Abstract
To determine if the 2005 Chinese outbreak strain of Streptococcus suis is circulating in the United States, three different PCR primer-pairs derived from the nucleotide sequences surrounding and internal to the unique pathogenicity island -like DNA segment of the Chinese outbreak strain (strain 05ZYH33) were used to screen 290 swine isolates of S. suis obtained from different locations. The first primer pair amplified an approximately 1000-bp fragment from 47 (16%) of the United States isolates and the second amplified an 1800-bp fragment from 23 (8%) of the isolates. Nucleotide sequences of the amplicons shared identity with those of strain 05ZYH33. The third primer pair amplified a 716-bp amplicon from the DNA of strain 05ZYH33 only. These observations demonstrated that the PAI homologue of strain 05ZYH33 is absent in the United States isolates tested and suggested that the PCR method may be useful for active surveillance to monitor possible spread of the highly invasive strain.
Keywords: Streptococcus suis, pathogenicity island, toxic shock, polymerase chain reaction (PCR), surveillance
Introduction
Streptococcus suis is commonly known as a veterinary pathogen that affect swine population causing fever, meningitis, septicemia, arthritis, polyserocitis, anorexia, endocarditis and sudden death [1]. In recent years, this bacterium has increasingly become an important zoonotic pathogen and an organism of food safety and general human health concern [2,3]. Human infections are often due to occupational exposure to pigs or pork products or consumption of under cooked pork [1-4]. Clinical manifestations in humans are similar to those observed in pigs such as fever, meningitis, septicemia and endocarditis. In addition, human infections can result in toxic shock, permanent hearing loss and colon carcinoma [1,2,5-10].
Recently, the incidence of S. suis infection in humans increased significantly world-wide lead ing to over 200 reported cases. Some of the countries with reported human infections include Austria, Canada, China, Croatia, Denmark, Germany, Greece, Hong Kong, Italy, Japan, Singapore, Spain, Taiwan, Thailand, New Zealand, Thailand, United Kingdom, United States and Vietnam [2,5,8,11, 12-25]. Two outbreaks of severe acute disease in humans with high morbidity and mortality were reported in China. The first occurred in 1998 in which 14 persons died out of 25 cases and the second occurred in 2005 and of 204 cases 38 eaths were reported [2,26]. The 2005 outbreak reportedly was caused by a more invasive strain of S. suis -serotype 2 that produced clinical symptoms characterized with deep-tissue infection, acute high-fever, vascular collapse, hypotension, multiple organ failure, toxic shock syndrome (TSS), as well as short course of disease [2]. By genome sequencing and comparative genomics, the 2005 outbreak strain (strain 05ZYH33) and the 1998 strain (strain 98HAH12) reportedly possess a unique pathogenicity island (PAI) of approximately 89-kb in length that are thought to be responsible for the highly virulent phenotype and induction of the toxic shock [2].
At present, there is no information available in the United States regarding the presence or absence of the Chinese-like outbreak strain. Due to the magnitude of the outbreak, severity of the disease, and concern that the strain may have crossed international boundaries it was of interest to screen S. suis isolates from regions within the United States in an effort to start monitoring its presence or lack of within the country. For this purpose, we employed the polymerase chain reaction (PCR) method using primers that targeted the PAI of the 2005 Chinese outbreak strain to screen isolates of S. suis recovered from naturally infected pigs in the Mid-Western region for the presence of the PAI homologue.
Materials and methods
Bacterial isolates and media
Two hundred and ninety veterinary clinical isolates of S. suis strains used in this study were obtained from the midwestern states of the United States. They were recovered from different body parts of infected pigs submitted to Veterinary Diagnostic Laboratories in the States of Iowa, Kansas, Wisconsin, and Minnesota for routine culture. The identity of the isolates was verified by standard and molecular methods [27]. The DNA of strain 05ZYH33 involved in recent Chinese human outbreaks [2] was obtained from Jiaqi Tang, East China Research Institute of Medical Biotechnics, Nanjing, China.
DNA extraction
Lysis by a boiling method was used to release DNA for the PCR screening as described elsewhere [27]. In some cases, BIO-RAD Aqua pure Genomic DNA Kit for Gram-positive bacteria (BIO-RAD, Hercules, CA) was used following overnight culture in Todd-Hewitt broth or on trypticase soy agar plates (TSA) containing 5% sheep blood.
PCR
To control for failure of DNA amplification, confirm reliability of the PCR assay and confirm the identity of the S. suis isolates the oligonucleotidesJP4 (5’-GCAGCGTATTCTGTCAAACG-3’) and JP5 (5’-CCATGGACAGATAAAGATGG-3’) were used as primers to target and amplify a 688-bp region of S. suis conserved housekeeping gene [27] in a multiplex PCR format. The oligonucleotides CH1 (5’-CACGCATCTCGTAGAGTTTGAC-3’) and CH2 (5’-AGATTGCGAGGCTTTTAGATTG-3’); CH3 (5’-TCGCCACTATGGTATCTGCTTA-3’) and CH4 (5’-GATTGTGGACCATGCTGTTTAG-3’); CH5 (5’-ATAAATAGCCCCATCCTCATCA-3’) and CH6 (5’GGGTAGCTGCTTAGTGCTACAA-3’) [2] were derived from portions of the PAI-like segment of strain 05ZYH33 involved in the 2005 Chinese outbreak. The CH1 and CH2 primer pair would result in the amplification of a 1800-bp fragment flanking the 5’ region of the PAI; CH3 and CH4 would result in the amplification of a 716bp fragment internal and unique to the PAI segment; and CH5 and CH6 would result in the amplification of a 1000-bp region flanking the 3’ end of the PAI [2]. Amplification reactions were performed in a total volume of 50 µl containing 10 mM Tris-HCL (pH 8.3); 1.5 mM MgCl2; 50 mM KCl; 0.001% gelatin; 200 µM of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, dTTP); 0.5 µM of each primer; 2.5 U of Taq polymerase (Applied Biosystems; Forster City, CA) and 5 µl of template. The PCR assay was carried out in an Applied Biosystems 9700 thermocycler, comprising 5 min of pre-incubation at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 55°C and 1 min at 72°C. Final extension was performed for 7 min at 72°C. DNA from strain 05ZYH33 was used as the positive control. The negative control was a reaction mixture containing all reagents but no DNA template. The PCR products were visualized by electrophoresis on a 0.7% agarose gel following standard procedures [28].
Nucelotide sequence determination
The nucleotide sequences of the PCR products of the United States isolates that were positive for any of the primers were determined using the Sanger di-deoxy chain termination method [28]. The primers used for PCR amplification were the same primers as those used for the sequencing reactions. Sequences were assembled, and multiple sequence alignment were performed against strain 05ZYH33 using Vector NTI AlignX (Invitrogen, Carlsbad, CA).
Nucleotide sequence accession number
The GenBank accession number for the nucleotide sequences of the PCR products of the United States isolates with primer combinations CH1 and CH2, CH5 and CH6 reported in this paper are HQ713676 and HQ713677 respectively.
Results
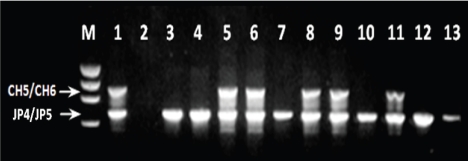
To be considered a match with strain 05ZYH33, DNA of test isolate must yield similar PCR product with primers CH1/CH2; CH3/CH4 and CH5/CH6. In addition, the nucleotide sequences of the PCR product must match that of strain 05ZYH33. Results are summarized in Table 1. All of the 290 isolates screened were positive using primers JP4 and JP5 (internal positive control and species specific primer) and produced the expected 688-bp product (Fig 1) in the multiplex PCR format. Elimination of JP4 and JP5 primers from the reaction mixture did not change results (data not shown). These observations indicated that the isolates tested were correctly identified as S. suis, and that the PCR assay contained no inhibitory substance.
Table 1.
Results summary of the 290 United States isolates of S. suis screened by PCR using the three different primer sets described in this study
| Number of Isolates Positive for CH1/CH2 primers | Number of Isolates Positive for CH3/CH4 primers | Number of Isolates Positive for CH5/CH6 primers | Number of Isolates Positive for CH1/CH2 and CH5/CH6 primers | Total Number of Isolates Screened |
|---|---|---|---|---|
| 23 (8%) | 0 (0%) | 47 (16%) | 22 (7%) | 290 |
Primers CH5 and CH6 amplified the predicted 1000-bp fragment from strain 05ZYH33 (positive control) and from 47 (16%) of the United States isolates (Table 1; Figure 1). Nucleotide sequence determination and analysis of the amplicon of 10 of the 47 positive isolates selected randomly revealed minor base-pair substitutions resulting in approximately 98% identity with the nucleotide sequence of the positive control strain (data not shown). Primers CH1 and CH2 amplified an 1800-bp fragment from strain 05ZYH33 and from 23 (8%) of the United States isolates. As in the previous case nucleotide sequence determination and analysis of 10 of the 23 positive isolates showed 98% identity with the nucleotide sequence of the positive control strain (data not shown). Taken together, 22 (7%) of the United States isolates and strain 05ZYH33 gave positive results with the two primer sets. Unlike the CH1/ CH2 and CH5/CH6 primers, CH3 and CH4 amplified the expected 716-bp fragment from strain 05ZYH33 but failed to yield detectable product from the DNA of any of the United States isolates (Table 1). Changing stringency of the PCR assay or using southern blot and hybridization analysis with the PCR products of strain 05ZYH33 as probes did not change results (data not shown).
Figure 1.
Example of the multiplex PCR results. Lanes: M, molecular size standard; 1, strain 05ZYH33 (positive control); 2, negative control; 3-13, United States isolates of S. suis serotype 2.
Discussion
Streptococcus suis is an emerging zoonotic pathogen with incidences reported globally. Although variation in the degree of virulence amongst strains of S. suis have been shown, the nature and magnitude of a recent outbreak in China caused by strain 05ZYH33 has increased awareness and worldwide concern. Thus, development of strategies to track and contain the spread of this highly pathogenic strain is desirable.
We included primers JP4/JP5 in this study because they have been shown previously to be specific and conserved amongst strains of S. suis encompassing all capsular types [27]. Because the DNA of all isolates used in this study could be amplified by the set of primers in a multiplex PCR format, it indicated that the isolates tested were correctly identified as S. suis, and that the PCR assay contained no inhibitory substance. In our previous study we demonstrated the existence of genetic heterogeneity amongst S. suis isolates [29]. Our findings in this work also revealed genetic differences amongst the isolates tested. In total, 22 (7%) of the United States isolates gave positive results with two of the three primers, indicating some genetic similarity between those United States isolates and the Chinese outbreak strain in the DNA region tested.
Strain 05ZYH33 reportedly possess a unique pathogenicity island-like (PAI) segment of approximately 89-kb in length that are thought to be responsible for the highly virulent phenotype and induction of the toxic shock in infected humans and high mortality in pigs. Primers CH3/ Ch4 are internal to the PAI DNA segment and specific to strain 05ZYH33, the cause of the 2005 outbreak in China as determined by whole genome sequencing [2]. Thus, the lack of amplification of DNA with these primers indicated the absence of the PAI-like DNA homologue in S. suis isolates of the United States origin screened.
In conclusion, the approach we used in this study may facilitate epidemiological investigations and surveillance of the highly pathogenic strain of S. suis.
Acknowledgments
We thank Jiaqi Tang for proving the DNA of the Chinese outbreak strains (05ZYH33), M. M. Chengappa for providing Minnesota strains, Tanya J. Purvis, D. Anderson, M. M. Chengappa for the Kansas strains and Joann M. Kinyon for the Iowa strains.
References
- 1.Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/a:1005870317757. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Tang J, Dong W, Wang C, Feng Y, Wang J, Zheng F, Pan X, Liu D, Li M, Song Y, Zhu X, Sun H, Feng T, Guo Z, Ju A, Ge J, Dong Y, Sun W, Jiang Y, Wang J, Yan J, Yang H, Wang X, Gao GF, Yang R, Wang J, Yu J. A glimpse of Streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE. 2007;2((3)):e315. doi: 10.1371/journal.pone.0000315. 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung P Y, Lo KL, Cheung TT, Yeung WH, Leung PH, Kam KM. Streptococcus suis in retail markets: How prevalent is it in raw pork. Int J Food Microbiol. 2008;127:316–320. doi: 10.1016/j.ijfoodmicro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Robertson ID, Blackmore DK. Occupational exposure to Streptococcus suis type 2. Epidem Inf. 1989;103:157–164. doi: 10.1017/s0950268800030454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidt MC, Mohamed W, Hain T, Vogt PR, Chakraborty T, Domann E. Human infective endocarditis caused by Streptococcus suis serotype 2. J Clin Microbiol. 2005;43:4898–4901. doi: 10.1128/JCM.43.9.4898-4901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y-T, Teng L-J, Ho S-W, Hsueh PR. Strepto-coccus suis infection. J Microbiol Immunol Infect. 2005;38:306–313. [PubMed] [Google Scholar]
- 7.Nghia HDT, Hoa NT, Linh LD, Campbell J, Diep TS, Chau NVV, Mai NTH, Hien TT, Spratt B, Farrar J, Schultz C. Human case of Streptococcus suis serotype 16 infection. Emer Infect Dis. 2008;14:155–157. doi: 10.3201/eid1401.070534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sriskandan S, Slater JD. Invasive disease and toxic shock due to zoonotic Streptococcus suis: an emerging infection in the East. PLoS Med. 2006;3((5)):e187. doi: 10.1371/journal.pmed.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Wang C, Feng Y, Yang W, Song H, et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006;3((5)):e151. doi: 10.1371/journal.pmed.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voutsadakis IA. Streptococcus suis endocarditis and colon carcinoma: a case report. Clin Colorectal Cancer. 2006;6:226–228. doi: 10.3816/CCC.2006.n.041. [DOI] [PubMed] [Google Scholar]
- 11.Aspiroz C, Vela AL, Pascual MS, Aldea MJ. Acute infective endocarditis due to Streptococcus suis serotype 2 in Spain. Enferm Infecc Microbiol Clin. 2009;27:370–371. doi: 10.1016/j.eimc.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Chang B, Wada A, Ikebe T, Ohnishi M, Mita K, Endo M, Matsuo H, Asatuma Y, Kuramoto S, Sekiguchi H, Yamazaki M, Yoshikawa H, Watabe N, Yamada H, Kurita S, Imai Y, Watanabe H. Characteristics of Streptococcus suis isolated from patients in Japan. Jpn J Infect Dis. 2006;50:397–399. [PubMed] [Google Scholar]
- 13.Dickie AS, Bremner DA, Wong PY, North JD, Robertson ID. Streptococcus suis bacteraemia. N Z Med J. 1987;100:677–678. [PubMed] [Google Scholar]
- 14.Ip M, Fung KSC, Chi F, Cheuk ESC, Chau SSL, Wong BWH, Lui S, Lai MRW, Chan PKS. Strepto-coccus suis in Hong Kong. Diagn Microbiol Infect Dis. 2007;57:15–20. doi: 10.1016/j.diagmicrobio.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Kopic J, Paradzik MT, Pandak N. Streptococcus suis infection as a cause of severe illness: 2 cases from Croatia. Scand J Infect Dis. 2002;34:683–684. doi: 10.1080/00365540210147769. [DOI] [PubMed] [Google Scholar]
- 16.Mazokopakis EE, Kofteridis DP, Papadakis JA, Gikas AH, Samonis GJ. First case report of Streptococcus suis septicaemia and meningitis from Greece. Eur J Neurol. 2005;12:487–489. doi: 10.1111/j.1468-1331.2005.00998.x. [DOI] [PubMed] [Google Scholar]
- 17.Mai NT, Hoa NT, Nga TV, le Linh D, Chau TT, Sinh DX, Phu NH LV, Chuong TS, Diep, Campbell J, Nghia HD, Minh Chau NV, de Jong MV, Chinch NT, Hien TT, Farrar J, Schultsz C. Streptococcus suis meningitis in adults in Vietnam. Clin Infect. Dis. 2008;1(46):659–657. doi: 10.1086/527385. [DOI] [PubMed] [Google Scholar]
- 18.Perseghin P, Bezzi G, Troupioti P, Gallina M. Streptococcus suis meningitis in an Italian blood donor. Lancet. 1995;346((8985)):1305–1306. doi: 10.1016/s0140-6736(95)91912-0. 11. [DOI] [PubMed] [Google Scholar]
- 19.Poggenborg R, Gaini S, Kjaeldgaard P, Christensen JJ. Streptococcus suis: meningitis, spondylodiscitis and bacteremia with a serotype 14 strain. Scand J Infect Dis. 2008;40:346–349. doi: 10.1080/00365540701716825. [DOI] [PubMed] [Google Scholar]
- 20.Spiss HK, Kofler M, Hausdorfer H, Pfausler B, Schmutzhard E. Streptococcus suis meningitis and neurophysiology of the acoustic system. First case report from Austria. Nervenarzt. 1999;70:738–741. doi: 10.1007/s001150050503. [DOI] [PubMed] [Google Scholar]
- 21.Tambyah PA, Kumarasinghe G, Chan HL, Lee KO. Streptococcus suis infection complicated by purpura fulminans and rhabdomyolsis: case report and review. Clin Infect Dis. 1997;24:710–712. doi: 10.1093/clind/24.4.710. [DOI] [PubMed] [Google Scholar]
- 22.Wangkaew S, Chaiwarith R, Tharavichitkul P, Supparatpinyo K. Streptococcus suis infection: a series of 41 cases from Chiang Mai University Hospital. Infect J Infect. 2006;52:455–460. doi: 10.1016/j.jinf.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Wertheim HFL, Nguyen HN, Yaylor W, Lien TTM, Ngo HT, Nguyen TQ, Nguyen BNT, Nguyen HM, Nguyen CT, Dao TT, Nguyen TV, Foxx A, Farrer J, Schultsz C, Nguyen HD, Nguyen KV, Horby P. Streptococcus suis, an important cause of adult bacterial meningitis in Northern Vietnam. PloS ONE. 2009;4((6)):e5973. doi: 10.1371/journal.pone.0005973. 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willenburg KS, Sentochnik DE, Zadoks RN. Human Streptococcus suis meningitis in the United States. N Engl J Med. 2006;354:12. doi: 10.1056/NEJMc053089. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Lou L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Xu J, Yang W, the Streptococcus suis study groups Human Strep-tococcus suis outbreak, Sichuan, China. Emer Infect Dis. 2006;12:914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. Outbreak associated with Streptococcus suis in pigs in China. 2005. Update. [PubMed]
- 27.Okwumabua O, O’Connor M, Shull E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol Lett. 2003:79–84. doi: 10.1111/j.1574-6968.2003.tb11501.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Okwumabua O, Staats J, Chengappa MM. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping) J Clin Microbiol. 1995;33:968–972. doi: 10.1128/jcm.33.4.968-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]