Abstract
Gene encoding heat shock protein (Hsps) are induced following a thermal stress thanks to the activation of heat shock transcription factor (HSF) which interacts with heat shock elements (HSE) located within the sequence of Hsp promoters. This cellular and protective response (heat shock response (HSR)) is well known and evolutionarily conserved. Nevertheless, HSR does not function in all the cells produced during the life of a multicellular organism, e.g., early mouse embryos. Taking advantage of mouse transgenic and knockout models, we investigated the roles of trans (HSF 1 and 2) and cis (HSE) regulatory elements in the control of Hsp70.1 (Hspa1b) through several developmental steps from oocytes to blastocysts. Our studies confirm that, even in absence of any stress, HSF1 regulates Hsp70.1 in oocytes and early embryos. Our data emphasize the role of maternal and paternal HSFs in the developmentally regulated expression of Hsp70.1 observed when the zygotic genome activation occurs. Furthermore, in this unstressed developmental condition, affinity and binding to HSEs might be more permissive than in the stress response. Finally, submitting blastocyst to different stress conditions, we show that HSF2 is differentially required for Hsp expression and cell survival. Taken together, our findings indicate that the role of heat shock trans and cis regulatory elements evolve along the successive steps of early embryonic development.
Keywords: Hsp, HSF, HSE, Embryos, Transgenic lines, Knockout
Introduction
Hsp genes encode heat shock proteins (HSPs), which are grouped in multigenic families based on their molecular weight. Expression and function of the multiple Hsp genes have been extensively studied and several of them (e.g., Hsp25, Hsp70) are best known for their stress-dependent induction by transcription factors named heat shock factors (HSFs) (Kampinga 2006; Lindquist and Craig 1988). Their promoters contain HSF DNA binding sites named HSE for heat shock element. HSF1 and HSF2, which share 70% of homology at the level of their DNA binding side domain, are the two main HSFs expressed in mammalian cells. Additional information on HSFs can be found in several reviews (Akerfelt et al. 2010a and references therein). HSF1 is the key regulator of the stress response elicited, e.g., by heat shock (heat shock response (HSR)) while in this context, HSF2 might exert a complex role of modifier of HSF1 function through different mechanisms such as bookmarking or formation of heterotrimer (Loison et al. 2006; Ostling et al. 2007; Sandqvist et al. 2009; Xing et al. 2005).
It was previously reported that despite the presence of HSF1 (Christians et al. 1997), fully grown oocytes and early mammalian embryos (see description of mouse early development in Fig. 1a) are unable to induce Hsp expression following heat stress as it is usually observed in adult somatic cells (Curci et al. 1987 and 1991; Hahnel et al. 1986; Morange et al. 1984). Using Hsp70.1Luc transgenic lines, we observed that heat shock provoked a different response of the transgene at each stage of the preimplantation development leading to the progressive establishment of the typical HSR characterized by a strong, rapid, and transient induction. In murine embryos, the classical heat-inducible expression can be observed at the blastocyst stage (3.5 days after fertilization, Fig. 1a). Based on work done on somatic cells, this inducible response is regulated by HSF1 and thus, abolished in HSF1-deficient cells (Inouye et al. 2004; McMillan et al. 1998; Zhang et al. 2002). In addition and in contrast to somatic cells, the embryonic cells at the blastocyst stage exhibit a constitutive DNA binding activity due to HSF2 (Mezger et al. 1994a, b). The role of this HSF2 activity remains to be clarified.
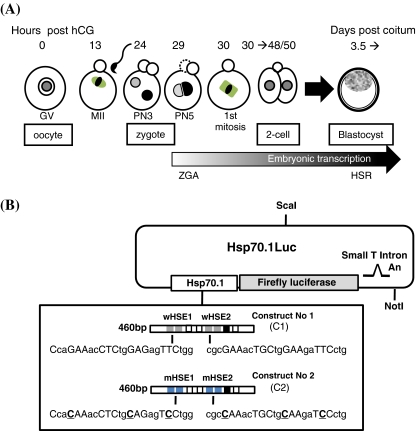
Fig. 1.
Biological and molecular materials used to study Hsp70.1 expression in oocytes and preimplantation embryos. a Description of the oocyte and embryonic developmental stages. Fully grown ovarian oocytes are noted germinal vesicle (GV) oocytes from the name given to their voluminous nucleus. Around ovulation, GV oocytes resume meiosis and progress until the metaphase stage of the 2nd meiotic division (MII) awaiting fertilization. The zygote is produced by the fusion between MII oocyte and spermatozoon. The zygotic genome activation (ZGA) is initiated during the G2 phase of the first cell cycle and transcriptional activity is first observed at the level of the paternal genome (male pronucleus) (Bouniol-Baly et al. 1997; Christians et al. 1995). The first mitosis gives an embryo with two cells (two-cell stage) which cleaves about four times before the formation of the blastocyst (3.5 days post-coitum) which are able to elicit the classical heat shock response (HSR). The timing of oocyte and early embryonic development is counted as hours post-hCG (for additional description of mouse oocyte and embryo development, see Kubiak et al. 2008). b Constructs used in transgenic experiments. Constructs No. 1 (C1) and No. 2 (C2) were linearized and microinjected to generate several independent transgenic lines (see also Wirth et al. 2002: line C1.101)
Before Hsp70 (Hsp70.1 and Hspab1) can be induced by heat shock at the blastocyst stage, this gene is transiently expressed from the end of the one-cell to the beginning of the long two-cell stage, in absence of any experimental stress (Bensaude et al. 1983; Christians et al. 1995). Based on competition experiments, this so-called constitutive, developmentally regulated expression was shown to involve HSF1 (Christians et al. 1997). More recently, it was claimed that HSF1 and HSF2 would modified the chromatin organization of Hsp70.1 in spermatozoa and make the gene accessible to rapid transcription after fertilization at the time of the zygotic genome activation (ZGA) (Wilkerson et al. 2008). Although other studies emphasized the role of HSF1 and HSF2 in the compaction of sperm genome (Akerfelt et al. 2008; 2010b), it remained to demonstrate how this intervention of HSFs functionally impacts gene expression at ZGA.
Those observations raised the question of the HSF–HSE interactions as well as the respective role of HSF1 and HSF2 existing in gametes and early embryos. To address the different aspects of this question, we took advantage of two panels of transgenic lines and two knockout mouse models (Hsf1 and Hsf2) and we focused our attention on Hsp70.1 regulation. In this paper, we chose to present our experiments as performed along the successive steps of early development from oocytes to blastocysts (Fig. 1a). Taken together, our data show that the roles exerted by both trans acting factors (HSF1 and HSF2) and cis acting regulatory element (HSE) evolve during development.
Materials and methods
Transgenic and knockout lines
Some of the transgenic lines (TgN(HSEw460)Hel; TgN(HSEm460)Hel) were described elsewhere (Wirth et al. 2002). The transgene includes the firefly luciferase reporter gene driven by a 460 bp Hsp70.1 promoter which contains all the consensus binding sites so far identified: two HSE sites (HSE1, CcaGAAacCTCtgGAGagTTCtgg; HSE2, cgcGAAacTGCtgGAAgaTTCctg), three Sp1 (Sp1.1, aaGGGGGGaac; Sp1.2, gaGGCGGGaagc; Sp1.3, gagtgGGCGGGgCCgg), one CAAT box, and one TATA box (Hunt and Calderwood 1990) (Fig. 1b). Five transgenic lines (named here C1.10 (copy number, 120), C1.18 (copy number, 105), C1.19 (copy number, 2), C1.89 (copy number, 55), and C1.101 (copy number, 20)) were established with the construct 1 containing wild-type HSE and are named TgN(HSEw460)Hel. Four transgenic lines (named here C2.11, C2.13, C2.2210, and C2.223) received the construct 2 with mutated HSE and are named TgN(HSEm460)Hel). Mutated mHSE1 and mHSE2 sequences correspond to CcaCAAacCTCtgCAGagTCCtgg and CgcCAAacTGCtgCAAgaTCCctg, respectively, where point mutations are rendered in bold and underlined (Christians et al. 1997) (Fig. 1b).
Mice from both knockout lines (Hsf1 (Hsf1tm1Ijb) and Hsf2 (Hsf2tm1Ijb)) were a gift from Dr. IJ Benjamin (Salt Lake City, UT) (McMillan et al. 1998 and 2002). They were maintained in a mixed genetic background. Protocols for animal breeding and experiments were approved by the Departmental Veterinary Office (Haute-Garonne) according to French legislation.
Production and manipulation of oocytes or embryos
Procedures followed to produce, collect and, if required, cultured oocytes or embryos were described elsewhere (Christians et al. 1995, 1997; Metchat et al. 2009). Briefly, ovarian fully grown oocytes were obtained from unprimed females by puncture of ovarian follicles. Preimplantation embryos (from zygotes/1-cell stage to blastocyst stage) were collected from superovulated females mated with males of defined genotype as indicated for each experiment. According to the experiment, oocytes or embryos were directly used for experiments or maintained in culture (M16, Sigma; see Bierkamp et al. 2010). Oocyte and embryonic stages with their timing of development are described in Fig. 1a.
When indicated, embryos were submitted to heat shock (43°C, 30 or 60 min; 42°C, 120 min) followed by recovery at 37°C. Heat shock conditions were adapted from Christians et al. (1997) and Paslaru et al. (2003).
Fluorescent staining of blastocysts: propidium iodide, Oregon green 488 phalloidin
To analyze cell morphology and viability after different levels of heat stress, blastocysts were incubated in a propidium iodide (PI; BD Pharmingen) solution (40 μg/ml in M2 (Sigma)). PI can only cross the plasma membrane of non viable cells and the number of stained cells/number of blastocyst cells was counted to evaluate the effect of heat stress. Oregon Green 488 phalloidin was used to stain the actin network as described previously (Bierkamp et al. 2010). Embryos were observed by confocal microscopy (CLSM Leica Sp5, Toulouse RIO-imaging).
Luciferase assay
Luciferase assay was performed as previously described (Christians et al. 1999) or using a kit following manufacturer’s protocol (Promega). Briefly, single or groups of oocytes or embryos were stored in 0.5-ml Eppendorf type tubes in 50 μl 1× reporter lysis buffer (Promega) and stored at −80°C. In order to measure luciferase activity, samples were thawed, transferred to a 96-well plate and 50 μl of luciferase substrate were added in each well. The light emission was integrated for the first 10 s. Background level for each series of samples to be assayed was measured under the same conditions with lysis buffer (blank), and the value obtained was subtracted from each measurement displayed by the luminometer (in order to exclude non responder samples).
Real-time RT-qPCR
Real-time experiment was performed as previously described (Metchat et al. 2009). The following primers were used : Hsp70.1 forward, 5′-TTGTCCATGT TAAGGTTTTGTGGTATA-3′, Hsp70.1 reverse, 5′-GTTTTTTTCATTAGTT TGTAGTGATGCAA-3′, Hsp105 forward, 5′-TCCTCTCTCGAGGCAGACAT-3′, Hsp105 reverse, 5′-GGCTTCTACAAGGCAGCTCAA-3′, 16S RNA forward, 5′-AGGAGCGATTTGCTGGTGTGG-3′, 16S RNA reverse, 5′- GCTACCAGGGCCTTTGAGATG-3′.
Statistical analysis
All experiments were repeated at least three times, including oocytes or embryos from at least N = 3 females in each category or genotype. Mean values were compared using Student’s t test and were considered as significantly different at a P value of less than 0.05. Data are presented as mean ± SEM.
Results
HSF1 regulates Hsp70.1 promoter in oocytes
Fully grown GV oocytes were obtained by puncturing large antral follicles. They are developmentally competent, meaning that they can successfully go through meiotic maturation and produce viable embryos after fertilization (Fig. 1a). Their transcriptional activity is reduced in comparison to growing oocytes and they are not able to respond to heat shock (Curci et al. 1987 and 1991).
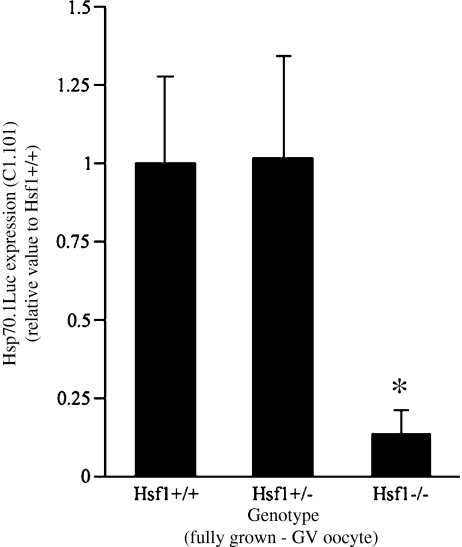
Previous report in the literature and our own data indicated that inducible Hsp70, Hsp70.1 (Hspa1b), and Hsp70.3 (Hspa1a) were expressed at very low levels in fully grown oocytes (Bensaude et al. 1983; Metchat et al. 2009). Using a Hsp70.1Luc transgene, we showed that the transgene was unexpectedly highly expressed in fully grown oocytes (Christians et al. 1999). In order to determine whether HSF1, which is abundant in fully grown oocytes could be involved in this peculiar transgenic expression, we measured the luciferase activity in fully grown oocytes collected from ovaries of either Hsf1+/+, Hsf1+/−, and Hsf1−/− females. Figure 2 shows that loss of HSF1 function was significantly affecting the level of Hsp70.1Luc activity (line C1.101, Fig. 1b) in oocytes totally depleted in HSF1.
Fig. 2.
HSF1 regulates Hsp70.1Luc expression in fully grown GV oocytes. Females homozygous for the transgene Hsp70.1Luc (line C1.101, Fig. 1b) and either WT (Hsf1+/+), heterozygotes (Hsf1+/−), or NULL (Hsf1−/−) were used to collect ovarian fully grown oocytes at GV stage (see Fig. 1a). Luciferase assay was performed on groups of oocytes (see “Materials and methods”) and data pooled from oocytes collected from at least N = 3 females from each genotype. The values are presented as normalized to the mean luciferase activity measured in Hsf1+/+. Hsf1−/− compared with Hsf1+/+, *P = 0.013
Maternal HSF1 and paternal HSF1 and HSF2 significantly impact the zygotic expression of Hsp70.1
To investigate the role played by the HSE trans interacting factors, HSF1 and HSF2 in the regulation of the zygotic expression of Hsp70.1, we first used the transgene Hsp70.1Luc as we had shown previously that it was expressed as early as the G2 phase of the 1-cell stage (about 30 h post-hCG) when included in the paternal genome (Christians et al. 1995).
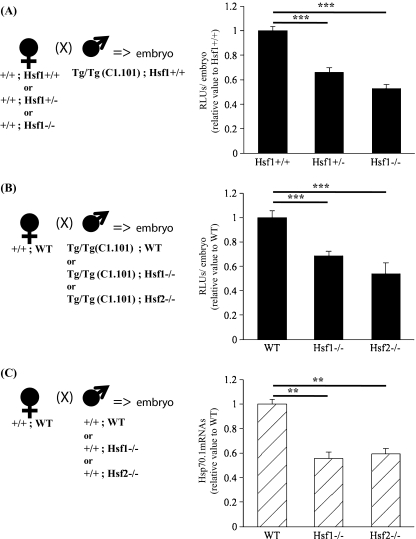
Maternal factors, which are stored in the oocyte cytoplasm are expected to remodel the paternal genome and trigger the expression of the genes brought by the spermatozoon (Chastant et al. 1996). As HSF1 is a major maternal factor present in oocytes while HSF2 is barely detectable (Christians et al. 1997), we examined the role of HSF1 by crossing Hsf1+/+, Hsf1+/−, and Hsf1−/− females with homozygous transgenic WT males. Embryos resulting from these matings were all heterozygous for the transgene and either Hsf1+/+, a mixture of Hsf1+/+or Hsf1+/−, respectively, according to the genotype of the mothers. Figure 3a shows that Hsp70.1 Luc expression is significantly reduced in embryos obtained from Hsf1+/− females. Similar observation could be made with embryos from homozygous Hsf1−/− females even if it was difficult to collect properly fertilized embryos from these females due to the maternal effect of the HSF1 knockout (Christians et al. 2000; Bierkamp et al. 2010). These data confirmed that maternal HSF1 significantly contributes to the zygotic expression of the Hsp70.1Luc transgene (C1.101).
Fig. 3.
Maternal (a) and paternal (b–c) HSFs impact the zygotic activity of Hsp70.1 promoter. a–b Luciferase activity was measured on single or groups of five embryos. The values are presented as normalized to the mean luciferase activity measured in Hsf1+/+. a Embryos were produced as presented by the scheme (left) using females from different genotypes (listed as x-axis legends on the graph) and homozygous transgenic WT males. Graph (right) shows that Hsf1+/− and Hsf1−/−females produced embryos exhibiting reduced transgenic Hsp70.1 activity. b Embryos generated with HSF1- or HSF2-deficient spermatozoa as shown on the scheme (left) had a lower level of luciferase activity in comparison to WT ones. The x-axis legends indicate the genotype of the fathers. c The amount of Hsp70.1 transcripts produced by embryos (30 h post-hCG) obtained from WT, Hsf1−/− or Hsf2−/− fathers was determined by RT-qPCR. Relative quantities of mRNA were normalized against the quantity of the ribosomal S16 transcripts and the relative expression was compared with the Hsf1+/+ sample which was arbitrarily given the value 1. The x-axis legends indicate the genotype of the fathers. **P < 0.01; ***P < 0.001
The paternal genome is first shaped during the last phase of spermatogenesis before being transmitted to the embryo through oocyte fertilization. It was shown that not only HSF1 and HSF2 are expressed during spermatogenesis (Alastalo et al. 1998; Fiorenza et al. 1995) but also that HSF1 or HSF2 loss of function have consequences on the chromatin organization of the spermatozoa (Akerfelt et al. 2008; 2010b). Furthermore it was hypothesized that the binding of HSF1 and HSF2 to the Hp70.1 promoter in spermatozoa would allow the early zygotic expression of Hsp70.1 (Wilkerson et al. 2008). Therefore, to test this hypothesis, we combined transgenic (C1.101, Fig. 1b) and Hsf1 or Hsf2 knockout lines to determine the expression of Hsp70.1 by measuring the luciferase activity in embryos at 30 h post-hCG, a very early stage of zygotic genome activity in mice. WT females were mated with transgenic, WT or HSF knockout males so that the transgene was brought by spermatozoa, which had undertaken differentiation in presence or absence of HSF1 or HSF2. Data presented in Fig. 3b indicate that absence of HSF1 or HSF2 during spermatogenesis reduced the level of Hsp70.1Luc transcriptional activity. Because early zygotic transcriptional activity is mainly due to the expression of the paternal genome (Bouniol-Baly et al. 1997; Christians et al. 1995), we also measured the amount of endogenous Hsp70.1 transcripts by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in embryos generated after fertilization with Hsf1−/− or Hsf2−/− spermatozoa. The same conclusion could be drawn from the data obtained with the transgene or the endogenous Hsp70.1 gene (Fig. 3c).
Hsp70.1 Luc expression reveals different effect of HSE mutations at the 2-cell and blastocyst stage
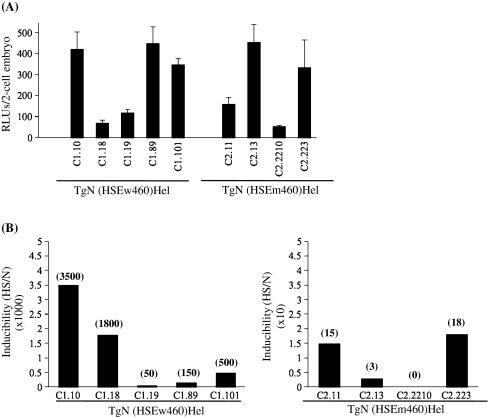
To assess the role of HSF–HSE interactions in Hsp70.1Luc expression, we exploited transgenic lines of mice with a construct including the luciferase reporter gene driven by the proximal part of the Hsp70.1 promoter containing either wild type HSE sequences or mutated ones (Fig. 1b). As previously described (Christians et al. 1995), two-cell embryos were produced by crossing wild type females with homozygous transgenic males. Figure 4a shows that the level of expression of the transgene was variable among several different independent transgenic lines and there was no correlation with the type of transgene including either a wild type promoter (construct C1; Fig. 1b) or a promoter with mutated HSEs (construct C2; Fig. 1b).
Fig. 4.
Functional consequences of point mutations introduced in HSE 1 and 2 sequences of Hsp70.1 promoter. a Hsp70.1 Luc transgenic lines were generated with either C1 or C2 constructs (Fig. 1b) and luciferase activity was assayed in two-cell embryos produced by crossing WT females with homozygous transgenic males. Mutations in HSE1 and 2 did not significantly modify the expression of the transgene at the two-cell stage. A minimum of 30 embryos was measured for each transgenic lines. Mean RLUs/single embryo ± SEM. b Stress inducibility of Hsp70.1Luc transgene at the blastocyst stage. Embryos were produced as in (a) using homozygous males bearing the construct C1 or C2 (see Fig. 1b). Heat shock (HS) was performed at 43°C during 30 min followed by a 5 h recovery at 37°C. Untreated transgenic blastocysts were maintained at 37°C (N). Fold induction (HS/N) value is indicated above each corresponding bar. Transgenic embryos collected from the transgenic lines with wHSE exhibited a strong induction—up to over 103 times the constitutive level of expression. Transgenic embryos collected from the transgenic lines with mHSE exhibited a very low level of induction—less than 20 times the constitutive level of expression
To determine whether HSF1-HSE interactions are similar in nature when Hsp70.1 expression is activated at the 2-cell stage in physiological conditions or to trigger stress inducible expression at the blastocyst stage, we used the same transgenic lines (C1 and C2 lines) as above and we measured the luciferase activity following heat shock.
In contrast to the data obtained at the two-cell stage, we observed a significant difference between transgenic blastocyst bearing mutated HSE Hsp70.1Luc transgene and wild type ones. Inducibility was on average 130-fold higher in blastocysts from C1 lines than from C2 lines with mHSE. (Fig. 4b). This demonstrated that the point mutations introduced in HSE1/2 had more drastic functional consequences for the stress-induced HSF–HSE interactions than for the constitutive developmentally regulated ones.
Hsp transcription and cell survival at blastocyst stage are modulated by HSF2 according to stress conditions
Classical HSR develops progressively during preimplantation and, based on experiments exploiting an Hsp70.1 Luc transgene, seems to be established at the blastocyst stage (Christians et al. 1997). In contrast to very early steps of preimplantation development, embryonic cells at the blastocyst stage contain both HSF1 and HSF2 raising the question of the respective role of these two factors in this emerging HSR ability (Christians et al. 1997, Mezger et al. 1994a).
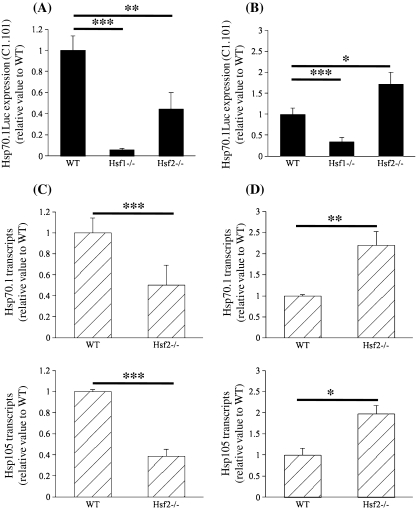
HSF1 and HSF2 knockout mice lines allowed us to investigate this question. By intercrossing the transgenic line C1.101 with WT and Hsf2 knockout lines, we collected groups of embryos, which were all, heterozygous for the transgene and either WT or Hsf2−/−. Due to the infertility of Hsf1−/− females, it was impossible to set up productive Hsf1−/− intercross, thus only groups of mixed Hsf1+/− and Hsf1−/− embryos could be obtained (noted as Hsf1−/− in Fig. 5ab). Because it was previously shown that, in mouse embryonic fibroblasts, HSF2 could modulate HSR according to stress conditions (Paslaru et al. 2003), we used the same parameters for the thermal stress applied here on blastocysts. Heat shock was performed during 30 min at 43°C (which could be qualified as a short and strong stress) or during 120 min at 42°C (which could be qualified as a long and mild stress) followed by recovery at 37°C. In WT embryos, expression of the transgene was strongly induced by the thermal stress as expected (Fig. 5a, see also Fig. 4b). In comparison to WT embryos, the group of mixed partial and totally HSF1 deficient embryos exhibited a markedly reduced level of induction. It can be suggested that the remaining induction was due to the heterozygotes existing in the group. HSF2-deficient embryos were also unable to reach the same level of induction as the WT ones after heat shock carried out at 43°C during 30 min (Fig. 5a). Unexpectedly Hsf2−/− embryos response was much stronger than WT one when the heat shock conditions were 42°C during 120 min (Fig. 5b).
Fig. 5.
Stress response in HSF2 deficient blastocysts. Two heat shock protocols were used: 43°C, 30 min (a–c) and 42°C, 120 min (b–d). a–b Hsp70.1 transgenic expression was measured as before. The values are presented as normalized to the mean luciferase activity measured in WT (Hsf1+/+). c–d Hsp70.1 and Hsp105 RT-qPCR was performed as in Fig. 3c. Relative quantities of mRNA were normalized against the quantity of the ribosomal S16 transcripts and the relative expression was compared with the Hsf1+/+ sample which was arbitrarily given the value 1. A reduced level of expression of the transgene as well as of the endogenous Hsp70.1 gene was observed in HSF2 deficient blastocysts. HSF2 seemed to be required to get the full HSR response after a thermal stress as applied in (a and c). This suggests that HSF2 could act as an activator. In contrast HSF2 deficiency led to a higher expression after a milder and longer thermal stress as applied in (b and d). This suggests that HSF2 could act as a repressor under these stress conditions
To further assess this observation, we repeated the same experimental conditions for thermal stress, using non-transgenic embryos and RT-qPCR to measure the level of endogenous Hsp70.1 expression (Fig. 5c, d (upper panel)). Similarly we observed a different response of HSF2 deficient embryos according to the parameters of the thermal stress. Despite a lower level of significance, we found a similar effect of increased expression of Hsp105 in Hsf2−/− embryos stressed at 42°C during 120 min (Fig. 5c, d (lower panel)).
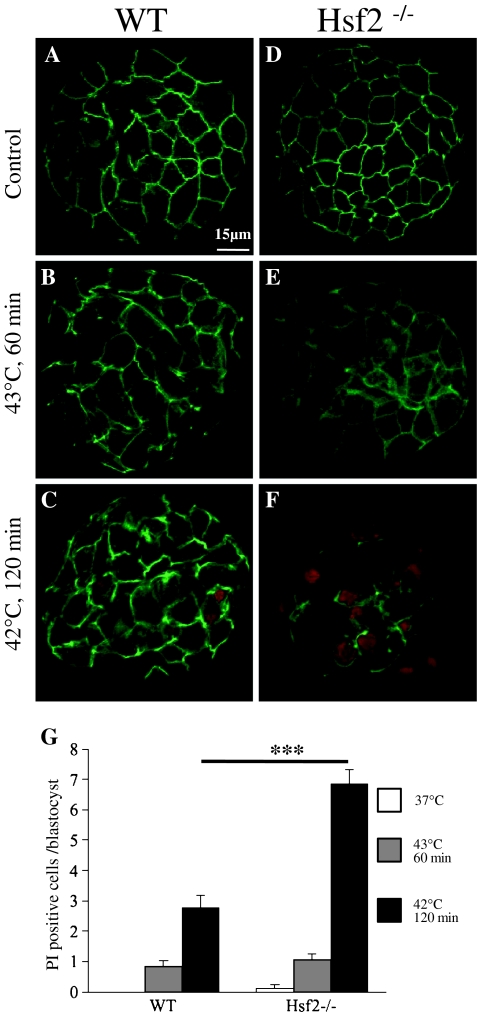
To determine whether HSF2 was required in maintaining the viability of stressed embryos, we used the membrane permeability test on blastocysts either kept at normal temperature or submitted to two different thermal stress (Fig. 6a–f). HSF2-deficient embryos exhibited a higher number of dead cells after the mild and long heat shock indicating that they were less resistant that WT ones under such conditions (Fig. 6g). Furthermore these observations indicated that Hsp expression could not be directly associated to cell survival.
Fig. 6.
HSF2-dependent cell survival is correlated to stress conditions. a–f Representative images of fluorescent staining of blastocysts (green, actin; red, nucleus of dead cells PI-positive cells) maintained at normal temperature or heat stressed as indicated. See “Materials and methods” for experimental procedure. g Graph showing the number of dead cells counted based on the PI staining of their nucleus (red). After a thermal stress at 43°C (60 min), there is no significant difference in the number of dead cells between Hsf2+/+ and Hsf2−/− blastocysts indicating a similar survival rate (b, e, and g). In contrast after a longer heat shock at 42°C (120 min), Hsf2−/− blastocysts were more affected than the WT ones (c, f, and g). ***P < 0.001
Discussion
A wealth of literature has been devoted to the regulation of the HSR since the initial discovery of this protective mechanism by Ritossa in the sixties (reviewed in Akerfelt et al. 2010a). From the study of this regulative paradigm emerged a dogma describing HSF1, the heat shock transcription factor as inactive at normal temperature or in unstressed conditions. Nevertheless, accumulating data suggest that HSF1 is involved in multiple—probably—networked functions in absence of any experimental or significant stress. This was exemplified not only by the developmental phenotypes exhibited by Hsf1 knockout mouse lines (reviewed in Christians and Benjamin 2006) but also by the genome wide changes in the transcriptome of HSF1 deficient MEF (mouse embryonic fibroblasts) (Trinklein et al. 2004) or by the HSF1 occupancy on multiple chromosomal sites (Akerfelt et al. 2010b). The molecular mechanisms by which those stress-unrelated functions of HSF1 can be exerted remain to be better understood.
In this paper we further demonstrate that HSF1 is involved in gene regulation in absence of stress. RT-qPCR experiments had shown that the very low level of Hsp70.1 transcripts was reduced in HSF1-deficient oocytes (Metchat et al. 2009). Expression of Hsp70.1 transgene measured in several transgenic lines (see also Christians et al. 1999) was high in comparison to the endogenous gene (data not shown). This enabled us to confirm the maternal role of HSF1 as a regulator of the transgenic as well as the endogenous Hsp70.1 promoter in fully grown oocytes. From these data, we can also speculate that the transgenic promoter is more accessible to HSF1 and chromatin—genomic environment is exerting a repressive action on the endogenous Hsp70.1 gene in fully grown oocytes. After fertilization, Hsp70.1 expression was previously presented as a landmark of early zygotic activity and the present study confirms that maternal HSF1 is involved in stimulating the transcription of this Hsp gene (Christians et al. 1997).
In addition to the maternal role of HSF1, we were also able to demonstrate the effects exerted by the paternal HSF1 and HSF2 on the early zygotic expression of genes brought by the spermatozoon genome. This provides a functional dimension to the previous observations showing that HSF1 and HSF2 bind Hsp70.1 promoter in spermatozoa and are required for the proper chromatin organization in those gametes (Wilkerson et al. 2008; Akerfelt et al. 2008, 2010b). We first tested this paternal effect of HSF1 and HSF2 using one transgenic line (C1.101, Hsp70.1Luc), and it should be mentioned that transgene expression could be affected by the insertion site, which is, by itself, characterized by a defined chromatin environment. It was not possible to analyze several transgenic lines in those experiments due the required time and animal resources. Nevertheless, we could expect to find similar results as those presented in Fig. 3b, i.e., reduction of the zygotic expression of Hsp70.1 when zygotes are produced by spermatozoa from HSF1- or HSF2-deficient males, excepted that this reduction could have variable amplitude according to the transgenic lines and the corresponding insertion site of the transgene. Furthermore, besides the transgenic analysis, we also observed that paternal HSF1 and HSF2 modified the endogenous Hsp70.1 expression, which is due mainly to the transcriptional activity initiated in the paternal counterpart of the zygotic genome.
Regarding cis regulatory element, our work revealed that the same mutated version of HSEs had different effect on Hsp70.1 Luc expression measured either in non stressed embryos at the time of the zygotic genome activation or in blastocysts after heat stress. Those data could be interpreted as reflecting two different types of HSF1–HSE interactions which are potentially integrating the following molecular elements. Firstly, HSF1 can recognize multiple forms of HSE sequence with some preferences as previously analyzed (Kroeger and Morimoto 1994, Trinklein et al. 2004; Yamamoto et al. 2009). Secondly, multiprotein complex and post-translational modifications (acetylation, sumoylation, and phosphorylation) of HSF1 can be changed according to the stress (or developmental) conditions and this impacts HSF1 DNA binding activity (reviewed in Akerfelt et al. 2010a). Finally, HSF1-HSE interactions could be modified by the chromatin environment and our data compared embryos which undertake important chromatin remodeling along preimplantation development (Albert and Peters 2009). Thus, it could be speculated that the combination of both HSF1 and chromatin modifications allows the recognition of mutated HSE only at the time of zygotic genome activation but not in stressed blastocysts.
In addition, in contrast to early embryos, blastocysts contain significant amount of HSF2 and exhibit a constitutive DNA binding activity due to HSF2, which could affect HSF1-HSE interactions in those embryos (Christians et al. 1997; Mezger et al. 1994a). Our data suggest that this activity of HSF2 was differently involved in Hsp regulation depending on stress conditions. Such modulating role for HSF2 was previously illustrated in Hsf2−/− MEFs which were able to induce Hsp70 expression in response to heat stress at 42°C (120 min) while WT MEFS were not (Paslaru et al. 2003). It was also apparent in the study carried out by Ostling et al. (2007) and seemed to be gene-specific as the lack of HSF2 provoked a stronger induction of Hsp40 and Hsp1110 but a lower one for Hsp70. All together, those data could be also interpreted as revealing a dual function for HSF2 acting as a transcriptional repressor or an activator downstream of HSF1.
Although HSF2 activity did not seem to be important for normal embryonic development as Hsf2−/− blastocysts progess as expected (McMillan et al. 2002), HSF2 deficient blastocysts displayed higher sensitivity and cell death after some defined stress. This could not be directly correlated to the transcriptional response of Hsp70.1 or Hsp105, if it is expected that higher induction of Hsps exerts a protective role. Similar observation was recently made in a different cellular and stress context: proteasome inhibition provokes a proteotoxic stress which induces Hsp70 expression at a very high level in Hsf2−/− MEFs but still those cells are more prone to death than WT MEFs (Lecomte et al. 2010). Thus it could be suspected that HSF2 is participating to other protective mechanisms and this should reinforce our interests in studying the different pathways, which are involved in protecting the cells against the deleterious effects of stress, e.g., HSR and ER stress response (Liu and Chang 2008, Fujikawa et al. 2010).
Finally, beyond the fundamental aspects of HSR study, a better understanding of the cellular mechanisms and related genes affected by stress conditions is important for biomedical reasons. As the number of in vitro procedures carried out to obtain human embryos is increasing, further experiments should determine whether these procedures affect the expression of genes involved in HSR and identify the still unknown and probably long-term consequences on organism health.
Acknowledgments
We want to thank E Michel, M Morange, and JP Renard for their contribution to the first phase of this study. We are grateful to C Charry and C Audouard for their invaluable technical help at various steps of this study. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Ministère de l’Education Nationale et de la Recherche (Ph.D. fellowship to F. LM), the University Toulouse III (to F. LM and E.S.C), and the Fondation pour la Recherche Médicale (FRM) (to E.S.C.).
References
- Akerfelt M, Henriksson E, Laiho A, Vihervaara A, Rautoma K, Kotaja N, Sistonen L. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci USA. 2008;105:11224–9. doi: 10.1073/pnas.0800620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–55. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Vihervaara A, Laiho A, Conter A, Christians ES, Sistonen L, Henriksson E (2010b). Heat shock transcription factor 1 localizes to the sex chromatin during meiotic repression. J Biol Chem. doi:10.1074/jbc.M110.157552 [DOI] [PMC free article] [PubMed]
- Alastalo TP, Lonnstrom M, Leppa S, Kaarniranta K, Pelto-Huikko M, Sistonen L, Parvinen M. Stage-specific expression and cellular localization of the heat shock factor 2 isoforms in the rat seminiferous epithelium. Exp Cell Res. 1998;240:16–27. doi: 10.1006/excr.1997.3926. [DOI] [PubMed] [Google Scholar]
- Albert M, Peters AH. Genetic and epigenetic control of early mouse development. Curr Opin Genet Dev. 2009;19:113–21. doi: 10.1016/j.gde.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Bensaude O, Babinet C, Morange M, Jacob F. Heat shock proteins, first major products of zygotic gene activity in mouse embryo. Nature. 1983;305:331–3. doi: 10.1038/305331a0. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Luxey M, Metchat A, Audouard C, Dumollard R, Christians E. Lack of maternal heat shock factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev Biol. 2010;339:338–53. doi: 10.1016/j.ydbio.2009.12.037. [DOI] [PubMed] [Google Scholar]
- Bouniol-Baly C, Nguyen E, Besombes D, Debey P. Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res. 1997;236:201–11. doi: 10.1006/excr.1997.3708. [DOI] [PubMed] [Google Scholar]
- Chastant S, Christians E, Campion E, Renard JP. Quantitative control of gene expression by nucleocytoplasmic interactions in early mouse embryos: consequence for reprogrammation by nuclear transfer. Mol Reprod Dev. 1996;44:423–32. doi: 10.1002/(SICI)1098-2795(199608)44:4<423::AID-MRD1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development. 1995;121:113–22. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- Christians E, Michel E, Adenot P, Mezger V, Rallu M, Morange M, Renard JP. Evidence for the involvement of mouse heat shock factor 1 in the atypical expression of the HSP70.1 heat shock gene during mouse zygotic genome activation. Mol Cell Biol. 1997;17:778–88. doi: 10.1128/mcb.17.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Boiani M, Garagna S, Dessy C, Redi C, Renard J, Zuccotti M. Gene expression and chromatin organization during mouse oocyte growth. Dev Biol. 1999;207:76–85. doi: 10.1006/dbio.1998.9157. [DOI] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–4. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- Christians ES, Benjamin IJ. Heat shock response: lessons from mouse knockouts. Handb Exp Pharmacol. 2006;172:139–52. doi: 10.1007/3-540-29717-0_6. [DOI] [PubMed] [Google Scholar]
- Curci A, Bevilacqua A, Mangia F. Lack of heat-shock response in preovulatory mouse oocytes. Dev Biol. 1987;123:154–60. doi: 10.1016/0012-1606(87)90437-4. [DOI] [PubMed] [Google Scholar]
- Curci A, Bevilacqua A, Fiorenza MT, Mangia F. Developmental regulation of heat-shock response in mouse oogenesis: identification of differentially responsive oocyte classes during Graafian follicle development. Dev Biol. 1991;144:362–8. doi: 10.1016/0012-1606(91)90428-6. [DOI] [PubMed] [Google Scholar]
- Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–74. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa T, Munakata T, Kondo S, Satoh N, Wada S. Stress response in the ascidian Ciona intestinalis: transcriptional profiling of genes for the heat shock protein 70 chaperone system under heat stress and endoplasmic reticulum stress. Cell Stress Chaperones. 2010;15:193–204. doi: 10.1007/s12192-009-0133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnel AC, Gifford DJ, Heikkila JJ, Schultz GA. Expression of the major heat shock protein (hsp 70) family during early mouse embryo development. Teratog Carcinog Mutagen. 1986;6:493–510. doi: 10.1002/tcm.1770060603. [DOI] [PubMed] [Google Scholar]
- Hunt C, Calderwood S. Characterization and sequence of a mouse hsp70 gene and its expression in mouse cell lines. Gene. 1990;87:199–204. doi: 10.1016/0378-1119(90)90302-8. [DOI] [PubMed] [Google Scholar]
- Inouye S, Izu H, Takaki E, Suzuki H, Shirai M, Yokota Y, Ichikawa H, Fujimoto M, Nakai A. Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem. 2004;279:38701–9. doi: 10.1074/jbc.M405986200. [DOI] [PubMed] [Google Scholar]
- Kampinga HH. Chaperones in preventing protein denaturation in living cells and protecting against cellular stress. Handb Exp Pharmacol. 2006;172:1–42. doi: 10.1007/3-540-29717-0_1. [DOI] [PubMed] [Google Scholar]
- Kroeger PE, Morimoto RI. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol Cell Biol. 1994;14:7592–603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak JZ, Ciemerych MA, Hupalowska A, Sikora-Polaczek M, Polanski Z. On the transition from the meiotic to mitotic cell cycle during early mouse development. Int J Dev Biol. 2008;52:201–17. doi: 10.1387/ijdb.072337jk. [DOI] [PubMed] [Google Scholar]
- Lecomte S, Desmots F, Masson F, Goff P, Michel D, Christians ES, Drean Y. Roles of heat shock factor 1 and 2 in response to proteasome inhibition: consequence on p53 stability. Oncogene. 2010;29:4216–24. doi: 10.1038/onc.2010.171. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–77. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27:1049–59. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loison F, Debure L, Nizard P, Goff P, Michel D, Drean Y. Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1-HSF2 heterocomplexes. Biochem J. 2006;395:223–31. doi: 10.1042/BJ20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–8. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- McMillan DR, Christians E, Forster M, Xiao X, Connell P, Plumier JC, Zuo X, Richardson J, Morgan S, Benjamin IJ. Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol. 2002;22:8005–14. doi: 10.1128/MCB.22.22.8005-8014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezger V, Rallu M, Morimoto RI, Morange M, Renard JP. Heat shock factor 2-like activity in mouse blastocysts. Dev Biol. 1994;166:819–822. doi: 10.1006/dbio.1994.1361. [DOI] [PubMed] [Google Scholar]
- Mezger V, Renard JP, Christians E, Morange M. Detection of heat shock element-binding activities by gel shift assay during mouse preimplantation development. Dev Biol. 1994;165:627–38. doi: 10.1006/dbio.1994.1281. [DOI] [PubMed] [Google Scholar]
- Metchat A, Akerfelt M, Bierkamp C, Delsinne V, Sistonen L, Alexandre H, Christians ES. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90alpha expression. J Biol Chem. 2009;284:9521–8. doi: 10.1074/jbc.M808819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange M, Diu A, Bensaude O, Babinet C. Altered expression of heat shock proteins in embryonal carcinoma and mouse early embryonic cells. Mol Cell Biol. 1984;4:730–5. doi: 10.1128/mcb.4.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paslaru L, Morange M, Mezger V. Phenotypic characterization of mouse embryonic fibroblasts lacking heat shock factor 2. J Cell Mol Med. 2003;7:425–35. doi: 10.1111/j.1582-4934.2003.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostling P, Bjork JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem. 2007;282:7077–86. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- Sandqvist A, Bjork JK, Akerfelt M, Chitikova Z, Grichine A, Vourc’h C, Jolly C, Salminen TA, Nymalm Y, Sistonen L. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell. 2009;20:1340–7. doi: 10.1091/mbc.E08-08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15:1254–61. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson DC, Murphy LA, Sarge KD. Interaction of HSF1 and HSF2 with the Hspa1b promoter in mouse epididymal spermatozoa. Biol Reprod. 2008;79:283–8. doi: 10.1095/biolreprod.107.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth D, Christians E, Munaut C, Dessy C, Foidart JM, Gustin P. Differential heat shock gene hsp70-1 response to toxicants revealed by in vivo study of lungs in transgenic mice. Cell Stress Chaperones. 2002;7:387–95. doi: 10.1379/1466-1268(2002)007<0387:DHSGHR>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–3. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Takemori Y, Sakurai M, Sugiyama K, Sakurai H. Differential recognition of heat shock elements by members of the heat shock transcription factor family. FEBS J. 2009;276:1962–74. doi: 10.1111/j.1742-4658.2009.06923.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi N. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem. 2002;86:376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]