Abstract
Many proteins have been isolated from eukaryotes as redox-sensitive proteins, but whether these proteins are present in prokaryotes is not clear. Redox-sensitive proteins contain disulfide bonds, and their enzymatic activity is modulated by redox in vivo. In the present study, we used thiol affinity purification and mass spectrometry to isolate and identify 19 disulfide-bond-containing proteins in Pseudomonas putida exposed to potential oxidative damages. Among these proteins, we found that a typical 2-Cys Prx-like protein (designated PpPrx) displays diversity in structure and apparent molecular weight (MW) and can act as both a peroxidase and a molecular chaperone. We also identified a regulatory factor involved in this structural and functional switching. Exposure of pseudomonads to hydrogen peroxide (H2O2) caused the protein structures of PpPrx to convert from high MW complexes to low MW forms, triggering a chaperone-to-peroxidase functional switch. This structural switching was primarily guided by the thioredoxin system. Thus, the peroxidase efficiency of PpPrx is clearly associated with its ability to form distinct protein structures in response to stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-010-0243-5) contains supplementary material, which is available to authorized users.
Keywords: Peroxiredoxin, Molecular chaperone, Peroxidase, Functional switch, Pseudomonas putida
Introduction
Bacteria have various biological mechanisms to protect against oxidative stress and the resultant protein unfolding and aggregation mediated by reactive oxygen species (ROS) (Butterfield et al. 1999; Kim et al. 2002). During the course of investigations into these protective mechanisms, a wide variety of antioxidant proteins have been discovered including superoxide dismutase, catalase, many types of peroxidases (Storz et al. 1990), and various forms of molecular chaperones such as heat shock proteins (Hendrick and Hartl 1993). Among these proteins, the peroxiredoxins (Prxs) have received considerable attention in recent years as a new and expanding family of thiol-specific antioxidant proteins (Kristensen et al. 1999). They are also known as thioredoxin (Trx)-dependent peroxidases (Jeong et al. 2000; Jang et al. 2004) and alkyl hydroperoxide reductase-C22 (AhpC) proteins (Kitano et al. 1999; Chauhan and Mande 2001; Chuang et al. 2006).
Prxs are abundant proteins in Escherichia coli, and many organisms produce more than one isoform of Prx. At least six Prxs have been identified in mammalian cells. The Prxs use redox-active cysteines (Cys) to reduce peroxides and were originally divided into two categories, the 1-Cys Prxs and 2-Cys Prxs, based on the number of conserved cysteine residues directly involved in catalysis (Chae et al. 1994). Structural and mechanistic data have further divided the 2-Cys Prxs into two classes known as the “typical” and “atypical” 2-Cys Prxs. The 2-Cys Prxs contain both peroxidatic and resolving cysteine residues, and the sulfenic acid product of the peroxidatic cysteine is attacked by the ‘resolving cysteine residue’ located at the carboxy-terminus of the protein, which forms either intermolecular (typical 2-Cys) or intramolecular (atypical 2-Cys) disulfide bonds between the two cysteines. Crystal structures of Prxs have revealed that, whereas atypical 2-Cys enzymes are monomers, typical 2-Cys Prxs exist as either dimers or large toroid-shaped complexes consisting of a pentameric arrangement of dimers [an (α2)5 decamer] (Wood et al. 2003). The latter has fivefold symmetry orthogonal to the ring and bear a striking resemblance to the electron microscopic images of high-molecular weight (HMW) species of Prx II (Wood et al. 2003; Jang et al. 2004).
Prxs exhibit dual physiological functions as peroxidases and molecular chaperones (Jeong et al. 2000; Wood et al. 2003; Jang et al. 2004; Moon et al. 2005; Jang et al. 2006). The molecular chaperone function has received considerable attention in recent years as a new role for these thiol-specific antioxidant proteins (Jeong et al. 2000; Jang et al. 2004; Moon et al. 2005; Jang et al. 2006). In general, upon exposure to oxidative stress or heat shock, the Prx protein structure switches from a low-molecular weight (LMW) form with peroxidase activity to a HMW complex with molecular chaperone activity (Jang et al. 2004; Moon et al. 2005). The molecular chaperone recognizes and binds nascent polypeptide chains and partially folded protein intermediates, preventing their aggregation and misfolding (Hartl 1996). The oligomerization of Prx leads to an increase in surface hydrophobicity, which allows chaperone activity to increase (Chauhan and Mande 2001).
ROS can induce a variety of biological responses. In addition, emerging evidence indicates that ROS can cause specific protein modifications that may lead to changes in the activity or function of the oxidized protein (Maher and Schubert 2000). Protein sulfhydryls can be oxidized to protein disulfides and sulfenic acids, as well as more highly oxidized states such as sulfinic and sulfonic acids (Cumming et al. 2004). Several disulfide-containing proteins that are involved in oxidative stress defense have been identified, and some of these proteins are induced under conditions of oxidative stress (Brown et al. 1995). In bacteria, disulfide bonds are required for the stability and function of many proteins (Choi et al. 2001; Graumann et al. 2001). In this study, we isolated 19 disulfide-bonded proteins from Pseudomonas putida KT2440, including a Prx-like protein (PP1084) that we renamed PpPrx. This prokaryotic Prx-like protein contains two conserved catalytic sites similar to the 2-Cys Prxs of other organisms and is capable of self-association to form HMW complexes; we therefore tested it for peroxidase and chaperone activity. Our genetic and biochemical studies revealed that PpPrx can act alternately as a Trx-dependent peroxidase and as a molecular chaperone. Furthermore, we show that the reversible switch between the dual functions of this protein is triggered by oxidative stress in connection with substantial structural changes.
Materials and methods
Bacterial strains, media, and materials
The bacterial strains P. putida KT2440 and E. coli were grown aerobically at 30°C in luria-bertani (LB) medium (DB, Franklin Lakes, NJ, USA) and were used for the cloning of the PRX gene. Yeast Trx and thioredoxin reductase (TR) were prepared as described (Chae et al. 1994). Protein molecular size standards, used in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS_PAGE), were purchased from ELPIS (ELPIS, Daejeon, Korea). Ampicillin, l-rhamnose, bovine serum albumin (BSA), H2O2 (30% v/v), nicotinamide adenine dinucleotide phosphate (NADPH), and trichloracetic acid (TCA) were obtained from sigma (Sigma, St. Louis, MO, USA).
Isolation of disulfide proteins
P. putida KT2440 was grown to mid-exponential phase (OD600 = 0.5) in 400 ml LB medium and split into 200 ml aliquots for stress and recovery sample preparation. H2O2 was added to a final concentration of 0.5 mM; after 30 min, irradiation was performed with a gamma irradiator (60Co, ca. 150 TBq of capacity; Atomic Energy of Canada Limited, Canada) at a dose rate of 30 gray (Gy)/h for 30 min. The stressed sample was immediately mixed with 1 volume of 20% TCA/acetone. After recovery, the sample was inoculated into fresh LB medium at 30°C and, after 30 min, was mixed with 1 volume of 20% TCA/acetone. Isolation of disulfide bond proteins was performed as described (Lee et al. 2004). Disulfide bond proteins were separated by 7.5-17.5% linear gradient SDS-PAGE and stained with Coomassie Brilliant Blue R-250. Protein bands were excised and digested with trypsin as described (Lee et al. 2004). Protein identification was performed using a QSTAR Pulsar-i MS system (AB/MDS Sciex, Toronto, Canada) equipped with a nanoelectrospray ion source (MDS Protana, Odense, Denmark) as previously described (Lee et al. 2004). The data were processed and interpreted with BioAnalyst software. The results of the peptide sequencing were submitted for analysis to the Protein-Info tool (http://prowl.rockefeller.edu/prowl/proteininfo.html) and the NCBInr database (http://www.ncbi.nlm.nih.gov/) for identification. MASCOT™ (http://WWW.matrixscience.com) was used for searching and interpreting the raw MS/MS data. Proteins were grouped according to the cellular localizations predicted by the PSORTb tool (http://www.psort.org/psortb/).
Cloning of the PpPrx gene and expression in E. coli
The PpPrx gene was cloned from P. putida KT2440 genomic DNA by the polymerase chain reaction (PCR). Briefly, specific PCR reactions were carried out in 20 μl mixtures containing 10 ng of genomic DNA, 0.2 μM deoxyribonucleoside triphosphates, 20 pmol of each primer set for PpPrx (XhoI, 5′-ccgctcgagatgagcgtactc-3′; SacI, 5′-cgagctcttacagcttgccagc-3′), and 1 unit of Taq DNA polymerase (Takara Bio, Otsu, Japan) in a standard PCR buffer under the following conditions: denaturation for one cycle at 94°C for 60 s; 35 cycles at 94°C for 30 s, 50°C for 45 s, and 72°C for 45 s; and one cycle at 72°C for 10 min. The PCR products were then analyzed by 1% agarose gel electrophoresis.
After PCR amplification, the products (615 bp) were collected and purified, treated with T4 DNA ligase (Takara), and subcloned into the pGEM-T vector to produce PpPrx, which was then transformed into E. coli DH5α cells. These constructs were confirmed by nucleotide sequencing, and then the PpPrx fragment from pGEM-T was transferred to the pRSETa expression vector containing His6 tag to create pRSETa::PpPrx.
E. coli KRX cells were transformed with pRSETa::PpPrx; cultured at 30°C overnight in 5 ml of LB medium supplemented with ampicillin (100 μg/ml), and then transferred to 500 ml of fresh LB medium in a shaking incubator. When the absorbance of the culture at 600 nm reached 0.4, expression was induced by adding 20% l-rhamnose to the medium to obtain a final concentration of 0.2%. After incubation for an additional 8 h, the cells were collected by centrifugation, frozen in liquid nitrogen, and stored at −70°C until used. The His6-fused PpPrx was purified by using a Ni2+-nitrilotriacetate-agarose (Ni-NTA) column (Peptron, Daejeon, Korea) and eluted with a linear gradient of 200–500 mM imidazole in phosphate buffered saline (PBS) buffer. After dialysis against 50 mM HEPES buffer (pH 8.0), the protein concentration was measured using the Bradford (1976) method , with BSA as the standard. The purity of the purified recombinant PpPrx was determined with SDS-PAGE to be >99%.
Assays for peroxidase and chaperone activities
The Trx-dependent, NADPH oxidation-linked peroxidase activity of PpPrx was measured by the decrease in absorbance at 340 nm (A340), and the chaperone activity of the protein was measured by using malate dehydrogenase (MDH) and citrate synthase (CS) as a substrates, as previously described (Hendrick and Hartl 1993; Lee et al. 1997; Cheong et al. 1999). Turbidity due to substrate aggregation indicated a lack of chaperone activity and was monitored in a DU800 spectrophotometer equipped with a thermostatic cell holder (Beckman, Fullerton, CA, USA).
Size exclusion chromatography
Size exclusion chromatography (SEC) was performed at 25°C by fast protein liquid chromatography (AKTA; Amersham Biosciences, Piscataway, NJ, USA) using a Superdex 200 10/300 GL column equilibrated at a flow rate of 0.5 ml/min at 25°C with 50 mM HEPES buffer (pH 8.0) containing 100 mM NaCl. Protein peaks (A280) were isolated and concentrated using a Centricon YM-30 (MILLIPORE, Billerica, MA, USA).
Electron microscopy and single-particle image processing
For negative staining, fractionated proteins were diluted 50-fold with 50 mM HEPES buffer (pH 8.0). Following dilution, 5 μl of the final mixture was applied to a carbon-coated grid that had been glow-discharged (Harrick Plasma, Ithaca, NY, USA) for 3 min in air, and the grid was negatively stained using 1% uranyl acetate. The same procedure was used for all specimens. For metal shadowing, the proteins were diluted ∼10-fold with 50 mM HEPES buffer (pH 8.0), then mixed with an equal volume of glycerol. The resulting mixture was sprayed onto freshly cleaved mica, then rotary shadowed with platinum at an angle of 6°. Grids were examined in a Technai G2 Spirit Twin TEM (FEI, USA) operated at 120 kV. Images were recorded at a magnification of 65,000 (0.37 nm/pixel). Electron microscopy (EM) methods were described in Burgess et al. (2004).
Species-dependent structural switch of P. putida KT2440 PpPrx in response to oxidative stress
To investigate the structural switch of PpPrx in cells exposed to H2O2, methyl viologen (MV), or gamma rays, cells were grown to an OD600 of 0.5, and then split into 10 ml aliquots for stress and recovery samples. H2O2 and MV were added to yield the indicated final concentrations for 30 min, and irradiation was performed at the indicated doses for 30 min. Each batch of stressed cells was harvested by centrifugation and resuspended in PBS. Crude extracts (3 μg) were dissolved in sample loading buffer and then resolved by native and reducing PAGE. Proteins were transferred to a nitrocellulose membrane and then analyzed by western blot using a mouse anti-PpPrx antibody. Immunoreactive proteins were detected using a horseradish peroxidase-conjugated goat anti-mouse secondary antibody.
In vivo observation of the structural switch caused by oxidative stresses
P. putida cells were cultured in LB medium at 30°C in a shaking incubator. Cells were grown to mid-exponential phase (OD600 = 0.5) in 100 ml LB medium and split into 50 ml aliquots for stress and recovery sample preparation. H2O2 was added to a final concentration of 20 mM for 30 min. The stressed sample was immediately harvested. The recovery sample was inoculated into fresh LB medium at 30°C for 30 min and then immediately harvested. Crude cell lysates were subjected to native, non-reducing, or reducing PAGE. Their structural properties were analyzed by immunoblotting using an anti-PpPrx antibody.
Results
Isolation of antioxidant proteins containing disulfide bonds from a pseudomonad
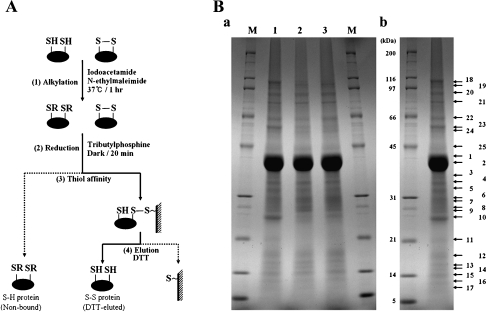
We used a thiol-affinity purification method (Fig. 1A) and mass spectrometry analysis (Lee et al. 2004) to isolate and identify disulfide-bonded proteins (DSBPs) from P. putida KT2440 exposed to the potential oxidative damages H2O2 and gamma rays. The proteins from exposed cells separated into 25 major bands in a 15 cm gel with a 7.5–17.5% acrylamide gradient (Fig. 1B). The expression of P. putida DSBPs was similar in the control and stress-treated samples, although the intensity of some protein bands increased after oxidative stress (Fig. 1B). Single bands contained more than one protein, which hampered their subsequent identification by the QSTAR pulsar-i MS system, so that a total of 19 candidate DSBPs were identified and were grouped according to the cellular localizations predicted by the PSORTb tool (http://www.psort.org/psortb/) (Jennifer et al. 2003; Gardy et al. 2005). Of the 19 DSBPs, 10 were predicted to be cytoplasmic proteins, four to be cytoplasmic membrane proteins, and five to be outer membrane or periplasmic proteins (Table 1). Here, we report on the characterization of the protein PP1084, which encodes a 21 kDa protein that is similar to members of the AhpC/thiol-specific antioxidant protein family and contains three-thiol groups. PP1084 was predicted to function as an AhpC protein and a member of the Prx family by the Clusters of Orthologous Groups prediction tool (Jennifer et al. 2003; Gardy et al. 2005). The PSORTb tool predicted that PP1084 which we renamed PpPrx is a cytoplasmic protein (Table 1).
Fig. 1.
Isolation of disulfide proteins from P. putida KT2440. A Schematic drawing of the process used to isolate disulfide proteins. (1) Proteins extracted with SDS/Tris were treated with both iodoacetamide and N-ethylmaleimide to alkylate free sulfhydryls, and are designated by “R”. (2) Disulfide bonds were converted into free thiols by reduction. (3) Proteins containing thiol groups were captured by thiol-disulfide exchange between the free thiol of the protein and the resin. (4) Proteins captured on the affinity resin were eluted with dithiothreitol (DTT). B SDS-PAGE analysis of fractions from the isolation procedure. DTT-eluted proteins were separated in a 15-cm gel with a 7.5–17.5% acrylamide gradient and stained with Coomassie Brilliant Blue R-250 Ba. The same amount of eluate from each sample was loaded. Lane 1 proteins eluted from unstressed cells after recovery conditions; line 2 proteins eluted after recovery from treatment with 0.5 mM H2O2; lane 3 proteins eluted after recovery from treatment with 30 Gy gamma rays. BbNumbers indicate the protein bands analyzed by LC/MS/MS
Table 1.
Identification and subcellular localization of putative disulfide proteins
| No.a | Protein name (PP no.) | SC (%) | MS/MS (sequence)b | Subcellular localizationsc |
|---|---|---|---|---|
| 1 (5) | Elongation factor Tu (0440) | 20 | R.VQDPLEIVGLR.D | Cytoplasmic |
| 2 (7) | Succinate dehydrogenase catalytic subunit (4190) | 30 | R.LASLDDPFSVFR.C | |
| 3 (8) | Ribosomal protein L3 (0454) | 59 | R.LEEGDFQAGDLIK.A | |
| 4 (10) | Alkyl hydroperoxide reductase, C subunit (2439) | 19 | R.GTFVINPEGQIK.I | |
| *5 (10) | Antioxidant, AhpC/Tsa family (1084) | 53 | K.AYDVESEGGVAFR.G | |
| 6 (12) | Hypothetical protein (0425) | 19 | S.CEAFLEAER.A | |
| 7 (13, 14) | Sensory box protein (0216) | 17 | G.AVEDREYR.I | |
| 8 (15) | Hypothetical protein (2462) | 15 | S.AELLEQAR.K | |
| 9 (18) | Ribonucleotide-diphosphate reductase alpha subunit (1179) | 28 | K.LNAVSSGGDSAPVQAAGPAPVPK.A | |
| 10 (24) | Dihydrolipoamide dehydrogenase (4187) | 22 | R.RPVTTDLLASDSGVTIDER.G | |
| 11 (3, 4) | Hypothetical protein (1245) | 52 | R.VTPAVNLDELDVAASK.E | Cytoplasmic membrane |
| 12 (9) | Polyhydroxyalkanoate granule associated protein GA2 (5007) | 15 | K.TAAEKPAAKPAAKPAAAK.P | |
| 13 (11) | Thiol peroxidase (3587) | 28 | R.EFLENYGVAIADGPLAGLAAR.A | |
| 14 (22) | Hypothetical protein (3699) | 46 | R.GNVTELSEFEYGK.L | |
| 15 (1, 2) | Outer membrane protein OprF (2089) | 38 | P.SVGVGLNFGGSPK.Q | Outer membrane or periplasmic |
| 16 (6) | Basic amino acid ABC transporter (4486) | 48 | R.LDGTVADATLLEDGFLK.T | |
| 17 (8) | Basic amino acid ABC transporter (0282) | 25 | T.QENAYLDLVSGR.I | |
| 18 (15) | Hypothetical protein (0587) | 22 | R.VFQEQSTSFNLPPGTVDVR.L | |
| 19 (19) | Outer membrane protein, bacterial surface antigen family (1599) | 26 | K.TGFFQDIQLSR.D |
SC sequence coverage
aNumbers in parenthesis indicate the band number (Figure 1B, b)
bOnly a single representative peptide sequence is shown, even in cases where multiple peptide sequences were obtained
cPrediction of subcellular localization
PpPrx functions as both a peroxidase and a molecular chaperone
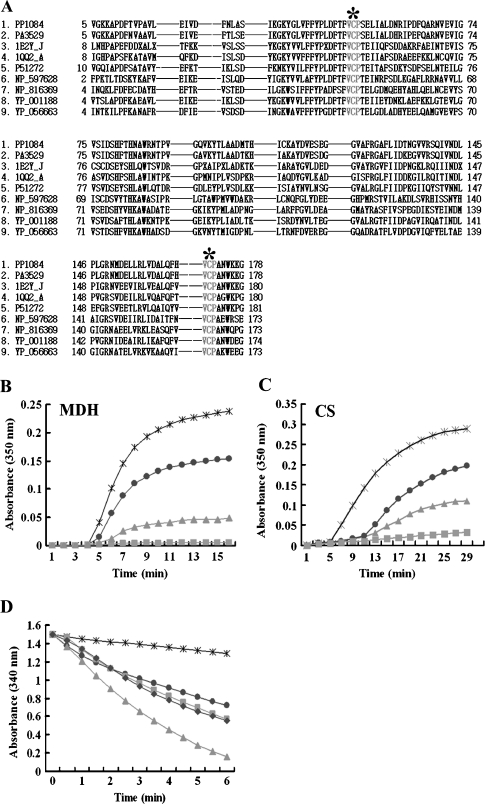
We aligned the PpPrx sequence with eight protein sequences representative of AhpC proteins and Prxs from other prokaryotic and eukaryotic organisms. The alignment showed that these proteins each contained two perfectly conserved Val-Cys-Pro (VCP) tripeptides (Fig. 2A). The sequence homology between PpPrx and PA3529 of Pseudomonas aeruginosa PAO1 was 89%. Most importantly, PpPrx has a high degree of sequence homology and shares similar biochemical properties with other Prxs. Recent evidence has indicated that Prx proteins have dual physiological functions as peroxidases and molecular chaperones (Jeong et al. 2000; Wood et al. 2003; Jang et al. 2004; Moon et al. 2005).
Fig. 2.
Investigation of PpPrx enzymatic functions. a Alignment of the amino acid sequences of P. putida PpPrx (2-Cys Prx) with homologous AhpC and Prxs from several representative prokaryotes and eukaryotes. The encoded amino acid sequences were aligned, and gaps (dashes) were introduced to optimize the sequence alignment. Two highly conserved VCP tripeptides, which are related to the catalytic function, are designated by gray letters and asterisks. The abbreviations for the amino acid sequences of Prxs from various species are as follows: 1 PP1084 of P. putida KT2440, 2 PA3529 of P. aeruginosa PAO1, 3 1E2Y_J of Crithidia fasciculata, 4 1QQ2_A of Rattus norvegicus, 5 P51272 of Porphyra purpurea, 6 NP_597628 of Encephalitozoon cuniculi GB-M1, 7 NP_816369 of Enterococcus faecalis, 8 YP_001188 of Leptospira sp., and 9 YP_056663 of Propionibacterium acnes. b The chaperone activity of recombinant PpPrx was measured using aggregation of MDH at 43°C at different molar ratios: 1 MDH to 0 PpPrx (asterisk), 1 MDH to 0.1 PpPrx (dots), 1 MDH to 0.5 PpPrx (squares), and 1 MDH to 1 yeast TPx (triangles). c The chaperone activity of recombinant PpPrx was measured using aggregation of CS at 43°C at different molar ratios: 1 CS to 0 PpPrx (asterisk); 1 CS to 0.2 PpPrx (dots); 1 CS to 0.5 PpPrx (triangles), and 1 CS to 1 PpPrx (squares). d Peroxide reductase activity of recombinant PpPrx from P. putida was measured with the yeast Trx system at different concentrations: without PpPrx (asterisk) in the reaction buffer; 10 μM PpPrx (dots); 20 μM PpPrx (squares); 40 μM PpPrx (triangle); and 5 μM yeast TPx (diamond; control). The data shown are the means of at least three independent experiments
To investigate whether the 2-Cys Prx from P. putida possesses both peroxidase and molecular chaperone activities, we performed a series of experiments in vitro. PpPrx suppressed the thermal aggregation of the model substrate MDH at 43°C in a concentration-dependent manner (Fig. 2B). At a subunit molar ratio of PpPrx to MDH of 0.5 vs. 1, MDH aggregation was completely suppressed. PpPrx can also efficiently protect the thermal aggregation of CS (Fig. 2C), suggesting that PpPrx can indeed act as an efficient molecular chaperone. However, foldase activity as another chaperone activity was not detected (data not shown). To test the ability of PpPrx to reduce H2O2, we monitored the rate of NADPH oxidation promoted by H2O2 and the change in A340 with yeast Trx and TR acting as a reducing system. PpPrx exhibited low H2O2 catabolic peroxidase activity (Fig. 2D), substantially lower than that of yeast Trx-dependent Prx (yTPx), but PpPrx exhibited five times higher the chaperone activity than yTPx, used as a positive control.
Recombinant ppprx with differently sized HMW structures in vitro
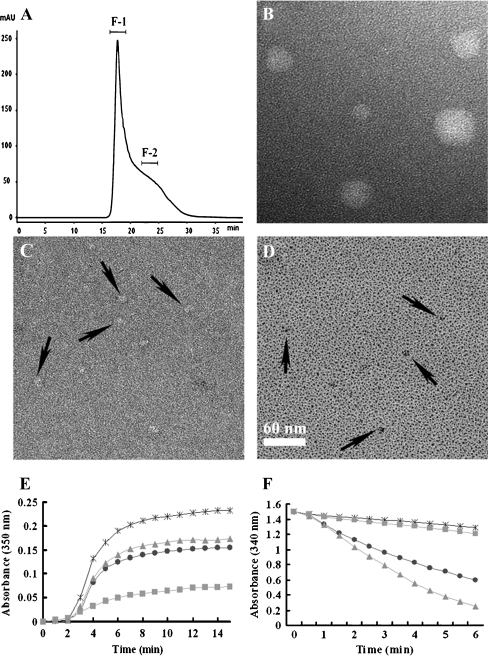
Several Prxs, including human NKEF, AhpC, calpromotin, HBP23, and Prx-B, form HMW complexes with masses of 230–500 kDa (Hirotsu et al. 1999; Schröder et al. 2000). To test whether PpPrx also forms large complexes, we determined the molecular mass of purified PpPrx by using SEC. Only one major peak was present (Fig. 3A). The majority of the PpPrx molecules were contained in the first fraction (F-1) and a minority in the second fraction (F-2). The molecular sizes of the proteins in the F-1 SEC fraction, which contained the largest multimer complexes, were too great to penetrate the pores of a 10% native polyacrylamide gel, and thus were retained at the top of the separating gel (Supplementary Fig. 1A, F-1). In contrast, the second fraction, which consisted of proteins with molecular masses ranging from about 80–230 kDa, contained partial multimer complexes (Supplementary Fig. 1A, F-2). In addition, the two fractions (F-1 and F-2) exhibited different structural patterns under non-reducing condition (Supplementary Fig. 1B).
Fig. 3.
Protein structure-dependent regulation of the peroxidase and chaperone functions. A SEC analysis of PpPrx protein. SEC was performed using a Superdex 200•10/300 GL column as described in the “Materials and methods” section. The separated proteins were divided and pooled into two fractions (F-1 and F-2) for further analysis. B–D Appearances of fractionated PpPrx visualized by electron microscopy: B and C negatively stained images of HMW (F-1) and LMW (F-2), respectively. D Metal shadowed images of LMW (F-2). Black arrows in C and D indicate irregularly shaped small oligomers found in F-2. Scale bar in D represents 60 nm in all fields (B–D). E The chaperone activity of the two separated fractions (F-1 and F-2) of recombinant PpPrx protein measured as aggregation of MDH at 43°C at different molar ratios: 1 MDH to 0 PpPrx (asterisk), 1 MDH to 0.2 total PpPrx (dots), 1 MDH to 0.2 F-2 fraction of PpPrx (triangle), and 1 MDH to 0.2F-1 fraction of PpPrx (square). F Peroxidase activity of the two separated fractions (F-1 and F-2) of recombinant PpPrx protein was measured using the yeast Trx system at different PpPrx concentrations: 0 μM PpPrx (asterisk), 20 μM total PpPrx (dots), 20 μM F-1 fraction of PpPrx (squares), and 20 μM F-2 fraction of PpPrx (triangles). The data shown are the means of at least three independent experiments
The dual functions of recombinant PpPrx are regulated by protein structure in vitro
A conserved feature of molecular chaperones is their tendency to associate into dimers, trimers, and higher oligomers in a reversible fashion (Hendrick and Hartl 1993). In particular, many small heat shock proteins (sHSPs) are known to form HMW complexes in vivo, which is a prerequisite for their chaperone activity (Haley et al. 1998). PpPrx exhibits stronger chaperone activity than a yTPx-positive control (Fig. 2B), and HMW complex formation of a chaperone protein is a typical feature (Supplementary Fig. 1A, F-1). We therefore investigated the specific peroxidase and chaperone activity in the F-1 and F-2 PpPrx fractions (Fig. 3E and F). The HMW complex fraction (F-1) exhibited high chaperone activity about fivefold compare to LMW fraction (F-2) and total protein fraction (Fig. 3E), indicating that the chaperone activity of PpPrx was significantly affected by the structural composition. In contrast, the F-2 fraction had more peroxidase activity than the total protein, and the F-1 fraction showed lower peroxidase activity than total protein (Fig. 3F). These results suggested that the formation of a HMW PpPrx protein can reduce the efficiency of peroxidase activity under normal conditions, whereas dissociation of the HMW complexes into LMW species can promote peroxidase activity under oxidative stress. Thus, the dual functions of PpPrx are clearly associated with their ability to form distinct protein structures.
Structural analysis of fractionated PpPrx with electron microscopy
To further investigate whether the dual functions of PpPrx are associated with the distinct protein structures, we used EM to visualize HMW complex structures (F-1 fraction) and LMW structures (F-2 fraction) of PpPrx. EM of negatively stained protein fractions revealed that fractionated proteins from F-1 and F-2 are in two different configurations, spherical and irregularly shaped small particles, respectively. It showed that spherical particles are with diameters ranging from 30 to 60 nm, possibly reflecting the number of assembled Prx molecules in each particle. In LMW species of PpPrx from the F-2 fraction, observed particles vary in size and in shape with diameters ranging from 8 to 12 nm that were distinct from the background (Fig. 3C and D). It was hard to detect ring-shaped structures as shown in other reports of Prxs (Wood et al. 2003; Jang et al. 2004). Combinations of SEC and EM results suggested that PpPrx was predominantly in the form of HMW complex structures. Unlikely previous results from 2-Cys Prxs where the chaperone activities are increased by their oligomerization (Jang et al. 2004; Moon et al. 2005), our results showed that chaperone activity of PpPrx was also affected by their oligomerization (Fig. 3E). In contrast, the peroxidase activity was enhanced by dissociation of oligomeric HMW complex structures (Fig. 3F).
Structural switch of ppprx due to oxidative stress in P. Putida
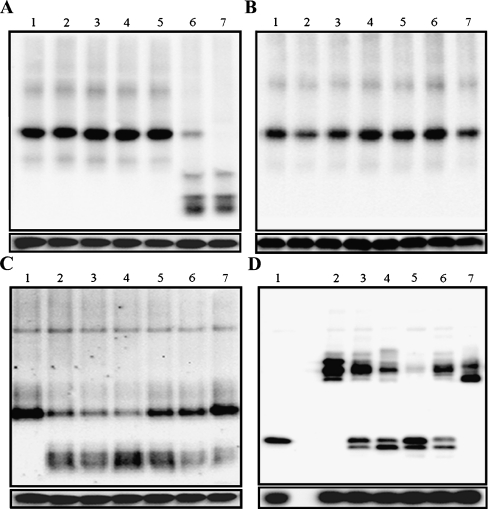
The typical 2-Cys Prxs are the largest class of Prxs (Wood et al. 2003) and are identified by the conservation of their two redox-active cysteines, the peroxidatic cysteine (near residue 50), and the resolving cysteine (near residue 170) (Hofmann et al. 2002). The typical 2-Cys Prxs are obligate homodimers containing two identical active sites (Alphey et al. 2000; Schröder et al. 2000; Wood et al. 2003). Recently, studies of several typical 2-Cys Prxs revealed that their oligomeric states change dramatically between dimers and decamers in response to changes in the redox state that occur during the catalytic cycle (Wood et al. 2003). We analyzed the structural changes of PpPrx that occurred in vivo in P. putida cells exposed to various oxidative stresses. After culturing cells with or without various oxidative stresses, crude extracts prepared from the cells were subjected to western blot analysis on native PAGE gels (Fig. 4A and B). The PpPrx proteins obtained from P. putida cells not treated with H2O2 consisted of only one type of oligomeric structure as HMW complexes. When these cells were challenged with different H2O2 concentrations for 30 min, most of the HMW complexes were immediately converted into LMW forms; however MV did not affect structural switching of PpPrx (Fig. 4A). The expression of PpPrx increased in response to gamma rays in a dose-dependent manner up to 150 Gy. At 200 Gy, the expression level was lower than in control cells, although gamma rays did not affect structural switching of PpPrx (Fig. 4B). We analyzed the structural changes of PpPrx after culturing cells with or without 20 mM H2O2 by subjecting crude extracts prepared from the cells to western blot analysis on a native PAGE gel. When these cells were challenged with 20 mM H2O2 for 30 min, most of the HMW complexes were converted into LMW proteins. However, the LMW proteins returned to their original structures within 30 min following the removal of H2O2 (Fig. 4C).
Fig. 4.
Oxidative stress-dependent structural changes between functional states of Prxs The structural changes of PpPrx in cells exposed to H2O2, MV, or gamma rays were investigated. a H2O2 and MV were added to the indicated final concentrations for 30 min: 1 no additions, 2 0.1 mM MV, 3 10 mM MV, 4 50 mM MV, 5 0.5 mM H2O2, 6 10 mM H2O2, and 7 50 mM H2O2. b Irradiation was performed at the indicated doses for 30 min: 1 no irradiation, 2 30 Gy, 3 60 Gy, 4 90 Gy, 5 120 Gy, 6 150 Gy, and 7 200 Gy. Crude extracts (3 μg) were dissolved in sample loading buffer and then resolved by native and reducing PAGE. Proteins were transferred to a nitrocellulose membrane and then analyzed by western blot using a mouse anti-PpPrx antibody. Immunoreactive proteins were detected using horseradish peroxidase-conjugated goat anti-mouse IgG. cP. putida was challenged with 20 mM H2O2 for 30 min and then allowed to recover for 60 min without H2O2. The structural changes in the PpPrx were then analyzed by native PAGE and western blotting every 15 min. P. putida cells were exposed to different conditions and then detected with an anti-PpPrx antibody: 1 normal conditions, 2 and 3 exposure to 20 mM H2O2 for 15 and 30 min, and 4–7 recovery without H2O2 for 15, 30, 45, and 60 min. d To understand the mechanism of the structural change, crude proteins were extracted from the P. putida cells under normal conditions and then exposed to various reagents; the structural changes in the PpPrx were analyzed by non-reducing PAGE and western blotting with an anti-PpPrx antibody. 1 Cell extract with 100 mM DTT; 2 cell extract without added reagents; 3 cell extract with 1 mM NADPH; 4 cell extract with 1 mM NADPH and 10 mM H2O2; 5 cell extract with 1 mM NADPH, 10 mM H2O2, and yeast Trx system; 6 cell extract with 1 mM NADPH, 10 mM H2O2, and yeast GSH system; 7 cell extract with 10 mM H2O2. The Trx and GSH systems contained Trx-thioredoxin reductase (TR) and GSH-glutathione reductase (GR). The lower panels indicate the amount of PpPrx in each sample under reducing conditions
To further investigate dissociation mechanism of PpPrx, we performed a series of experiments in vivo (Fig. 4D). Although we were unable to detect any H2O2-induced structural changes in PpPrx directly (Fig. 4D, lane 7), the adding of dithiothreitol or NADPH promoted conversion of monomer forms on non-reducing-PAGE (Fig. 4D, lanes 1 and 3). The dissociation of PpPrx suggested that PpPrx could be reduced using NADPH as an electron donor. In fact, we were able to detect structural changes in PpPrx indirectly, using the Trx system containing Trx and TR (Fig. 4D, lane 5). H2O2 stimulated a Trx system-dependent structural change in PpPrx in vivo (Fig. 4D, lane 4). These results suggest that Trx is essential for the formation of LMW forms of PpPrx in vivo (Fig. 4D, lane 5); in contrast, the glutathione (GSH) system is not essential for the structural switching for PpPrx in vivo (Fig. 4D, lane 6).
Discussion
The 2-Cys Prx proteins are members of a ubiquitous family of peroxidases that participate in redox-sensitive signaling and act both as peroxidases with antioxidant activity and as molecular chaperones (Wood et al. 2003; Jang et al. 2004; Chuang et al. 2006). Most 2-Cys Prxs form condition-dependent oligomeric structures, although the physiological relevance of the association, or dissociation, of these proteins has been unclear. The PpPrx we investigated in this study is found in P. putida and belongs to the typical prokaryotic 2-Cys Prxs. Like other 2-Cys Prxs, PpPrx exhibits dual functions as a peroxidase and a molecular chaperone (Fig. 2C to D). Although, PpPrx had about fourfold lower peroxidase activity than yTPx as positive control, PpPrx exhibited about 3∼4-fold higher chaperone activity than yTPx (Fig. 2B). In addition, PpPrx suppressed the thermal aggregation of MDH and CS at 43°C; however, foldase activity as another chaperone activity did not observed. These functions are regulated by dynamic exchanges in its oligomeric structures. These functions are regulated by dynamic exchanges in its oligomeric structures like sHSPs. The sHSPs have been previouly reported that molecular chaperone activity of sHSPs was modulated by their oligomerization (Leroux et al. 1997). A common feature of sHSPs is their formation of large oligomeric complexes. The simplest and best characterized sHSP quaternary structure is Methanococcus jannaschii sHSP16.5 which is a hollow spherical complex composed of 24 subunits generated by a threefold crystallographic symmetry operation of an asymmetric unit containing eight subunits (Kim et al. 1998). Plant sHSPs assemble into complexes of 200–300 kDa (Waters et al. 1996) and the typical oligomeric size of α-crystallins and sHSPs from mammals and yeast is between 400 and 800 kDa (Bentley et al. 1992; Groenen et al. 1994). Thus, the HMW complex structure of PpPrx could also exhibit high chaperone activity about fivefold compare to LMW forms, whereas LMW forms predominately exhibited high peroxidase activity (Fig. 3E and F). In addition, we also discovered that these structural changes are sensitively regulated by H2O2 in vivo, which in the presence of the Trx system dissociates the HMW PpPrx complexes into LMW forms (Fig. 4D, lane 5). This differs from previous reports in which the Trx system and oxidative stresses restructured the LMW forms of several 2-Cys Prxs into HMW complexes (Jang et al. 2004; Moon et al. 2005). This structural change is specific for H2O2 because the oxidative stresses of gamma rays and MV did not affect the dissociation of PpPrx from HMW complexes into LMW forms (Fig. 4A and B). Furthermore, although PpPrx formed a regular oligomeric structure that was converted into LMW forms by H2O2 in vivo. However, the direct regulation of the structural change by H2O2 was not occurred (Fig. 4A, lane 7). PpPrx was only reduced by the Trx system, resulting in disassociation of the oligomeric structure and transition to LMW forms in vivo (Fig. 4D).
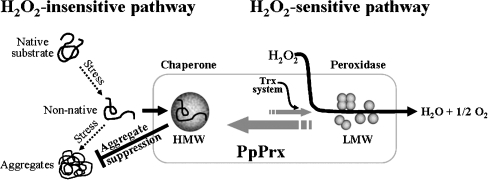
Our observations allow us to develop a comprehensive model of how PpPrx functions as both a peroxidase and a chaperone during oxidative stress (Fig. 5). This model shows that PpPrx can reversibly change its protein structure in vivo from HMW complexes to LMW species through two different pathways, H2O2-insensitive and H2O2-sensitive. At low concentrations of ROS generated under normal conditions, PpPrx exists principally in oligomers and HMW complexes in cells. In the H2O2-insensitive pathway, the HMW complexes act as chaperones, which offer a high level of protection to substrate proteins against oxidative stress. In the H2O2-sensitive pathway, Trx switches most of the HMW complexes to LMW forms in vivo. The LMW PpPrx function acts as a Trx-dependent peroxidase that catalyzes the removal of H2O2.
Fig. 5.
A model of oxidative stress-dependent structural and functional switching of PpPrx from a molecular chaperone to a peroxidase In the normal condition, PpPrx exists in cells principally in oligomeric and HMW complex forms. The structures are formed by two independent pathways, one H2O2-insensitive and the other H2O2-sensitive. In the H2O2-insensitive pathway, the HMW complexes act as superchaperones, which offer a high level of protection to substrate proteins against oxidative stress. In the H2O2-sensitive pathway, most HMW complexes are converted to LMW forms by the Trx system in vivo to remove the oxidative stress of H2O2. The Trx system containing Trx-TR is also necessary for the dissociation of HMW complexes into LMW protein species
Our results suggest that H2O2 reversibly changes PpPrx protein structures in vivo from HMW complexes into LMW forms. This dissociation of the complexes into LMW species enhances the peroxidase activity. When P. putida cells are exposed to H2O2 stress, cells need a powerful H2O2 scavenger. To meet this need, the HMW complexes of PpPrx are converted into LMW forms (Fig. 4C), which exhibit effective peroxidase activity (Fig. 3F) in proportion to the H2O2 concentration.
In conclusion, the dual functions of 2-Cys Prxs in modulating ROS concentrations and preventing protein aggregation may act as potent stress sensors and chaperones to protect P. putida cells against various stresses, enabling them to survive and persist in extreme environments.
Electronic supplementary material
Below is the link to the electronic supplementary material.
A Each SEC fraction was separated by 10% native-PAGE. B Each SEC fraction was separated by 12% non-reducing-PAGE. PpPrx recombinant protein was stained with Coomassie Brilliant Blue R-250. F-1 lane F-1 fraction of PpPrx, F-2 lane F-2 fraction of PpPrx (GIF 61 kb)
Acknowledgments
This project was carried out under the Nuclear R&D Program of the Ministry of Science and Technology (http://WWW.mest.go.kr), Republic of Korea. EM work was supported by KBSI grant T3021A to Jung, HS.
References
- Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN. The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2 Cys-peroxiredoxins. J Mol Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Fitch IT, Tuite MF. The small heat-shock protein Hsp26 of Saccharomyces cerevisiae assembles into a high molecular weight aggregate. Yeast. 1992;8:95–106. doi: 10.1002/yea.320080204. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Yatin SM, Varadarajan S, Koppal T. Amyloid beta-peptide-associated free radical oxidative stress, neurotoxicity, and Alzheimer’s disease. Methods Enzymol. 1999;309:746–768. doi: 10.1016/S0076-6879(99)09050-3. [DOI] [PubMed] [Google Scholar]
- Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan R, Mande SC. Characterization of the Mycobacterium tuberculosis H37Rv alkyl hydroperoxidase AhpC points to the importance of ionic interactions in oligomerization and activity. J Biochem. 2001;354:209–215. doi: 10.1042/0264-6021:3540209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong NE, Choi YO, Lee KO, Kim WY, Jung BG, Chi YH, Jeong JS, Kim K, Cho MJ, Lee SY. Molecular cloning, expression, and functional characterization of a 2Cys-peroxiredoxin in Chinese cabbage. Plant Mol Biol. 1999;40:825–834. doi: 10.1023/A:1006271823973. [DOI] [PubMed] [Google Scholar]
- Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/S0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Hiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming RC, Andon NL, Haynes PA, Park MK, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FSL. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. Activation of the redox-regulated molecular chaperone hsp33-a two-step mechanism. Structure (Camb) 2001;9:377–387. doi: 10.1016/S0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- Groenen PJTA, Merck KB, Jong WW, Bloemendal H. Sturcture and modifications of the junior chaperone alpha-crystallin. From lens transparency to molecular pathology. Eur J Biochem. 1994;225:1–19. doi: 10.1111/j.1432-1033.1994.00001.x. [DOI] [PubMed] [Google Scholar]
- Haley D, Horwitz J, Stewart PL. The small heat shock protein, αB-crystallin, has a variable quaternary structure. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperone in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hirotsu S, Abe Y, Okada K, Nagahara N, Hori H, Nishino T, Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci USA. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann B, Hecht HJ, Flohé L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Jang HH, Chi YH, Park SK, Lee SS, Lee JR, Park JH, Moon JC, Lee YM, Kim SY, Lee KH, Lee SY. Structural and functional regulation of eukaryotic 2-Cys peroxiredoxins including the plant ones in cellular defense signaling mechanisms against oxidative stress. Physiol Plant. 2006;126:549–559. doi: 10.1111/j.1399-3054.2006.00685.x. [DOI] [Google Scholar]
- Jennifer LG, Cory S, Ke W, Martin E, Gabor ET, Istvan S, Sujun H, Katalin D, Christophe L, Kenta N, Brinkman FSL. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Research. 2003;31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WJ, Cha MK, Kim IH. A new member of human Tsa/AhpC as thioredoxin-dependent thiol peroxidase. J Biochem Mol Biol. 2000;33:234–241. [Google Scholar]
- Kim KK, Kim R, Kim S-H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- Kim KS, Choi SY, Kwon HY, Won MH, Kang TC, Kang JH. Aggregation of α-synuclein induced by the Cu, Zn-superoxide dismutase and hydrogen peroxide system. Free Radic Biol Med. 2002;32:544–550. doi: 10.1016/S0891-5849(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Kitano K, Niimura Y, Nishiyama Y, Miki K. Stimulation of peroxidase activity by decamerization related to ionic strength: ahpC protein from Amphibacillus xylanus. J Biochem (Tokyo) 1999;126:313–319. doi: 10.1093/oxfordjournals.jbchem.a022451. [DOI] [PubMed] [Google Scholar]
- Kristensen P, Rasmussen DE, Kristensen BI. Properties of thiol-specific anti-oxidant protein or calpromotin in solution. Biochem Biophys Res Commun. 1999;262:127–131. doi: 10.1006/bbrc.1999.1107. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. J EMBO. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee J, Kim Y, Bae D, Kang KY, Yoon SC, Lim D. Defining the plant disulfide proteome. Electrophoresis. 2004;25:532–541. doi: 10.1002/elps.200305677. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Melki R, Gordon B, Batelier G, Candido EPM. Structure-function studies on small heat shock protein oligomeric assembly and ineraction with unfold polypeptides. J Biol Chem. 1997;272:24646–24656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- Maher P, Schubert D. Signaling by reactive oxygen species in the nervous system. Cell Mol Life Sci. 2000;57:1287–1305. doi: 10.1007/PL00000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MK, Kim CW, Cho MJ, Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- Schröder E, Littlechild JA, Lebedev AA, Errington N, Vagin AA, Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 Å resolution. Structure. 2000;8:605–615. doi: 10.1016/S0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- Storz G, Tartaglia LA, Farr SB, Ames BN. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-E. [DOI] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. doi: 10.1093/jxb/47.3.325. [DOI] [Google Scholar]
- Wood ZA, Schröder E, Robin HJ, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
A Each SEC fraction was separated by 10% native-PAGE. B Each SEC fraction was separated by 12% non-reducing-PAGE. PpPrx recombinant protein was stained with Coomassie Brilliant Blue R-250. F-1 lane F-1 fraction of PpPrx, F-2 lane F-2 fraction of PpPrx (GIF 61 kb)