER stress.
Conclusions
HCV infection can induce ER stress, with NS2 protein being a major mediator. The stress can be relieved by a feedback mechanism.
Introduction
Hepatitis C virus (HCV) infects approximately 3% of the world population, leading to liver steatosis, cirrhosis, and eventually hepatocellular carcinoma [1,2]. The molecular mechanisms of viral persistence and pathogenesis are not well understood. HCV translates a large polyprotein, from a single open reading frame through an internal ribosomal entry site (IRES), which is processed at the endoplasmic reticulum (ER) by both host- and viral-derived proteases to generate structural proteins (core, E1, E2) and non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Transfection experiments and transgenic mouse models revealed the ability of individual viral proteins to modulate cellular signaling pathways implicated in proliferation / apoptosis, lipid metabolism, and host immune response [3,4]. The NS2 protein is a transmembrane protein of 217 amino acids, which harbors an autoprotease activity at its C-terminus, responsible for the release of the amino-terminus of the NS3 protein [5].
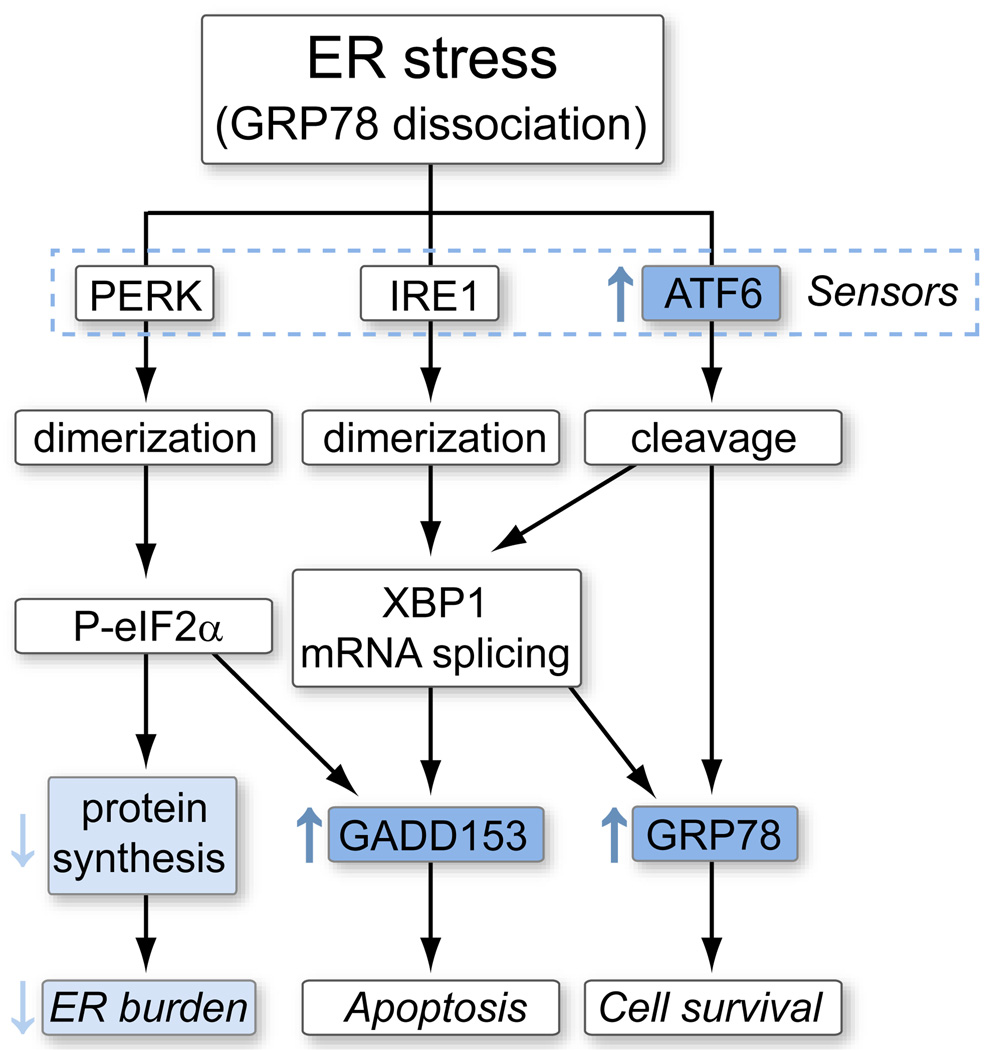
We recently reported that the NS2 protein of genotype 1a, but not the corresponding core, E1, or p7 protein, markedly inhibits luciferase and green fluorescent protein expression driven by cytomegalovirus (CMV), simian virus 40 (SV40), tumor necrosis factor alpha (TNFα), NFκB, and ferrochelatase promoters [6]. The NS2 protein also suppresses replication of hepatitis B virus (HBV) and secretion of its e and surface antigens. In this regard, global repression of protein synthesis is a hallmark of cellular response to ER stress induced by misfolded proteins, protein overexpression or viral infection (Fig. 1) [7]. PERK (PKR-like ER kinase), IRE1 (inositol-requiring enzyme 1), and ATF6 (activating transcription factor 6) are stress sensors located in the ER; each sensor is bound via its luminal domain to GRP78 (glucose regulated protein 78), a major molecular chaperone. ER stress leads to GRP78 disengagement, thus triggering the unfolded protein response characterized by eIF2α phosphorylation and general translational attenuation to reduce ER protein load; up regulation of enzymes involved in protein degradation to eliminate misfolded proteins; as well as up regulation of ER chaperones such as GRP78 to refold the proteins. These concerted efforts relieve ER stress and lead to cell survival unless the stress is severe or prolonged. In the present study, we measured promoter activities and transcription levels of GRP78, ATF6, and GADD152, protein level of GRP78, and phosphorylation status of eIF2α. To rule out the possibility that the ER stress observed is an artifact of isolated NS2 expression or attributed to the transfection procedure, we also co-expressed NS2 with core, E1, E2, and p7 (core-NS2 cassette), and induced NS2 expression from a stable transfectant. The biological relevance of these findings was further tested using the full-length HCV replicon and an infectious HCV clone.
Fig. 1. Diagram of the ER stress pathway.
Molecular chaperone GRP78 is normally bound to the three stress sensors: PERK, IRE1, and ATF6. Its dissociation during ER stress triggers dimerization or cleavage of the sensors, which unleashes a cascade of molecular signaling events as indicated by solid arrows. Dashed arrows indicate potential cross talks between different pathways. The components analyzed in the present study are shown in boldface.
Materials and Methods
Plasmid constructs
Promoter sequences were amplified from genomic DNA extracted from Huh-7 cells. Table 1 provides the primer sequences. The GRP78 promoter (122-bp) and GADD153 promoter (364-bp) were amplified according to previous reports [8, 9]. Since the promoter of ATF-6 has not been characterized, a 1 kb DNA fragment upstream of the coding sequence (nt −992 to +24) was amplified. The PCR products were cloned into a promoter-less luciferase reporter plasmid pGL3-Basic (Promega). The coding sequences for the NS2 protein of 1a, 1b, and 2a genotypes (Accession #AAA45677, AJ238799, and AB047639) preceded by an ATG codon were inserted into expression vector pcDNA3.1/Zeo(+) for transient transfection. The same sequence of the 2a genotype was cloned into the pTRE2Hyg vector for stable transfection. The HA-tagged NS2 expression constructs in the pcDNA3.1/Zeo(+) vector contain the HA tag immediately behind the initiating methionine. The NS2-null construct of genotype 1a contains a TGA stop codon immediately after the ATG codon. The phosphorylation site, S168, was mutated in the S168A mutant, and deleted in the ΔSQME mutant (residues 168–171). The coding sequences for core to p7 and core to NS2 of the 2a genotype were cloned into pcDNA3.1/Zeo(−) vector via EcoRI / BamHI sites. For a luciferase reporter under control of HCV 5' UTR, the cDNA of nucleotide 1–398 of the HCV genome (GenBank accession #AJ238799) was cloned into pGl3-basic vector via KpnI and HindIII sites. The KpnI – BamHI fragment of this construct was cloned into pcDNA3.1/Zeo+ vector and digested with BamHI for run-off mRNA transcription in vitro [10]. HCV replicon of genotype 2a (JFH) or a derivative with the luciferase reporter (JFH-luc) have been described [11]. The full-length (FL) and subgenomic (SG) replicons of 1b genotype, both containing an S2204I adaptive mutation in the NS5A region, as well as the full-length replicon with a defective polymerase (Pol−), were kindly provided by Dr. Charles Rice [10].
Table 1.
Primers used for PCR cloning and mutagenesis.
| Name | Sense | Anti-sense |
|---|---|---|
| GRP78 promoter | ggggtaccGGAGGGGGCCGCTTCGAATCGGC | ggctcgagTTATATACCCTCCCCCAGCCCCGT |
| GADD153 promoter | ggggtaccTGTCCCCTGCGCGTGCGCGTGCAGA | ggaagcttAGACTTAAGTCTCTGACCTCGGGA |
| ATF-6 promoter | ggggtaccTAATATACCCACGAGCAGTTTG | ggaagcttGGCAACCCCAGCCGGCTCCCCCAT |
| GRP78 qPCR | CTGTCCAGGCTGGTGTGCTCT | CTTGGTAGGCACCACTGTGTTC |
| GADD153 qPCR | GGTATGAGGACCTGCAAGAGGT | CTTGTGACCTCTGCTGGTTCTG |
| ATF6 qPCR | CAGACAGTACCAACGCTTATGCC | GCAGAACTCCAGGTGCTTGAAG |
| 18S qPCR | ATCTTGGGAGCGGGCGGGCG | GCGGTCCTATTCCATTATTCCTA |
| ΔSQME (5–576bp) | ggggatccATGCTGGACACGGAGGTGGCCGCG | CGTGATGAGCTTGGTGAAGACGACTGGCTCTACAGCCAC |
| ΔSQME (3–165bp) | GAGCCAGTCGTCTTCACCAAGCTCATCACGTGGGGGGCA | cggaattcCTACAGCAACCTCCACCCCTTGGAGACC |
| S168A (5–588bp) | ggggatccATGCTGGACACGGAGGTGGCCGCG | GAGCTTGGTCTCCATTTGAGCGAAGACGACTGGCTC |
| S168A (3–177bp) | GAGCCAGTCGTCTTCGCTCAAATGGAGACCAAGCTCATC | cggaattcCTACAGCAACCTCCACCCCTTGGAGACC |
| Core-NS2 (2a) | acagaattcACCATGGGAAGCACAAATCCTAAACCTCAAAGAAA | attggatccCTACCCCTTGGAGGTGTAGCCATCA |
| Core-p7 (2a) | acagaattcACCATGGGAAGCACAAATCCTAAACCTCAAAGAAA | attggatccTTACCGGGGCAGTGCCATGAGCAG |
Note: Italic represents irrelevant sequence and restriction sites (KpnI, XhoI, BamHI, EcoRI or HindIII) for cloning.
Cell culture and transfection
Huh-7 and Huh-7.5 cells were cultured as described [3]. DNA constructs were transfected using polyamine as a carrier [3], while RNA was transfected by electroporation. Briefly, Huh-7 or Huh-7.5 cells (~7×105) were suspended in 70 µl of Nucleofector T solution with supplement (Amaxa), and mixed with in vitro transcribed HCV RNA (5 µg). Similar procedures were used for electroporation of NS2 construct (1 µg) and in vitro transcribed RNA (5 µg) of HCV 5' UTR-luciferase reporter construct. A reporter plasmid for the secreted alkaline phosphatase (SEAP) (Clontech) was co-transfected to evaluate the transfection efficiency. For inducible NS2 protein expression, Huh-7 cells were first transfected with Tet regulatory vectors pTet-on and pTet-tTS (Clontech) at a ratio of 1:10. Following addition of G418 (200 µg/ml), individual clones were expanded. They were transiently transfected with a luciferase reporter plasmid (pTRE2hyg-luciferase, Clontech) to identify clones with tight regulation of expression. One such clone was further transfected with pTRE2hyg-NS2 plasmid, and grown in the presence of both G418 and hygromycin (100 µg/ml) to select for doubly transfected cells.
Promoter activity assay
Transfected cells were grown in 24-well plates and luciferase activity was measured according to manufacturer’s instruction (Luciferase Assay System, Promega). Light intensity was read in 96-well plates in a luminometer. Data are presented as mean ± SD (n = 4). Differences among the groups were examined via student’s t-test. All assays were repeated at least three times using HA-Tagged or non-tagged NS2 constructs with consistent observations.
Real time PCR
Cellular RNA was extracted and reverse transcribed to cDNA as described [3]. Quantitative real time PCR was performed using 2 µl of 1:50 diluted cDNA and QuantiTect SYBR Green PCR Mixture in 20 µl using Mastercycler ep realplex instrument and software (Eppendorf AG, Hamburg, Germany). Ribosomal 18S was measured in parallel to serve as an internal control.
Western blot, antibodies, and inhibitors
Proteins were detected by Western blot using incubation conditions recommended by the antibody manufacturers. Signals were revealed by enhanced chemiluminescence (ECL). The GRP78 protein was detected by immunoprecipitation (IP)-Western blot (H-29, Santa Cruz Biotechnology). Other antibodies were: HA Tag (Upstate), human albumin (Roche), eIF2α and phosphorylated eIF2α (Cell Signaling), β-actin (Sigma), glycine decarboxylase [12], HCV NS2 [6], and core (C-750) [3]. Inhibitors: IFN (Sigma) and HCV NS3 protease inhibitor Ac-DELE-Cha-C (AnaSpec, Fremont, CA) [13].
Detection of de novo protein synthesis
As described previously [14], Huh-7 cells grown in 6-well plates were starved in methionine/cysteine-free medium for 1 h before incubation for 4 h with the same medium supplemented with 35S-methionine/cysteine (Perkin Elmer) at 0.1 mCi/ml. Cells were lysed and 20 µg of proteins were separated in SDS- PAGE followed by fluorography.
Analysis of HCV RNA replication
RNA was extracted from transfected Huh-7.5 cells using Trizol solution (Invitrogen). Ten µg of RNA was separated in 1% agarose gel and transferred to Hybond N+ filter, which was incubated at 42°C overnight with a randomly primed HCV cDNA probe. The blot was washed twice with 2× SSC/0.1% SDS and twice with 1× SSC/0.1% SDS, at 65°C for 15 min, and exposed to X-ray films.
Results
NS2 protein enhanced eIF2α phosphorylation and reduced general protein translation
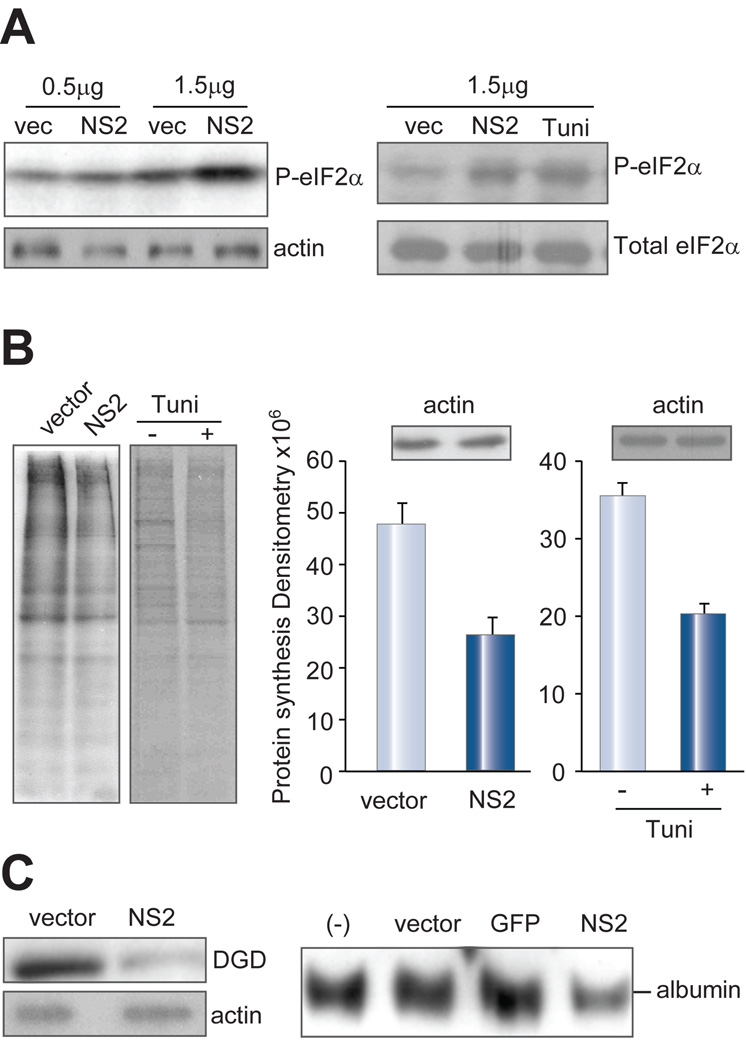
The activation status of the PERK pathway was monitored by Western blot analysis of eIF2α phosphorylation. Higher levels of eIF2α phosphorylation were observed in Huh-7 cells transfected with the NS2 expression construct than in cells transfected with the empty vector (Fig. 2A). Metabolic labeling followed by densitometric analysis revealed approximately 50% reduction in new protein synthesis in cells expressing the NS2 protein compared to those transfected with the empty vector (Fig. 2B). The effect was comparable to treatment with tunicamycin, a known ER stress inducer. Steady-state levels of glycine decarboxylase (DGD) and albumin, two liver-specific proteins, also declined (Fig. 2C). These findings demonstrate the global nature of translational repression and link this phenomenon to phosphorylation of eIF2α.
Fig. 2. NS2 increases eIF2α phosphorylation and reduces de novo protein synthesis.
(A) eIF2α phosphorylation. Huh-7 cells (7×105) were transfected with 0.5 or 1.5 µg of empty vector (pcDNA3.1/Zeo+) or NS2 cDNA, and harvested at day 2 post-transfection (p.t.) for Western blot analysis of the phosphorylated eIF2α. The right panel shows comparison of NS2 protein with tunicamycin (tuni) treatment. Duplicate blots were probed with antibodies recognizing total and phosphorylated eIF2α, respectively. (B) Metabolic labeling of de novo protein synthesis. Huh-7 cells (7×105) were labeled with 35S-Met/Cys 12 hrs post-transfection with 1 µg of NS2 construct or empty vector. Cell lysate (20 µg) was separated in SDS-PAGE and signals were revealed by fluorography. Tunicamycin (Tuni, 5 µg/ml) treatment served as a positive control. Western blot detection of β-actin, a protein with relatively long half life, served as a control for equal volume loading. Signals were quantified by densitometry using NIH image program and averaged from three experiments. (C) Endogenous proteins. Huh-7 cells (7×105) were transfected with 1 µg of empty vector or NS2 cDNA, and harvested at day 3 p.t. for Western blot analysis of glycine decarboxylase (DGD). Albumin was detected from culture supernatant.
NS2 protein increased GRP78, GADD153, and ATF6 mRNA transcription
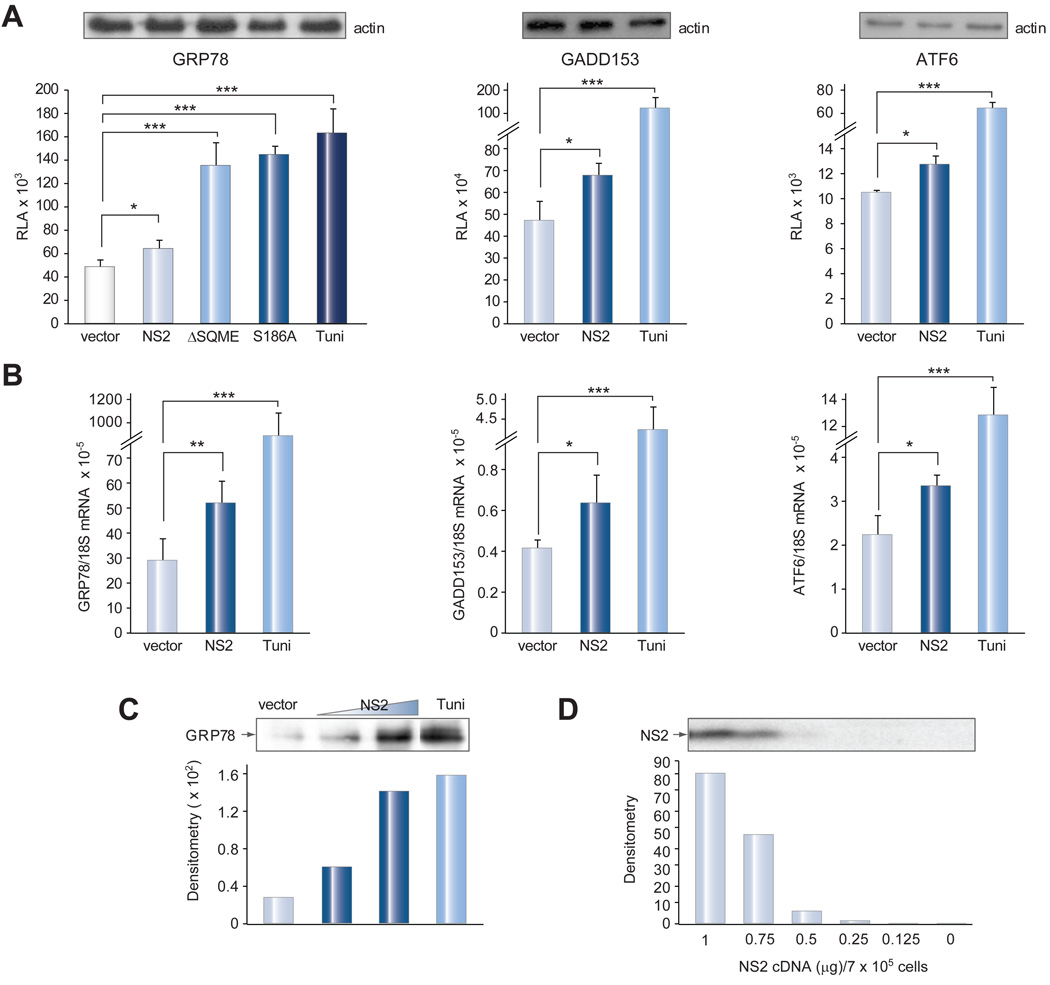
Three complementary approaches were taken. First, the promoter sequences of GRP78, ATF6, and GADD153 were cloned upstream of the luciferase reporter gene, and these reporters were then co-transfected with the NS2 construct or the empty vector. Luciferase expression driven by all three promoters was higher in Huh-7 cells co-transfected with the NS2 construct than with the empty vector (Fig. 3A). The effect was also dose-dependent (Supplementary Fig. 1). Moreover, preventing NS2 phosphorylation by mutating (S168A) or deleting the phosphorylation site (ΔSQME), which increases the steady-state level of NS2 [15], further enhanced GRP78 promoter activity (Fig. 3A, left panel). Second, real time PCR revealed higher levels of GRP78, ATF6, and GADD153 mRNAs in NS2 transfected Huh-7 cells than in vector transfected cells (Fig. 3B), consistent with transcriptional activation. Third, Western blot analysis revealed increased GRP78 protein level depending on the amount of NS2 cDNA transfected (Fig. 3C).
Fig. 3. NS2 protein activated three ER stress inducible genes.
(A) GRP78, GADD153, and ATF6 promoter activities. Huh-7 cells grown in 24-well plates (1.5 × 105) were transfected in quadruplicate with a luciferase reporter construct driven by GRP78, GADD153, or ATF6 promoter (35 ng/well), together with the empty vector or the expression construct for the full-length NS2 protein (70 ng/well). Cells were lyzed 2 days later for measurement of luciferase activity. Relative luciferase activity (RLA) is presented as the mean ± SD and adjusted by transfection efficiency according to a SEAP assay. For positive control, cells were incubated with 10 µg/ml of tunicamycin (Tuni) overnight. Actin served as a loading control (top). (B) GRP78, GADD153, and ATF6 mRNA levels. Huh-7 cells were transfected with empty vector or NS2 construct as described above. mRNA was quantified by reverse transcription-real time PCR. The relative mRNA abundance (ng) was calculated over 18S, and shown as the mean ± SD (n-4). *p <0.05, ** p <0.005, ***p <0.0005. (C) GRP78 protein level. Huh-7 cells (~7 × 105) were transfected with empty vector (1 µg) or the NS2 expression construct (0.5 µg or 1µg). GRP78 was detected by IP-Western blot. Tunicamycin treatment (10 µg/ml) served as a control. (D) Dose-dependent expression of NS2 protein (1a genotype). NS2 was detected at day 2 p.t. by a monoclonal antibody against the HA tag. This serves as a reference for relative NS2 expression level in different transfection experiments.
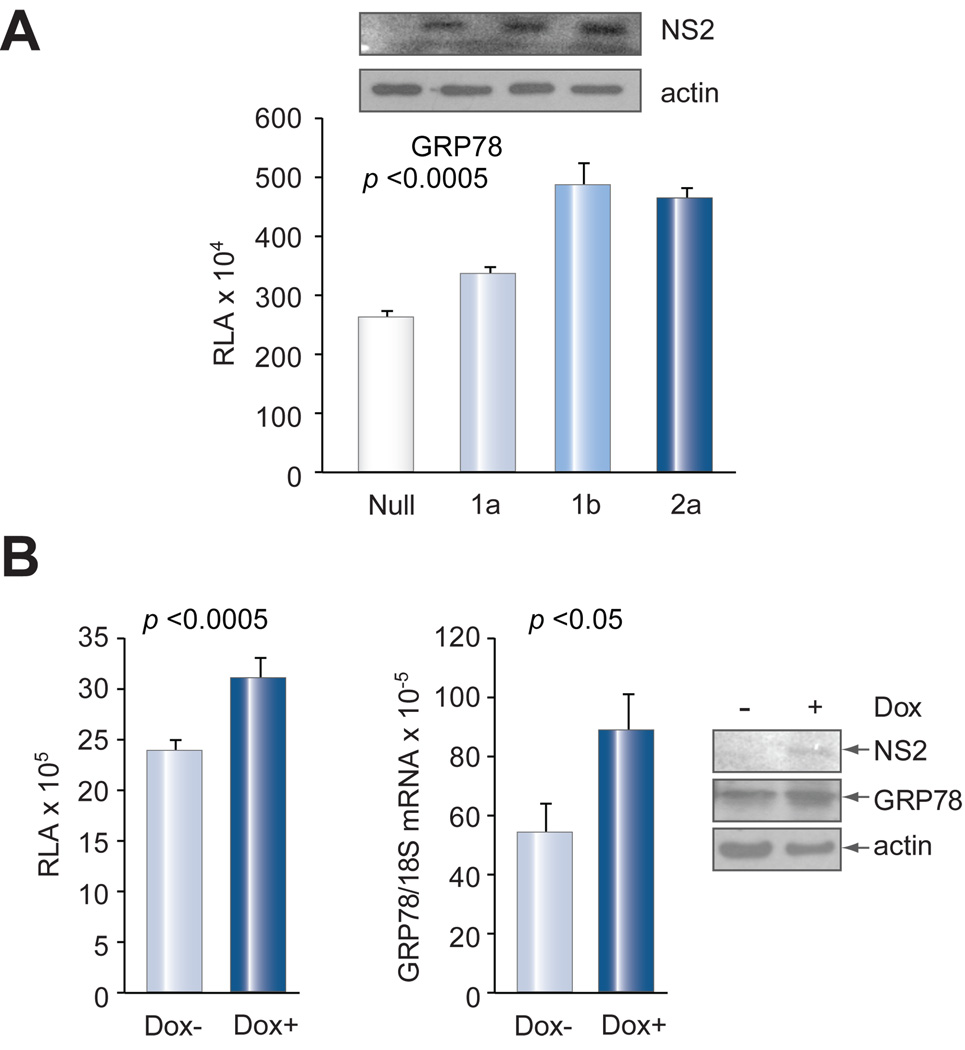
The study described so far was based on NS2 protein of the 1a genotype. Interestingly, NS2 protein of 1b and 2a genotypes stimulated GRP78 promoter activity more efficiently, despite comparable expression levels as shown by Western blot using the HA-tag antibody (Fig. 4A). Considering the possibility that the transfection procedure itself may trigger ER stress, we established a stable transfectant of the NS2 protein of the 2a genotype. Following transient transfection of GRP78 promoter reporter construct, cells were either untreated or treated with doxycycline to induce NS2 protein expression (Fig. 4B, right panel). Indeed, the promoter activity of GRP78 (Fig. 4B, left panel), and endogenous GRP78 mRNA and protein levels (Fig. 4B, middle & right panels), were significantly increased in doxycycline treated cells.
Fig. 4. (A) GRP78 promoter activity in response to NS2 protein of different genotypes.
All constructs were cloned in pcDNA3.1/Zeo+ with identical Kozak sequence and HA-tag. NS2 null mutant served as a control. Expression of NS2 protein of different genotypes is shown at the top, and actin served as a loading control. (B) Activation of GRP78 by inducible expression of NS2. Huh-7 cells stably transfected with the NS2 construct were transfected with the GRP78 promoter construct followed by addition of doxycycline (1 µg/ml). Luciferase activity (left panel) and GRP78 mRNA (middle panel) were measured two days later. Data are presented as mean ± SD (n-4). NS2 and GRP78 proteins were revealed by Western blot (right panel).
Transfection with the full-length HCV replicon or infection by HCV induced ER stress
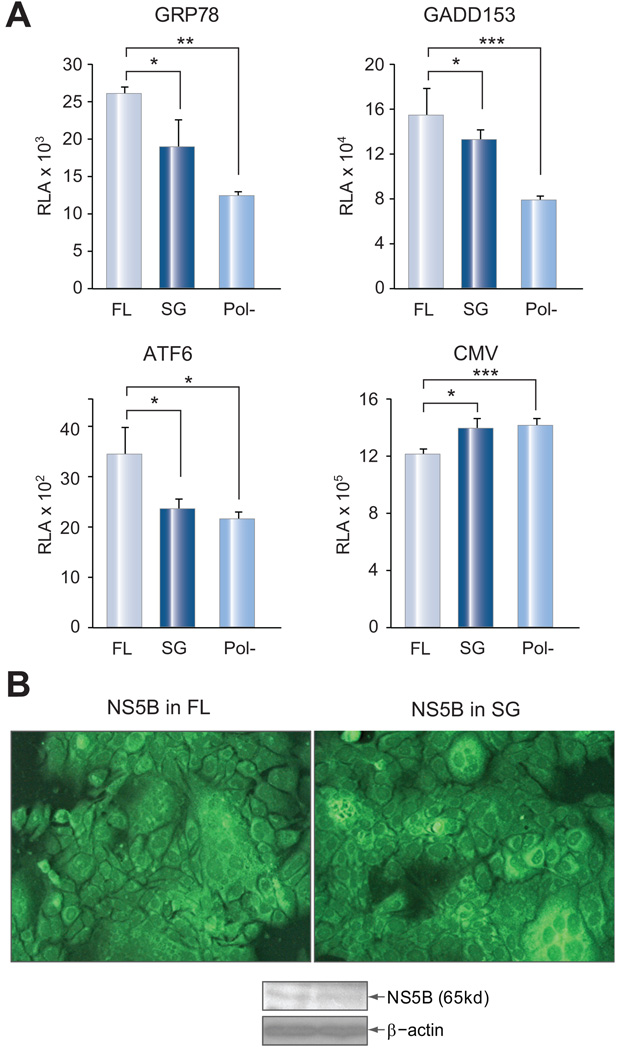
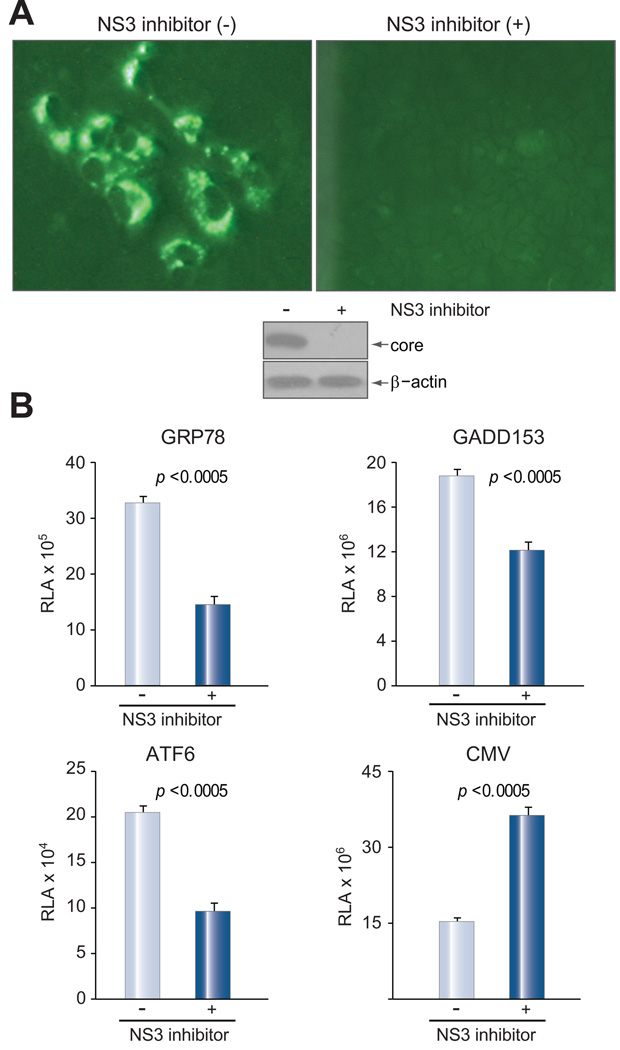
If NS2-mediated ER stress is not an artifact of its expression in isolation, then the full-length (FL) HCV replicon should be more efficient at inducing ER stress than the subgenomic (SG) replicon, which does not express the NS2 protein. A full-length construct deficient in replication because of mutation in the active site of the NS5B polymerase served as a negative control [10]. Cells were electroporated with replicon RNAs and subsequently transfected with the luciferase reporter construct driven by different promoters. Immunofluorescence staining and Western blot analysis revealed similar levels of steady state NS5B protein in FL and SG replicon transfected cells (Fig. 5B), but the FL replicon triggered higher level of luciferase expression driven by the GRP78, GADD153 or ATF6 promoter (Fig. 5A). In contrast, luciferase expression driven by the CMV promoter was higher in cells transfected with the SG replicon (Fig. 5A, right panel). These findings indicate that the FL replicon triggers greater ER stress than the SG replicon. A direct test for the role of NS2 protein has been unsuccessful, because deletion of the coding sequence for NS2 residues 5–130, or 5–217 from the full-length replicon abolished genome replication. In another approach, Huh-7.5 cells were transfected with an infectious full-length clone of 2a genotype (JFH). Continuous in vitro culture led to the establishment of chronic infection, as demonstrated by immunostaining and Western blot analysis of the core protein (Fig. 6A). Treatment with an NS3 protease inhibitor (or IFN) not only efficiently reduced HCV replication (Fig. 6A, middle and right panels), but also relieved ER stress as suggested by reduced GRP78, GADD153, and ATF6 promoter activities but enhanced CMV promoter activity (Fig 6B and Supplementary Fig. 2). Taken together, these results indicate that the full-length HCV genome induces ER stress whether through transfection or infection, although the role of NS2 protein cannot be established.
Fig. 5. Full-length HCV replicon induced severer ER stress than the subgenomic replicon.
Huh-7.5 cells (~2.8 × 106) were electroporated with 4 µg of RNA of the FL or SG replicon (Con1 strain), and seeded into 24-well plates. A replication deficient FL replicon was included as a negative control. After an overnight incubation, cells were transfected, in quadruplicate, with the promoter constructs of GRP78, GADD153, ATF6, and CMV (50ng/well). (A) Luciferase assays at day 2 p.t. Relative luciferase activity (RLA) is presented as the mean ± SD (n=4) and adjusted by transfection efficiency. (B) Immunofluorescent staining and Western blot analysis of NS5B protein. Note that the signals represent the steady state protein produced from input replicon RNA and newly synthesized RNA.
Fig. 6. Effect of an NS3 protease inhibitor on HCV-induced ER stress.
Huh.7.5 cells (6×106) were electroporated with 5 µg of HCV RNA of 2a genotype (JFH) and cultured under sub-confluent condition for 2 months. Duplicate samples were treated or not treated with an NS3 protease inhibitor (20 µM). (A) IF staining and Western blot analysis of core protein following three days of treatment. (B) Promoter activities in response to the protease inhibitor. Cells seeded in 24 well plates were transfected with the promoter reporter constructs (0.25 µg/well), followed by addition of the NS3 protease inhibitor. Cells were harvested 3 days later for luciferase assay. Data are presented as the mean ± SD (n = 4).
NS2 but not HCV structural proteins activated GRP78 promoter in the presence of SG replicon
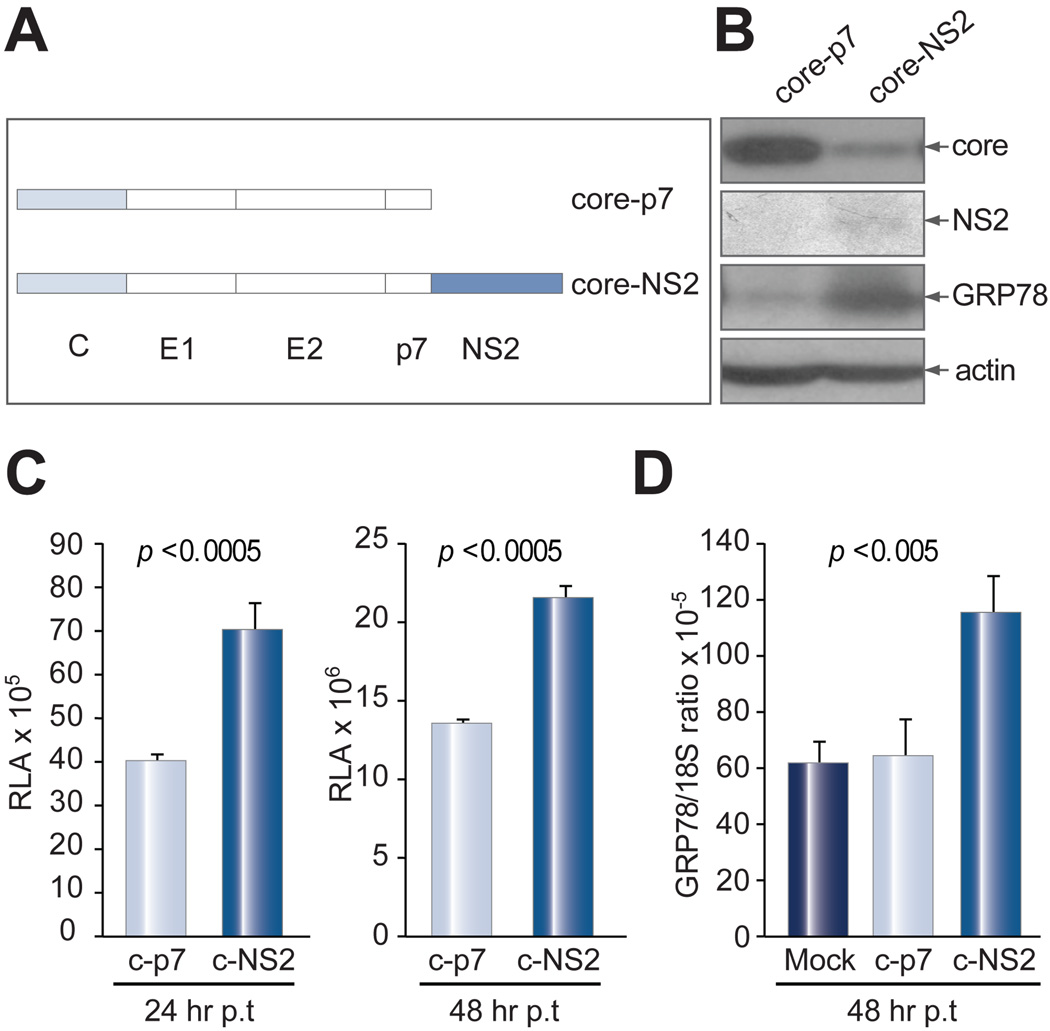
The SG replicon differs from the FL replicon in lacking the sequence coding for E1, E2, p7, and NS2 proteins. We electroporated Huh-7 cells with the GRP78 promoter construct together with 5 µg of cellular RNA derived from Huh-7 cells that had been transfected with core, E1, E2, E1-p7, NS2, or NS2 null constructs. Only the RNA derived from cells transfected with the NS2 construct markedly stimulated GRP78 promoter activity (Supplementary Fig. 3A). Similar results were obtained in Huh-7.5 cells in the presence of the SG replicon (Supplementary Fig. 3B). Furthermore, Huh-7 cells transfected with the core-NS2 cassette (expressing core, E1, E2, p7, and NS2) generated higher level of luciferase from the GRP78 promoter than cells transfected with the core-p7 cassette (expressing core, E1, E2, and p7) (Fig. 7C). The same is true with GRP78 protein and transcript levels (Fig. 7B and D). This result demonstrates that the NS2 protein, when translated and processed from a polyprotein, could trigger ER stress of the same order as during authentic infection. Interestingly, the core protein level was much reduced in cells expressing the NS2 protein (Fig. 7B), indicating feedback repression of protein translation. These results suggest that NS2 protein rather than core, E1, or E2 protein is a major mediator of cellular ER stress.
Fig. 7.
NS2 induced ER stress in the context of viral structural proteins. (A) Diagram of core - p7 and core - NS2 constructs. (B) Western blot analysis of core and NS2 proteins and IP-Western blot of GRP78. (C) Luciferase assay for GRP78 promoter activities at 24 and 48 hrs p.t. (D) Real time PCR assay for endogenous GRP78 mRNA level at 48 hrs p.t. Data are presented as mean ± SD (n = 4).
NS2 inhibited protein translation mediated by HCV IRES and suppressed HCV replication
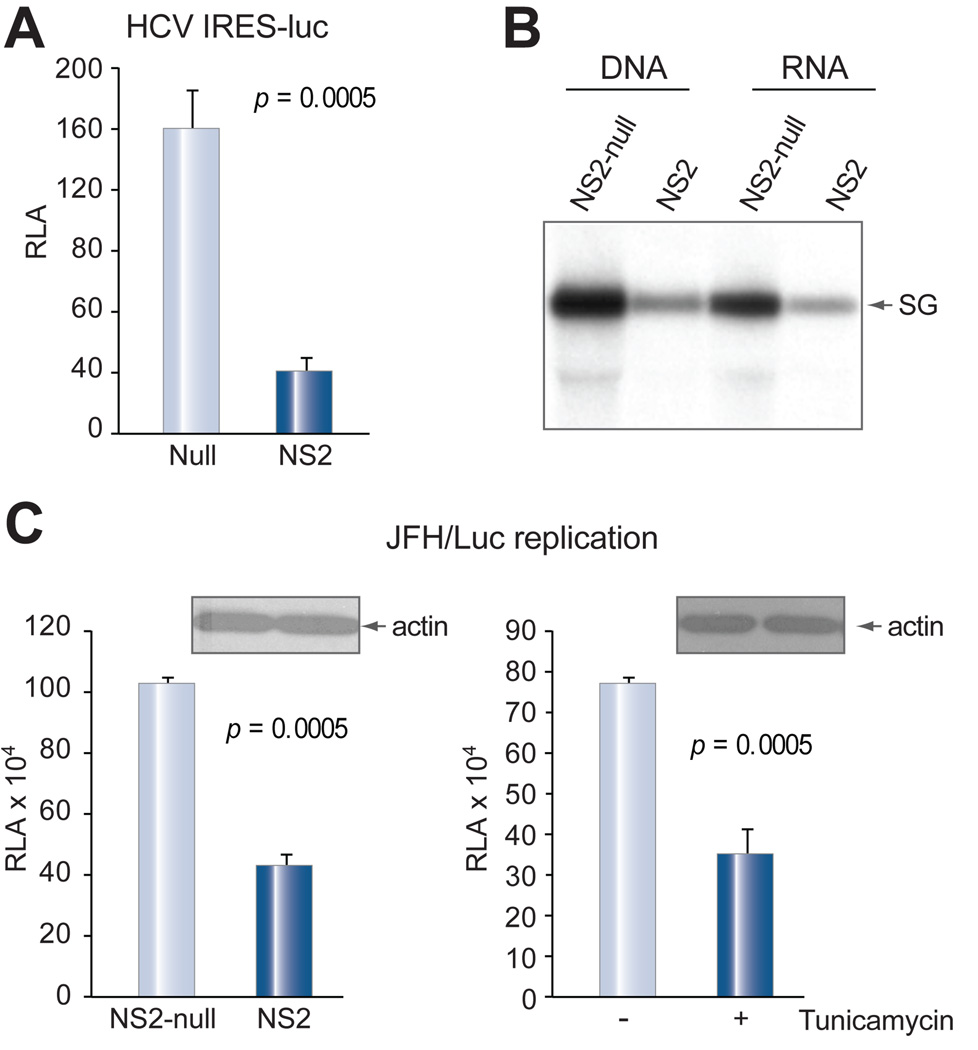
To determine whether translation of HCV polyprotein, which is mediated by internal ribosomal entry site (IRES), is also subject to translational repression, we generated a luciferase expression construct under the HCV IRES. The RNA transcribed in vitro was transfected into Huh-7.5 cells together with the NS2 expression construct or its null mutant. The NS2 protein markedly suppressed luciferase expression (Fig. 8A). Since replication of the HCV genome requires nonstructural proteins, we further examined the effect of NS2 on replication of the subgenomic replicon. The NS2 DNA construct reduced HCV replication by more than 50% compared to its null construct (Fig. 8B, two left lanes). A similar inhibitory effect was observed when RNA, derived from Huh-7 cells that had been transfected with NS2 construct, was transfected into Huh-7.5 cells (Fig. 5B, two right lanes). In another experiment, Huh-7.5 cells were electroporated with HCV replicon RNA (JFH-Luc) [11] together with the NS2 expressing construct, and viral replication readily measured by luciferase activity [11]. As shown in Fig. 8C, NS2 protein significantly reduced viral replication (left panel), similar to tunicamycin treatment (right panel).
Fig. 8. NS2 protein inhibited IRES-driven protein translation and HCV genome replication.
(A) Protein translation driven by the HCV 5' UTR. Huh-7 cells were electroporated with 5 µg of in vitro transcribed RNA of HCV 5’ UTR-luciferase construct, together with 1 µg of NS2 cDNA or its null mutant. Luciferase was measured 2 days later. Data are presented as mean ± SD (n = 4) and adjusted by transfection efficiency. (B) Replication of susbgenomic replicon (Con1). Huh-7.5 cells (6×106) were electroporated with 1 µg each of subgenomic replicon RNA and NS2 cDNA or its null mutant (left two lanes), or the replicon RNA together with 5 µg of total RNA derived from Huh-7 cells that had been transfected with the corresponding DNA for 2 days (right two lanes). HCV replication was detected 3 days later by Northern blot. (C) Replication of JFH-luc replicon. Huh-7.5 cells (6×106) were electroporated with 5 µg of replicon RNA together with NS2 cDNA or its null mutant (1 µg), and seeded in quadruplicate. Viral replication was measured at day 3 post electroporation by luciferase reporter assay. Alternatively, Huh-7.5 cells were electroporated with replicon RNA alone, and seeded in quadruplicate in two sets. One set was treated with tunicamycin (5 µg/ml) for 24 hrs. Viral replication was measured 3 days later.
Discussion
HCV infection may alter ER homeostasis. We previously reported that expression of NS2 but not HCV structural proteins suppressed luciferase or GFP reporter expression under the control of various cellular or viral promoters such as CMV, SV40, TNFα, and NFκB [6]. In the present study, we extended these observations to include endogenous cellular proteins such as albumin and glycine decarboxylase. This inhibition was not restricted to conventional cap-dependent translation but also applicable to translation initiated from HCV internal ribosomal entry site. Furthermore, metabolic labeling of NS2 transfected cells revealed approximately a 50% reduction in de novo protein synthesis suggesting the global nature of translational repression. The molecular basis for this repression is the increased phosphorylation of the translation initiation factor eIF2α. In contrast to general translational repression, the NS2 protein enhanced promoter activities and transcript levels of GRP78, ATF6, or GADD153, the three important ER stress responsive genes. We further validated the activation status of GRP78 at the protein level. These findings are consistent with the activation of all three branches of ER stress signaling including the PERK pathway leading to eIF2α phosphorylation and general translational suppression; the ATF6 cascade leading to transcriptional up regulation of GRP78; and the IRE1 pathway leading to transcriptional up regulation of GADD153 (Fig. 1). Failure of the NS2 null mutant to induce ER stress suggests that the NS2 protein rather than its DNA or RNA triggers ER stress. The effect of NS2 construct was dose-dependent (Fig. 2A and Supplementary Fig. 1A), and transient possibly due to the short half life of NS2 and feedback suppression of its translation by ER stress (Supplementary Fig. 1B). Preventing NS2 degradation through mutation or deletion of the phosphorylation site (S168A and ΔSQME) [15] induced even greater GRP78 promoter activity than the wild-type construct.
The E2 envelope protein of HCV has been reported to activate promoters of GRP78 and GRP94, two ER luminal chaperone proteins required for ER homeostasis [16]. Both E1 and E2 proteins were reported to activate IRE1-XBP1-GADD153 pathway in HepG2 and HeLa cells [17]. Another structural protein, core, was found to induce ER stress in transfected liver cells as well as in transgenic mice [18]. In addition, stable transfection of Huh-7 cells with an HCV subgenomic replicon led to the activation of ATF6/GRP78 pathway, although the IRE1-XBP1 signaling cascade was suppressed in this system [19,20]. In our hands, the NS2 protein is a more potent inducer of ER stress than the structural proteins. The NS2-induced ER stress appears biologically relevant. First, in addition to the 1a genotype that we originally studied, we could extend this finding to the 1b and 2a genotypes as well. Second, the effect could be achieved by both DNA construct and RNA extracted from cells transfected with such DNA construct. Third, besides transient transfection, inducible NS2 expression from stable transfectants also led to transcriptional up regulation of GRP78. Fourth, NS2 could trigger ER stress not only as a protein expressed alone, but also when processed from core-E1-E2-p7-NS2 fusion polypeptide. As anticipated from the proposed role of the NS2 protein in causing ER stress, full-length HCV replicon induced ER stress to a greater extent than the subgenomic replicon. Moreover, reduction of viral infection by an NS3 protease inhibitor or interferon was associated with reduced ER stress. In these cases the role of NS2 cannot be established unequivocally.
It is important to note that NS2 could suppress the expression of reporter protein driven by the HCV IRES element, suggesting that ER stress triggered by the NS2 protein (or together with other viral proteins) feeds back to reduce the expression of the precursor of NS2 itself, thus alleviating ER stress. Reduced production of viral nonstructural proteins essential for the replication complex also slows down genome replication. Whether that will facilitate viral persistence warrants further investigation.
Supplementary Material
SUPPLEMENTARY FIG. 1. (A) Dose-dependent effect of the NS2 protein on the GRP78 promoter. Huh-7 cells grown in 24-well plates were co-transfected with GRP78 promoter construct (35ng/well) and various amounts of NS2 construct or empty vector. Luciferase activity was measured two days later. Difference (P<0.05) was observed in cells transfected with 30 to 110 ng of NS2 vs. vector. (B) Kinetics of ER stress induced by the NS2 protein. Huh-7 cells stably transfected with the NS2 construct (Tet-on/NS2) were transfected with GRP78 promoter construct followed by addition of doxycycline (1µg/ml). Cells were harvested at 1, 2, 3 and 4 days later, and luciferase activity was measured accordingly. Data are presented as fold changes in Dox+ vs. Dox− cells.
SUPPLEMENTARY FIG. 2. Treatment of HCV infected cells with interferon reduced ER stress. Huh.7.5 cells (6×106) were electroporated with 5µg of HCV RNA of 2a genotype (JFH) and cultured under sub-confluent condition for 2 months. Cells were maintained for 4 more passages with or without 105/ml interferon (IFN, Sigma), before transfection in quadruplicate with the GRP78, GADD153, ATF6, and CMV promoter constructs (0.25µg/1.5× 105 cells). (A) HCV infection as detected by IF staining and Western blot of viral core protein. (B) Promoter activities based on luciferase assay at day 2 p.t.
SUPPLEMENTARY FIG. 3. NS2 but not HCV structural proteins stimulated GRP78 promoter activity. (A) Experiments performed in the absence of HCV replicon. Huh-7 cells (~7 × 105) were electroporated with GRP78 promoter construct (200ng) together with total RNAs (5µg) extracted from Huh-7 cells that had been transfected with core, E1, E2, E1-p7 or NS2 cDNA, or the NS2 null mutant. Cells were seeded in quadruplicate into 24-well plates and harvested two days later for luciferase assay. (B) Experiments performed in the presence of subgenomic replicon. Huh-7.5 cells (~1 × 106) were electroporated with SG replicon RNA (1µg) together with GRP78 promoter construct (200ng) and cellular RNAs (5µg) derived from Huh-7 cells that had been transfected with the NS2 or structural protein constructs. Tunicamycin (Tuni) added at a concentration of 10µg/ml posttransfection served as a positive control for ER stress. Luciferase assay was performed two days later. Data are presented as the mean ± SD (n=4) and adjusted by transfection efficiency. * P<0.05, **P<0.005. Actin served as protein loading control of luciferase assay, while ribosomal RNA (18 and 18S) served as cellular RNA control used for co-transfection.
Acknowledgments
We would like to thank Dr. Charles Rice, Rockefeller University, New York, for providing replicon constructs and Huh-7.5 cells. We also thank Dr. R. Bartenschlager for providing JFH-Luc construct. This work was supported, in part, by NIH grants DK066950, CA109733, CA95490, CA35711, AA08169, P20 RR15578 and DK28614. J. Li was a liver scholar of American Liver Foundation.
Abbreviations
- HCV
hepatitis C virus
- NS2
no-structural protein 2
- ER
endoplasmic reticulum
- eIF2α
eukaryotic translation initiation factor 2
- GRP78
glucose regulated protein 78
- ATF6
activating transcription factor 6
- GADD153
growth arrest and DNA damage induced gene-153
- CMV
cytomegalovirus
- SV40
simian virus 40
- TNFα
tumor necrosis factor alpha
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- HBV
hepatitis B virus
- FL
full-length
- SG
subgenomic
- SEAP
secreted alkaline phosphatase
- ECL
enhanced chemiluminescence
- PERK
PKR-like ER kinase
- IRE1
inositol-requiring enzyme 1
- IRES
internal ribosomal entry site
- DGD
glycine decarboxylase
- p.t.
post transfection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosure
None
REFERENCES
- 1.Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K, Nakano Y, et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 2.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- 4.Giannini C, Brechot C. Hepatitis C virus biology. Cell Death Differ. 2003;10 Suppl 1:S27–S38. doi: 10.1038/sj.cdd.4401121. [DOI] [PubMed] [Google Scholar]
- 5.Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci U S A. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumoulin FL, von dem Bussche JA, Li J, Khamzina L, Wands JR, Sauerbruch T, et al. Hepatitis C virus NS2 protein inhibits gene expression from different cellular and viral promoters in hepatic and nonhepatic cell lines. Virology. 2003;305:260–266. doi: 10.1006/viro.2002.1701. [DOI] [PubMed] [Google Scholar]
- 7.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 10.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JS, Tong SP, Wands JR. Characterization of a 120-Kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J Virol. 1996;70:6029–6035. doi: 10.1128/jvi.70.9.6029-6035.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingallinella P, Altamura S, Bianchi E, Taliani M, Ingenito R, Cortese R, et al. Potent peptide inhibitors of human hepatitis C virus NS3 protease are obtained by optimizing the cleavage products. Biochemistry. 1998;37:8906–8914. doi: 10.1021/bi980314n. [DOI] [PubMed] [Google Scholar]
- 14.Tong S, Li J, Wands JR. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franck N, Le Seyec J, Guguen-Guillouzo C, Erdtmann L. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J Virol. 2005;79:2700–2708. doi: 10.1128/JVI.79.5.2700-2708.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberman E, Fong YL, Selby MJ, Choo QL, Cousens L, Houghton M, et al. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J Virol. 1999;73:3718–3722. doi: 10.1128/jvi.73.5.3718-3722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SW, Egan PA. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. Faseb J. 2005;19:1510–1512. doi: 10.1096/fj.04-3455fje. [DOI] [PubMed] [Google Scholar]
- 18.Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–4933. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- 19.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279:17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 20.Tardif KD, Mori K, Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIG. 1. (A) Dose-dependent effect of the NS2 protein on the GRP78 promoter. Huh-7 cells grown in 24-well plates were co-transfected with GRP78 promoter construct (35ng/well) and various amounts of NS2 construct or empty vector. Luciferase activity was measured two days later. Difference (P<0.05) was observed in cells transfected with 30 to 110 ng of NS2 vs. vector. (B) Kinetics of ER stress induced by the NS2 protein. Huh-7 cells stably transfected with the NS2 construct (Tet-on/NS2) were transfected with GRP78 promoter construct followed by addition of doxycycline (1µg/ml). Cells were harvested at 1, 2, 3 and 4 days later, and luciferase activity was measured accordingly. Data are presented as fold changes in Dox+ vs. Dox− cells.
SUPPLEMENTARY FIG. 2. Treatment of HCV infected cells with interferon reduced ER stress. Huh.7.5 cells (6×106) were electroporated with 5µg of HCV RNA of 2a genotype (JFH) and cultured under sub-confluent condition for 2 months. Cells were maintained for 4 more passages with or without 105/ml interferon (IFN, Sigma), before transfection in quadruplicate with the GRP78, GADD153, ATF6, and CMV promoter constructs (0.25µg/1.5× 105 cells). (A) HCV infection as detected by IF staining and Western blot of viral core protein. (B) Promoter activities based on luciferase assay at day 2 p.t.
SUPPLEMENTARY FIG. 3. NS2 but not HCV structural proteins stimulated GRP78 promoter activity. (A) Experiments performed in the absence of HCV replicon. Huh-7 cells (~7 × 105) were electroporated with GRP78 promoter construct (200ng) together with total RNAs (5µg) extracted from Huh-7 cells that had been transfected with core, E1, E2, E1-p7 or NS2 cDNA, or the NS2 null mutant. Cells were seeded in quadruplicate into 24-well plates and harvested two days later for luciferase assay. (B) Experiments performed in the presence of subgenomic replicon. Huh-7.5 cells (~1 × 106) were electroporated with SG replicon RNA (1µg) together with GRP78 promoter construct (200ng) and cellular RNAs (5µg) derived from Huh-7 cells that had been transfected with the NS2 or structural protein constructs. Tunicamycin (Tuni) added at a concentration of 10µg/ml posttransfection served as a positive control for ER stress. Luciferase assay was performed two days later. Data are presented as the mean ± SD (n=4) and adjusted by transfection efficiency. * P<0.05, **P<0.005. Actin served as protein loading control of luciferase assay, while ribosomal RNA (18 and 18S) served as cellular RNA control used for co-transfection.