Abstract
Given strong evidence implicating an important role of altered microRNA expression in cancer initiation and progression, the genes responsible for microRNA biogenesis may also play a role in tumorigenesis. Exportin-5 (XPO5) is responsible for exporting pre-miRNAs through the nuclear membrane to the cytoplasm, and is thus critical in miRNA biogenesis. In the current study, we performed both genetic and epigenetic association studies of XPO5 in a case control study of breast cancer. We first genotyped two missense SNPs in XPO5, rs34324334 (S241N) and rs11544382 (M1115T), and further analyzed methylation levels in the XPO5 promoter region for blood DNA samples from a breast cancer case-control study. We found the variant genotypes of rs11544382 to be associated with breast cancer risk (OR=1.59, 95% CI: 1.06 −2.39), compared to the homozygous common genotype. When stratified by menopausal status, the variant alleles of both rs11544382 (OR=1.82, 95% CI: 1.09-3.03) and rs34324334 (OR=1.76, 95% CI: 1.10-2.83) were significantly associated with breast cancer risk in post-menopausal women. The methylation analysis showed that the “high” and combined “high/middle” tertiles of methylation index were associated with reduced risk of breast cancer (OR=0.34, 95% CI:0.15-0.81 and OR=0.47, 95% CI:0.24-0.94, respectively; Ptrend=0.015). These results were corroborated by data from a publicly available tissue array, which showed lower levels of XPO5 expression in healthy controls relative to tumor or adjacent tissues from breast cancer patients with tumor tissue exhibiting the highest expression levels. These findings support the hypothesis that variations in components of the miRNA biogenesis pathway, in this case XPO5, may affect an individual's risk of developing breast cancer.
Keywords: XPO5, breast cancer, microRNA biogenesis
Introduction
MicroRNAs (miRNAs) are single-stranded, 19-25 nucleotide long RNAs that have been found to contribute both to the translational repression and activation of target gene transcripts [1, 2]. miRNAs act on cellular transcripts through cleavage-dependent RNA degradation, or via other mechanisms, broadly referred to as “miRNA-mediated translational repression”. Current bioinformatics tools predict that each miRNA recognizes an average of 100-200 different mRNA targets [3]. Since each miRNA is thought to play a wide-ranging regulatory role, it holds that a disruption of this expression pattern could result in the onset of one or more disease processes, includingtumorigenesis [4].
The biogenesis of a functional miRNA involves several steps and multiple proteins [4]. Briefly, this process begins with the expression of a primary ∼1000 nt miRNA transcript known as the pri-miRNA, which is cleaved to produce a ∼60-70 nt precursor known as the pre-miRNA. Next, the pre-miRNAs are transported from the nucleus into the cytoplasm, where they are further processed into mature miRNAs. The gene responsible for nuclear export and stabilization is exportin-5 (XPO5), making it a critical element of miRNAformation [5].
XPO5 is a member of the importin-b family of proteins that comprise one major class of nu-cleo-cytoplasmic transporters. XPO5 binds directly to its pre-miRNA cargo in a RanGTP-dependent manner [6]. Additionally, XPO5 can recognize and export structured RNAs that are unrelated to pre-miRNAs, including viral mini-helix RNA and tRNA, along with certain other proteins, such as STAU2, ILF3, and JAZ [7, 8]. It has also been demonstrated that XPO5 plays a role in siRNA biogenesis and therefore is a key point of intersection between the si RNA and miRNA pathways [5]. The over-expression of XPO5 has been shown to result in enhanced miRNA activity, which suggests that XPO5-mediated nuclear export of pre-miRNAs may be a rate-limiting step in miRNA biogenesis [9]. Conversely, loss of XPO5 binding results in reduced pre-miRNA expression and function [10]. Although there is no direct link between XPO5 and cancer, the importance of XPO5 in the miRNA pathway suggests that structural alterations in this transporter could potentially impact global miRNA expression, thereby altering an individual's risk of developing cancer.
Although a fair amount of work has been conducted regarding variations, both genetic and epigenetic, in microRNAs and cancer susceptibility [11, 12], little work has been done regarding variations in the miRNA processing components and risk of breast cancer development. In the current study, we performed both genetic and epigenetic association studies of XPO5 in a case control study of breast cancer conducted in Connecticut. To the best of our knowledge, the role of XPO5 in breast cancer has not been examined, making this the first molecular epide-miological investigation to explore associations between XPO5 variants and breast cancer risk.
Materials and methods
Case-control study of breast cancer
The study population consisted of subjects (441 cases and 479 controls) enrolled in a previous breast cancer case-control study conducted in Connecticut. The study was approved by the Institutional Review Boards (IRB) at Yale University, the Connecticut Department of Public Health, and the National Cancer Institute. Participation was voluntary, and written informed consent was obtained. Details regarding subject recruitment and participant characteristics have been described in previous publications [13-15]. Cases were incident, histologically confirmed breast cancer patients (International Classification of Diseases for Oncology, 174.0 −174.9) between the ages of 30 and 80 with no previous diagnosis of cancer other than non-melanoma skin cancer. Cases were obtained either from computerized patient information at Yale-New Haven Hospital (YNHH) in New Haven County, Connecticut, or from nearby Tolland County, Connecticut via hospital records by the Rapid Case Ascertainment Shared Resource at the Yale Cancer Center. YNHH controls were patients who underwent breast-related surgery at YNHH for histologically confirmed benign breast diseases. Random digit dialing was used to obtain controls younger than 65 and the utilization of the Health Care Finance Administration files was employed to identify controls for those subjects age 65 and older at the Tolland county site. After approval from each participant's hospital and physician, potential subjects were contacted by letter and then by telephone, and those who agreed to participate were interviewed by a trained interviewer, resulting in participation rates of 71% for controls and 77% for cases among YNHH subjects, and 61% for controls and 74% for cases among Tolland County subjects. Numerous participant characteristics including family history of cancer, reproductive history, diet, and demographic factors were obtained via a standardized, structured questionnaire. At the conclusion of the interview, blood was drawn into sodium-heparinized tubes for immediate DNA isolation and subsequent analyses. Estrogen and progesterone receptor (ER and PR) status was determined immunohisto-chemically at YNHH, as previously described [16]. Cases were denoted receptor positive if they had an H-score greater than 75.
SNP selection and genotyping
Eight non-synonymous SNPs (nsSNPs) in XPO5 were identified in the NCBI SNP database (rs11544382, rs12173786, rs115544379, rs35794454, rs34324334, rs61739889, rs61762965, and rs61762966). Of these, five had no variation in the HapMap population (rs12173786, rs115544379, rs61739889, rs61762965, and rs61762966), and were thus excluded from the genotyping pool, leaving three SNPs for genotyping in the current study: rs34324334 (S241N), rs35794454 (A808V), and rs11544382 (M1115T). Genotyping for all SNPs was performed at Yale University's W.M. Keck Foundation Biotechnology Research Laboratory using the Sequenom MassARRAY multiplex genotyping platform (Sequenom, Inc., San Diego, CA) according to the manufacturer's protocol. All samples underwent multiplex PCR followed by a single base extension reaction, which results in unique primer extensions products for each allele of each SNP. Samples were then analyzed using MALDI-TOF Mass Spec-trometry and genotypes were assigned based on the presence of one or both allele-specific products for each SNP using Sequenom's proprietary software. Duplicate samples from 100 study subjects were interspersed throughout the genotyping assays. The concordance rates for QC samples were over 95% for all assays. All genotyping calls, including quality control data, were re-checked by different laboratory personnel and genotyping scores were reproduced with 100% accuracy.
Bioinformatic assessment of SNP functionality
nsSNP functionality was assessed by four widely-used computational tools: SIFT, PolyPhen, SNPs3D, and PMut. SIFT (blocks.fhcrc.org/~pauline/SIFT.html) assesses the conservation level of a particular amino acid position in a protein [17], while PolyPhen (http://genetics.bwh.harvard.edu/pph/) estimates the structural and functional impact of an amino acid substitution [18]. SNPs3D (www.snps3d.org) employs two methods for determining an nsSNP's functional impact. One prediction is based on the estimated impact on protein stability [19], while the other takes conservation of a given amino acid within a protein family into account [20]. The last computer modelingtool used was PMut (mmb2.pcb.ub.es:8080/PMut/), which uses neural networks trained using a database of disease-associated and neutral SNPs to determine the predicted impact of an amino acid substitution [21].
Three tools were used to assess the region in which the nsSNP resided: Pfam, ProDom, and NCBI's Conserved Domain Search. Pfam (pfam.sanger.ac.uk/) is a comprehensive collection of protein domains and families [22]. Pfam families are divided into two groupings, Pfam-A, which consists of curated seed alignment, and Pfam-B, which are automatically generated from the ProDom database. ProDom (prodom. prabi.fr/) is another comprehensive database of protein domain families created by the global comparison of all available protein sequences [23]. The last tool employed was NCBI's conserved domain search (ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which is a collection of multiple sequence alignments that represent domains conserved in molecular evolution [24].
CpG island identification and methylation analysis
Using the CpG Island Searcher web tool (www.cpgislands.com/), one CpG island was identified that spans the promoter region, the first exon, and part of the first intron of the XPO5 gene (−600 to +808). The MethPrimer program (www.urogene.org//methprimer) was then used to design methylation specific PCR primers within the identified CpG island region. To distinguish methylated and unmethylated DNA sequences, genomic DNA samples were bisulfite treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. Upon bisulfite treatment, unmethylated cytosines are converted into uracil, whereas methylated cytosines remain unchanged. After the conversion, the presence of methylation was determined by quantitative PCR using primers specific to the methylated or unmethylated sequence, and the Power SYBR Green Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The primers sequences used to detect unmethylated DNA were L: 5′-GGT GTG TTT AGT AAT GTA GTT GT-3′; R: 5′-CTA AAT AAA CAA AAC AAA AAA CAA A-3′ and the primers sequences used to detect methylated DNA were L: 5′-GGC GCG TTT AGT AAT GTA GTC-3′; R: 5′-CTA AAT AAA CGA AAC GAA AAA CGA A-3′. The methylation index was calculated as MI = [1/ (1+2-(CTu-CTme)] × 100%, as previously described [25], where CTu = the average cycle threshold obtained from duplicate qPCRs using the unmethylated primers, and CTme = the average cycle threshold obtained using the methylated primers. Since radio- and chemotherapy can affect DNA methylation, only patients from the breast cancer population who had not undergone these treatments were included in this portion of the analysis (n=81), along with an equal number of randomly selected controls. Each PCR reaction was performed in duplicate using both the un-methylated and methylated primers, for a total of four reactions per subject. One or more of these reactions failed in samples from three of the cases, leaving a final sample of 78 cases and 81 controls. Untreated cases were compared to treated cases using a chi-square test for a variety of patient characteristics to determine whether the untreated cases used in the methylation analysis were representative of all cases in the study population.
Expression analysis ofXPO5 in breast tumor and normal breast tissue
The Atlas of Gene Expression function, implemented in the ArrayExpress database (www.ebi.ac.uk/arrayexpress; accessed on May 20, 2009), was used to search for expression array comparisons involving breast tissue drawn from breast cancer patients and healthy controls. The keywords used were: Gene, “XPO5”; Conditions, “Breast Cancer”; and the results were filtered by species to include only Homo sapiens. One experiment was identified that examined gene expression in breast tumor tissue (N=23), adjacent tissue (N=28), and healthy control breast tissue obtained from individuals undergoing breast reduction mammo-plasty (N=10). The experimental protocol and additional information pertaining to tissue collection for this array can be found under accession number E-TABM-276 at the ArrayExpress database, or from the primary publication [26].
Statistical analysis
All statistical analyses were performed using the SAS statistical software (SAS Institute, Cary, NC), unless otherwise noted. For the case-control analyses, allelic distributions for all SNPs were tested by goodness-of-fit chi-square for compliance with Hardy-Weinberg equilibrium (HWE), and no departures from equilibrium were detected (P<0.05) among the controls. Odds ratios and 95% confidence intervals were determined for each SNP-disease association by unconditional multivariate logistic regression, including the following covariates: age (continuous), race, family history of cancer in a first-degree relative, BMI study site, menopausal status (for all only), and parity. Additional covariates, such as alcohol use and smoking did not alter the parameter estimates and were therefore excluded from the final model. For the tissue expression data, P-values were determined using the Wilcoxon two-sample test for comparisons of mean normalized XPO5 expression in normal tissue and tumor tissue or adjacent tissue, and the sign rank test for the paired comparison of tumor tissue to adjacent tissue.
Results
SNPs rs11544382 and rs34324334 are associated with breast cancer risk
No variation was found for rs35794454 and it was excluded from further analysis. Adjusted odds ratios were calculated for the remaining two SNPs (rs11544382 and rs34324334). The combined variant genotypes of rs11544382 were significantly associated with breast cancer risk (OR=1.59, 95% CI 1.06-2.39), as compared to the homozygous common genotype. Stratification by menopausal status showed that the association was strongest among postmeno-pausal women (OR=1.82, 95% CI 1.09-3.03) (Table 1). Similarly, the combined variant genotypes for rs34324334 were also found to be significantly associated with postmenopausal breast cancer risk (OR=1.76, 95% CI 1.10-2.83).
Table 1.
Association of XPO5 variants with breast cancer risk
| All | Premenopausal | Postmenopausal | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Cases | Controls | OR* | Cases | Controls | OR* | Cases | Controls | OR* |
| N (%) | N (%) | (95% CI) | N (%) | N (%) | (95% CI) | N (%) | N (%) | (95% CI) | |
| rs34324334 | |||||||||
| C/C | 353 (82.9) | 400 (86.2) | Ref. | 84 (83.2) | 125 (79.6) | Ref. | 269 (82.8) | 275 (89.6) | Ref. |
| C/T or T/T | 73 (17.1) | 64 (13.8) | 1.40 (0.96-2.03) | 17 (16.8) | 32 (20.4) | 0.89 (0.46-1.74) | 56 (17.2) | 32 (10.4) | 1.76 (1.10-2.83) |
| rs11544382 | |||||||||
| A/A | 351 (83.6) | 408 (89.1) | Ref. | 82 | (82.8) | 137 (86.2) | Ref. | 269 (83.8) | 271 (90.6) |
| A/G or G/G | 69 (16.4) | 50 (10.9) | 1.59 (1.06-2.39) | 17 (17.2) | 22 (13.8) | 1.17 (0.57-2.40) | 52 (16.2) | 28 (9.4) | 1.82 (1.09-3.03) |
Adjusted for age, race, family history of breast cancer, BMI, menopausal status (all only), parity, and study site
Protein domains affected by SNPs rs11544382 and rs34324334
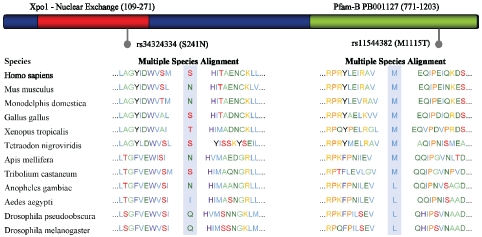
The locations of rs 11544382 and rs34324334 relative to the predicted XPO5 protein domains are indicated in Figure 1. Specifically, SNP rs11544382 is located in a Pfam-B PB001127 region and rs34324334 is located in an Ex-portin-1/Importin-b-like region, which raises the possibility that both mutations occur in a functionally conserved region. The results from the rs11544382 amino acid alteration were in line with four computer programming models we used (SIFT, PolyPhen, SNPs3D, Pmut), which all predicted such a substitution would be detrimental, suggesting that this polymorphism may alter XPO5′s protein structure. Utilizing another bioinformatics search, ProDom, we found a “good quality” (NorMD value=1.723) 266 amino acid alignment, PD329401, covering this region that is associated with nucleocytoplasmic transporter activity (precision=1.000, probabil-ity=0.32) and tRNA binding (precision=0.821, probability=0.32), lending more evidence to support a potential functional impact of rs11544382. For rs34324334, however, the abovementioned computer modeling programs predicted the effect of the SNP to be benign.
Figure 1.
Representation of Exportin-5 and location of nsSNPs. Pfam (pfam.sanger.ac.uk) predicts two functionally conserved domains in XPO5, the Xpo1 domain, which is important for nuclear exchange, and is located from residues 109 to 271 (depicted in red), and an automatically generated PfamB domain from residues 771 to 1203. The multiple species alignment generated by PolyPhen (genetics.bwh.harvard.edu/pph/) is also shown for each SNP. While rs34324334 is not highly conserved, and PolyPhen predicts this mutation to be benign, the amino acid altered by rs11544382 is fairly conserved, and the variant allele (T), is not observed in any species. PolyPhen predicts this SNP to be “probably damaging”.
Methylation of a CpG rich region upstream of XPO5 is associated with breast cancer
In addition to the genetic variation identified in XPO5, a CpG island was also identified which spans the promoter region, the first exon, and part of the first intron of the XPO5 gene (-600 to +808). The level of methylation in this region was determined by real-time methylation-specific PCR for 81 controls and 78 cases who had not undergone radio- or chemotherapy. Cases were classified as having “high”, “middle”, or “low” methylation, based on their methylation score in relation to the tertile distribution of methylation indices among the controls. Cases were significantly less likely to have high methylation indices (OR=0.34, 95% CI: 0.15-0.81) (Table 2). The combined “high” and “middle” tertiles were also associated with a protective role (OR=0.47, 95% CI: 0.24-0.94). Overall, there was a significant trend of decreasing breast cancer risk with each increase in methylation tertile (P=0.015). Further stratified analysis by menopausal status showed that these significant associations were only detected in post-menopausal women. The methylation indices were also compared using the Wilcoxon rank sum test, and again, methylation was significantly lower among cases (P=0.032). Since only cases that had not received radio- or chemotherapy were included in this analysis due to the potential for such treatments to influence DNA methylation, a comparison of untreated cases to treated cases was conducted that revealed no significant differences in any patient characteristics (data not shown).
Table 2.
XPO5 promoter methylation status among breast cancer cases and controls
| MI | All | Premenopausa | Postmenopausal | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Tertile | Controls (%) | Cases (%) | OR* (95% CI) | Controls (%) | Cases (%) | OR* (95% CI) | Controls (%) | Cases (%) | OR*(95% CI) |
| Low | 27 (33.3) | 36 (46.1) | Ref. | 11 (45.8) | 9(52.9) | Ref. | 16 (28.1) | 27 (44.3) | Ref. |
| Mid | 27 (33.3) | 25 (32.1) | 0.60 (0.27-1.33) | 5 (20.9) | 6 (35.3) | 1.16 (0.23-5.94) | 22 (38.6) | 19 (31.1) | 0.47 (0.18-1.20) |
| High | 27 (33.3) 17 (21.8) 0.34 (0.15-0.81) 8 (33.3) | 2 (11.8) | 0.38 (0.05-3.13) | 19 (33.3) | 15 (24.6) | 0.32 (0.11-0.88) | |||
| Mid+High | 54 (66.6) | 42 | 0.47 (0.24-0.94) | 13 (54.2) | 8 (47.1) | 0.77 (0.19-3.19) | 41 (72.9) | 34 (55.7) | 0.40 (0.17-0.91) |
| P for Trend | 0.015 | 0.469 | 0.024 | ||||||
Adjusted for age, race, family history of breast cancer, BMI, menopausal status (all only), parity, and study site
Overexpression ofXPO5 in tumor tissues: data from a public resource
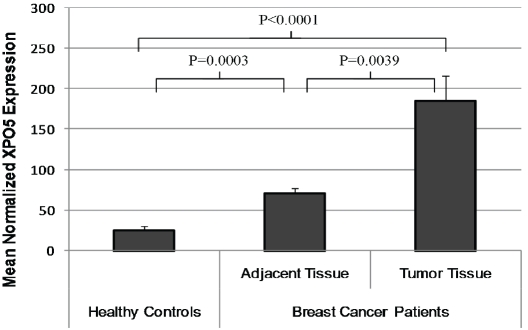
Since RNA was not available from subjects in our case-control population, we determined whether XPO5 expression was altered in breast cancer patients relative to controls by utilizing data from the ArrayExpress microarray database. One experiment (accession # E-TABM-276) was identified that included data on XPO5 gene expression in breast tumor tissue, adjacent breast tissue (with normal appearance or nonproliferative changes), and tissue from healthy controls obtained during breast reduction mammoplasty. Normalized mean XPO5 gene expression values for tumor tissue, adjacent tissues and healthy controls were: 185.3±30.9, 71.8±6.2, and 26.0±4.8, respectively (Figure 2). Breast tissue from healthy controls had significantly lower XPO5 gene expression in comparison to all tissue samples from breast cancer cases, including adjacent tissue (P=0.0003) and invasive carcinoma samples (P<0.0001). Additionally, among cases, XPO5 expression was significantly lower in adjacent tissue compared to invasive carcinoma tissue (P=0.0039).
Figure 2.
XPO5 gene expression in breast tissue from breast cancer patients and healthy controls. An array experiment was identified in the publicly available ArrayExpress database (accession #: E-TABM-276) that compared XPO5 expression in breast tissue from patients with breast carcinoma to XPO5 expression levels in breast tissue from healthy controls. Mean normalized expression (±SEM) was 185.3±30.9 for invasive carcinomas (N=23), 71.8±6.2 in tissues adjacent to the tumor (N=28), and 26.0±4.8 for healthy control breast tissue (N=10).
Discussion
There is increasing evidence showing that expression of miRNAs is deregulated in cancer. This deregulation is thought to occur via a number of different pathways, including transcriptional [27, 28] and epigenetic [29, 30] alterations. Additionally, mutations in the coding regions of miRNAs, as well as DNA copy number abnormalities, are thought to contribute to miRNA deregulation [31-33]. Finally, dysfunctional or deregulated proteins in the miRNA biogenesis pathway may also play a role in human tumorigenesis [34, 35].
The current study demonstrates that two non-synonymous mutations (rs11544382 (M1115T) and rs34324334 (S241N)) in an important mi-croRNA biogenesis gene, XPO5, have significant associations with breast cancer risk. These results are consistent with findings from two previous studies, which showed a borderline significant association between renal cell carcinoma and rs11070, a SNP in the 3′ untranslated region of XPO5, in addition to an association with esophageal cancer risk [12, 36]. This SNP, however, showed no association with bladder cancer risk in another recent study [37]. Nevertheless, these studies suggest a potential role for the microRNA biogenesis gene, XPO5, in human cancers.
The two nsSNPs examined in the current study may influence XPO5 functionality and may therefore be causally related to breast cancer development. Our bioinformatic searches indicate that both SNPs are located in XPO5 functional domains, and are strongly suggestive of the deleterious impact of an rs11544382 mis-sense variant. Interestingly, the effect of rs34324334 missense variation was predicted to be benign. However, the fact that rs34324334 is located in a highly conserved Exportin-1/Importin-b-like region is suggestive of its potential functional importance. Alternatively, it is possible that the rs34324334 mis-sense variant is itself not causal, but is linked to one or more polymorphisms associated with breast cancer risk. Unfortunately, the lack of supporting experimental evidence precludes the drawing of any definitive conclusions with respect to rs34324334's functional role in breast cancer at this point. Further functional analysis is required to determine with greater certainty whether either of these SNPs has a true functional impact on XPO5 or whether they are merely indicative of the influence of other polymorphisms.
It should be noted that a portion of the controls used for our genotypic analysis consists of women who underwent surgery for benign breast conditions. It is possible that some of these women may be at an increased risk of developing breast cancer, and assuming that the observed genetic associations are true, the inclusion of these women into the control population might bias our estimates toward the null (i.e., the true effect sizes are stronger than those observed).
In addition to genetic associations, our epigenetic analysis further demonstrates that alterations in the methylation pattern of XPO5′s promoter region could be a risk biomarker. To our knowledge, there have been no studies linking genetic or epigenetic variations in key components of the miRNA biogenesis pathway to breast cancer tumorigenesis. One potential concern regarding these results is whether the observed epigenetic changes in the surrogate tissue, peripheral blood lymphocytes (PBLs), accurately reflect the changes in the breast tissue. A study examining whether methylation status of IGF2 in PBLs was representative of the status in colon tissue found high agreement between the two tissue types (kappa statistic = 86.5%, p < 0.0001) [38]. Additionally, a large case-control study of breast cancer reported significant associations between methylation status of genes measured from PBLs and cancer risk, demonstrating that, in principle, methylation in PBLs may be useful biomarkers for risk prediction [39]. Although both studies support the concept that methylation status of PBLs may serve as a surrogate measure for epigenetic alterations at the target tissue, without paired RNA and DNA from patient tissue, it is difficult to definitively determine the phenotypic impact.
Despite the absence of RNA in our study, our methylation findings are consistent with the XPO5 tissue expression levels present in the ArrayExpress database (www.ebi.ac.uk/arrayexpress; [40]). Since hypermethylation is generally associated with decreased gene expression, and we found decreased methylation at the XPO5 promoter to be associated with increased breast cancer risk, we would expect greater expression of XPO5 in breast tissue from individuals with breast cancer. The tissue expression array data was consistent with this prediction, as breast tumor tissue had the highest level of XPO5 expression, followed by adjacent tissue from breast cancer patients, and tissue from healthy controls. This significant gradient in gene expression lends further support to the idea that aberrant XPO5 expression may play a role in early tumor development.
Furthermore, altered expression of XPO5 has been previously detected in human cancers. In a study of low-grade lung adenocarcinoma, expression of XPO5 was 2.8-3.0-fold down-regulated [34]. On the contrary, expressions of XPO5 was 1.6-fold up-regulated in high-grade prostate cancer [41], which may explain an almost global increase of microRNA expression in prostate adenocarcinoma. Overexpression of XPO5 has been shown to enhance not only miRNA expression, but also the inhibition of target gene expression [9]. Likewise, reduced XPO5 expression has been shown to limit miRNA biogenesis [10]. Alterations in miRNA levels resulting from aberrant XPO5 expression may have a similar impact on a cell's prolifera-tive rate, offering a possible mechanism by which altered XPO5 promoter methylation patterns could increase an individual's risk of developing breast cancer.
As previously mentioned, a key limitation in our investigation is the absence of an experimental functional component to complement our genetic association and bioinformatics analyses. The lack of empirical information on the functional nature of rs11544382 and rs34324334 variants precluded the possibility of corroborating our genetic association results with findings from our methylation and transcriptional profiling analyses. A more in-depth examination of the functional impact of these genetic variants is thus warranted to check for consistency between the impact of genetic and epigenetic XPO5 variations on breast cancer risk. Moreover, the small size of the subjects eligible for our methylation analysis prevented an examination of potential interactions between XPO5 polymorphisms and promoter methylation in modulating XPO5 function. As such, further efforts should be made to investigate whether, and to what degree, XPO5 methylation differs under disparate genotypic settings, and whether these differences can be informative in predicting XPO5 function and breast cancer risk. Conversely, future studies may also need to consider how different methylation settings can enhance or attenuate the phenotypic effects conferred by XPO5 polymorphisms.
Conclusions
Overall, our findings suggest that variations, both genetic and epigenetic, in the miRNA biogenesis gene exportin-5 may significantly influence breast cancer susceptibility. To our knowledge, no previous work has been conducted to investigate the role of XPO5 in breast cancer risk. Given increasing evidence implicating an important role of altered microRNA expression in cancer initiation and progression, the genes responsible for microRNA biogenesis may also play a central role in tumorigenesis.
Acknowledgments
We would like to thank Irina Tikhonova at Yale University's W.M. Keck Foundation Biotechnology Research Laboratory for Sequenom geno-typing analysis. This work was supported by the National Institutes of Health (grants CA122676 and CA110937).
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 5.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 6.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microR-NAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 7.Macchi P, Brownawell AM, Grunewald B, DesGroseillers L, Macara IG, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem. 2004;279:31440–31444. doi: 10.1074/jbc.C400226200. [DOI] [PubMed] [Google Scholar]
- 8.Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. Rna. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng T, Holford TR, Mayne ST, Tessari J, Ward B, Carter D, Owens PH, Boyle P, Dubrow R, Archibeque-Engle S, Dawood O, Zahm SH. Risk of female breast cancer associated with serum polychlorinated biphenyls and 1,1-dichloro-2,2'-bis(p-chlorophenyl)ethylene. Cancer Epidemiol Biomarkers Prev. 2000;9:167–174. [PubMed] [Google Scholar]
- 14.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
- 15.Hoffman AE, Yi C-H, Zheng T, Stevens RG, Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J, Zhu Y. CLOCK in Breast Tumorigenesis: Genetic, Epigenetic, and Transcriptional Profiling Analyses. Cancer Res. 2010;70:1459–1468. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS., Sr Estrogen receptor analyses. Correlation of biochemical and immunohisto-chemical methods using monoclonal antirecep-tor antibodies. Arch Pathol Lab Med. 1985;109:716–721. [PubMed] [Google Scholar]
- 17.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 19.Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in mono-genic disease. J Mol Biol. 2005;353:459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 22.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Servant F, Bru C, Carrere S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 26.Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 28.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with down regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 31.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 35.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in mi-croRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 38.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 39.Widschwendter M, Apostolidou S, Raum E, Rothenbacher D, Fiegl H, Menon U, Stegmaier C, Jacobs IJ, Brenner H. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, Holloway E, Lukk M, Malone J, Mani R, Pilicheva E, Rayner TF, Rezwan F, Sharma A, Williams E, Bradley XZ, Adamusiak T, Brandizi M, Burdett T, Coulson R, Krestyaninova M, Kurnosov P, Maguire E, Neogi SG, Rocca-Serra P, Sansone SA, Sklyar N, Zhao M, Sarkans U, Brazma A. ArrayExpress up-date—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]