Abstract
Reprogramming somatic cells to become induced pluripotent stem cells (iPSCs) by using defined factors represents an important breakthrough in biology and medicine, yet remains inefficient and poorly understood. We therefore devised synthetic factors by fusing the VP16 transactivation domain to OCT4 (also known as Pou5f1), NANOG and SOX2, respectively. These synthetic factors could reprogramme both mouse and human fibroblasts with enhanced efficiency and accelerated kinetics. Remarkably, Oct4–VP16 alone could efficiently reprogramme mouse embryonic fibroblasts (MEFs) into germline-competent iPSCs. Furthermore, episomally delivered synthetic factors could reproducibly generate integration-free iPSCs from MEFs with enhanced efficiency. Our results not only demonstrate the feasibility of engineering more potent reprogramming factors, but also suggest that transcriptional reactivation of OCT4 target genes might be a rate-limiting step in the conversion of somatic cells to pluripotent cells. Synthetic factor-based reprogramming might lead to a paradigm shift in reprogramming research.
Keywords: reprogramming factor, iPS, induced pluripotency, reprogramming, stem cells
Introduction
Induced pluripotent stem cells (iPSCs) have been generated using exogenous transcription factors introduced by methods including plasmid transfection (Okita et al, 2008; Yu et al, 2009) and protein transduction (Kim et al, 2009; Zhou et al, 2009), but with low efficiency (Yamanaka, 2009a, 2009b). This has hindered both basic research in stem-cell biology and therapeutic application of iPSCs. As the reprogramming factors OCT4, SOX2 and NANOG are natural transcription factors that have evolved to govern pluripotency, their activities might have been finely calibrated through evolution to control the delicate balance between self-renewal and differentiation in embryonic stem cells (Ralston & Rossant, 2010). We hypothesized that these factors might be rationally improved for the purpose of reprogramming. Given their crucial roles in targeting the expression of pluripotency genes (Jaenisch & Young, 2008; Silva & Smith, 2008), we further hypothesized that modified versions with greater transcriptional activity would be able to more efficiently overcome the barriers (Sridharan et al, 2009) to reprogramming.
Results And Discussion
We designed and constructed synthetic factors by fusing the potent transactivation domain of herpes simplex virus protein VP16 (Sadowski et al, 1988) with OCT4, SOX2 and NANOG (Fig 1A), and confirmed their expression as fusion proteins in mouse embryonic fibroblasts (MEFs; supplementary Fig S1 online). We then introduced these factors into MEFs and measured the reactivation kinetics of pluripotency marker genes in MEFs undergoing reprogramming. Endogenous genes, including Nanog and Oct4, were already expressed at day 6 upon transduction of the synthetic factors, whereas their expression was not detectable until day 12 in cells transduced with the unmodified transcription factors (Fig 1B), indicating an improved reprogramming kinetic.
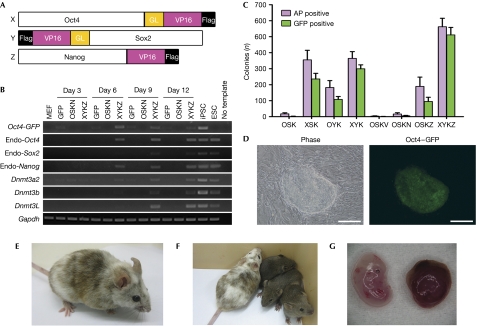
Figure 1.
Synthetic factors improve reprogramming efficiency. (A) The synthetic factors used to reprogramme MEFs. Full-length Oct4, Sox2 and Nanog were fused to the VP16 transactivation domain. Other fusions of various transcription factors and activators/repressors tested are summarized in supplementary Table S1 online. (B) Reactivation of pluripotency genes in transduced MEFs of the cell population. RT–PCR was performed on RNA samples prepared at the indicated time points on retroviral transduction of native (OSKN) and synthetic factors (XYKZ). Transduction of GFP-expressing retroviral constructs provided a negative control. Tg-GFP OG2 MEFs (C57/CBA) contain the Oct4-GFP transgene as a reprogramming reporter. The experiment was performed twice. (C) Comparison of the numbers of AP-positive and Oct4–GFP-positive embryonic stem cell-like colonies obtained at day 15 post-transduction. 5 × 104 MEFs were transduced with various combinations of retroviral constructs, as indicated. Mean colony numbers from triplicate experiments are shown with error bars indicating s.d. (D) Morphology of colonies induced with XYKZ. Representative images are shown from cells 14 days after viral infection. Scale bar, 200 μm. (E) An 8-week-old chimera mouse. Agouti coat colour and pigmented eyes indicate iPSC contribution. iPSC line OG2 XYKZ 4 was microinjected into blastocysts of mice obtained from the ICR. (F) Agouti coat colour of three offspring of the chimeric mother confirming germline competence of iPSC line 4 (C57/CBA). (G) An embryo at embryonic day 13.5 and placenta derived from tetraploid embryo complementation. iPSCs (XSKZ line 4) were injected into tetraploid blastocysts of mice obtained from the ICR. AP, alkaline phosphatase; ESC, embryonic stem cell; GFP, green fluorescent protein; GL, glycine-rich linker; ICR, Institute of Cancer Research; iPSC, induced pluripotent stem cell; K, Klf4; MEF, mouse embryonic fibroblast; O, OCT4; OSKN, OCT4, SOX2 and KLF4 plus NANOG; S, SOX2; V, VP16.
Next, we tested these new factors in selection-free reprogramming of MEFs on the basis of a three-factor (OCT4, SOX2 and KLF4; referred to as OSK) or a four-factor system (OSK plus NANOG; OSKN). When the native OSKs were introduced into MEFs carrying the Oct4-GFP transgene as a reprogramming reporter, we obtained 3±1 (mean±s.d.; n=3) green fluorescent protein (GFP)-positive colonies from 5 × 104 transduced fibroblasts at day 16 (Fig 1C). By contrast, when OCT4–VP16 was used to replace OCT4 in the co-transduction with SOX2 and KLF4 (XSK), we obtained 236±35 GFP+ colonies; a 78-fold increase. Similarly, replacement of SOX2 with SOX2–VP16 (OYK) resulted in 108±19 GFP+ colonies, a 36-fold increase. When NANOG–VP16 was used to replace NANOG (N) in the four-factor system (OSKZ), we obtained 95±27 colonies, 19 times the number obtained from the transduction with the native factors (5±3; OSKN). The combination of all three synthetic factors in the four-factor system generated 511±47 colonies (XYKZ), 100 times the number obtained with the natural factors (5±3; OSKN). Further comparison was performed by fluorescence-activated cell sorting analysis of the transduced cells (supplementary Fig S2A online). Around day 12, 6.8% of the synthetic factor-transduced cells were double positive for OCT4–GFP and SSEA-1, whereas almost no double-positive cells were observed with the native factors. We confirmed that synthetic factors induce reprogramming more quickly (iPSC colonies appeared earlier) and more efficiently (a higher number of iPSC colonies were seen at a given time point) by using single-cell seeding assay (supplementary Fig S2B online). The enhancement of reprogramming efficiency is dependent on VP16 fusion to Oct4, Nanog and Sox because co-expression of VP16 alone (OSKV, Fig 1C) or Klf4–VP16 (supplementary Table S1 online) had no stimulation effect. The obtained GFP+ colonies resembled normal embryonic stem cells morphologically (Fig 1D).
We could establish iPSC lines from colonies obtained by using the three synthetic factors. They showed similar colony morphology (supplementary Fig S3A online) and growth rate to mouse embryonic stem cells. They are positive for alkaline phosphatase, SSEA-1 and Nanog (supplementary Fig S3A,B online). Their gene expression patterns resembled those of embryonic stem cells, showing reactivation of endogenous Oct4, Sox2 and Nanog genes and repression of the MEF-lineage specific gene Thy1 (supplementary Fig S3C online). The transgenes were silenced (supplementary Fig S3D online) to a similar degree as in iPSCs established with the native factors (not shown). Reactivation of endogenous Oct4 and Nanog was accompanied by demethylation of their promoters (supplementary Fig S3E online). The overall gene expression profile of XYKZ factor-based iPSCs was similar to embryonic stem cells (supplementary Fig S3F–H online). To test developmental pluripotency, we examined the contribution to embryonic development and capacity of germline transmission. Mice with high coat-colour chimerism were efficiently generated by injecting the iPSCs into diploid blastocysts (Fig 1E; supplementary Table S2 online). Moreover, germline transmission was obtained for iPSC lines in two different genetic backgrounds (Fig 1F; supplementary Table S2 online). No tumours were observed among chimeras and germline-transmitted progenies of four generations over the period of 8 months. In addition, injection of the iPSCs into tetraploid blastocysts resulted in live embryos at embryonic day 13.5 (Fig 1G). These data demonstrate that iPSCs generated with synthetic factors have full developmental potential similar to embryonic stem cells and do not increase tumour risk.
The generation of integration-free iPSCs has been inefficient to date (Jia et al, 2010). We then attempted to produce non-integration iPSCs from MEFs with synthetic factors delivered with a single episomal vector, pCEP4–XKYZ (Fig 2A). After a single transfection, we observed 55−450 Oct4–GFP-positive embryonic stem-like colonies formed from 1 × 106 MEFs approximately 18 days post-transfection, whereas two or fewer GFP+ colonies appeared when using native factors. We randomly picked 24 colonies, all of which successfully established stable cell lines with embryonic stem morphology (Fig 2B). In screening by genomic polymerase chain reaction (PCR), none of them was found to contain plasmid insertion (Fig 2C). Southern hybridization with probes specific for transgenes further confirmed the lack of DNA integration in the iPSCs (supplementary Fig S4A online). The expression of embryonic stem cell markers OCT4, NANOG and SSEA-1 was detected by immunostaining (supplementary Fig S4B online). Quantitative reverse transcriptase–PCR (RT–PCR) and global gene expression analysis demonstrated that expression levels of the pluripotency genes examined were comparable to those of embryonic stem cells (supplementary Fig S4C–E online). These iPSCs had normal karyotypes (supplementary Fig S4F online) and formed chimeras with germline contribution (Fig 2D,E). Most previous attempts to generate integration-free iPSCs necessitated the use of c-Myc (Okita et al, 2008; Kim et al, 2009; Yu et al, 2009; Zhou et al, 2009) and the reprogramming efficiency from MEFs with several plasmid transfections was only 0.0007% (Okita et al, 2008). By comparison, the use of high-performance synthetic factors has allowed us to reach an efficiency of 0.03% without using c-Myc. The higher efficiency of our episomal iPSC induction compared with other reports can be explained by the possibility that VP16 fusions function to reactivate endogenous pluripotency genes at a lower protein concentration. It is known that the expression level of reprogramming factors is reduced when delivered with episome, compared with viral vectors.
Figure 2.
Generation of integration-free mouse induced pluripotent stem cells with an episomal vector. (A) A map of the episomal vector used for iPSC generation. Coding sequences for OCT4–VP16 (X), KLF4, SOX2–VP16 (Y) and NANOG–VP16 (Z) were linked as indicated with 2A elements and subcloned into pCEP4 (Invitrogen). Note that hygromycin selection was not used in our experiments. (B) Morphology representative of in situ iPSC colonies induced with pCEP4–XKYZ and an iPSC line (Passage 5) derived from these colonies. Scale bar, 200 μm. (C) Detection of plasmid integration by PCR. Genomic DNA from episome-induced iPSC lines (1–5, 11 and 14) and OG2-MEFs (lane ‘MEF’) were used as templates for PCR amplification with primers specific for transgenes and the plasmid backbone. The pCEP4–XKYZ plasmid mixed with genomic DNA of MEFs was used as a positive control for PCR template (lane ‘XKYZ’). The experiment was performed twice. (D) Chimeric mouse derived from injecting episomal iPSC line 2 into blastocysts of mice obtained from the ICR. Agouti coat colour and pigmented eyes indicate contribution from iPSCs. (E) Germline competence of episomal iPSCs. Episomal iPSCs of line 2 were injected into blastocysts of mice obtained from the ICR. Detection of GFP signals in the genital ridge of chimeric embryos at embryonic day 13.5 indicates germline contribution. GFP, green fluorescent protein; ICR, Institute of Cancer Research; iPSC, induced pluripotent stem cell; MEF, mouse embryonic fibroblast.
Furthermore, high-quality iPSCs could be induced from MEF cells by using Oct4–VP16 alone (supplementary Fig S5 online). Interestingly, the reprogramming efficiency and kinetics were drastically improved by increasing the copy number of VP16 fused to Oct4. GFP+ colonies emerged as early as day 9 upon the transduction of Oct4-3 × VP16 and the colony number reached 120 at day 17, a level (0.24%, supplementary Fig S5A online) comparable to or more efficient than those reported for the three-factor OKS (Nakagawa et al, 2008; Wernig et al, 2008) or four-factor OKSM (Takahashi & Yamanaka, 2006).
We then applied the synthetic factors to human foreskin fibroblasts (HFFs) through an inducible lentiviral vector for the preparation of human iPS cells (hiPSC). Consistent with our observations with MEFs, 3 weeks post-infection, more iPSC colonies were obtained with synthetic factors than with native ones in both the three- and four-factor systems (Fig 3A). The resulting hiPSCs displayed normal human embryonic stem cell morphology and were alkaline phosphatase-positive (Fig 3B). We picked nine colonies, all of which established stable cell lines. Immunofluorescence staining showed that hiPSCs from two examined lines uniformly expressed embryonic stem cell markers OCT4, NANOG, SOX2, SSEA-4, Tra-1-60 and Tra-1-81 (Fig 3C). Gene expression analysis showed that iPSCs and embryonic stem cells expressed common pluripotency marker genes at comparable levels (Fig 3D). Two examined iPSC lines had normal karyotype (supplementary Fig S6 online). When grown in a differentiation medium, the iPSCs could generate cell types of all three germ layers (Fig 3E). When injected into immunocompromised mice, these iPSCs formed teratomas consisting of differentiated derivatives of three primary germ layers (Fig 3F). Therefore, synthetic factor-induced human pluripotent stem cells can differentiate into all three germ lineages both in vitro and in vivo. These results demonstrate that these high-performance synthetic factors are effective for iPSC generation not only from mouse, but also from human somatic cells.
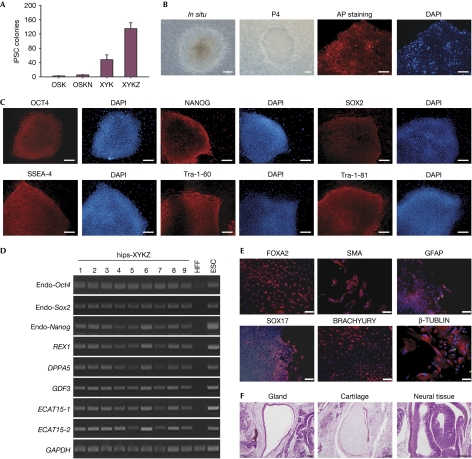
Figure 3.
Synthetic factors increased the efficiency of human induced pluripotent stem cell generation. (A) Numbers of AP-positive colonies with iPSC-like morphology obtained from 1 × 105 HFF cells transduced with lentiviral constructs for three or four reprogramming factors, as indicated. Error bars, s.d. (n=4). (B) Representative images of human XYKZ primary colony (in situ) and derived iPSC line colonies (P4) using phase contrast or AP staining as before. Scale bar, 200 μm. (C) Immunostaining for hiPSC line XYKZ 6 using antibodies against OCT4, NANOG, SOX2, SSEA-4, TRA-1-60 and TRA-1-81. Scale bar, 200 μm. (D) RT–PCR assays for the endogenous pluripotency markers indicated at the left in different hiPSC lines compared with HFFs or ESCs. Representative data for two independent experiments are shown. (E) Immunocytochemistry of differentiated human iPSCs with antibodies against markers for the three germ layers (FOXA2 and SOX17 for endoderm; SMA and BRACHYURY for mesoderm; and GFAP and β-TUBULIN for ectoderm). Scale bar, 100 μm. Nuclei were stained with DAPI (blue). (F) Hematoxylin and eosin-stained sections from teratomas derived from human iPSCs 8 weeks after transplantation into immunodeficient mice. Representative images show tissues from all three germ layers: gland (endoderm), cartilage (mesoderm) and neural tissue (ectoderm). Teratomas were obtained from the two iPSC lines (3 and 6) examined. Scale bar, 100 μm. AP, alkaline phosphatase; ESC, embryonic stem cell; HFF, human foreskin fibroblast; hiPSC, human induced pluripotent stem cell; iPSC, induced pluripotent stem cell.
Recent reports indicate that the p53 pathway suppresses cell replicative potential and that the inhibition of this pathway significantly increases the efficiency of iPSC generation (Zhao et al, 2008; Hong et al, 2009; Kawamura et al, 2009; Li et al, 2009; Marion et al, 2009; Utikal et al, 2009). We found that the enhanced reprogramming by the synthetic factors is unrelated to the p53 pathway in the transduced MEFs, as the p53 protein level was slightly increased in the transduced MEFs (supplementary Fig S7 online). Therefore, iPSCs derived from synthetic factors do not have increased tumour risk due to compromised genome integrity, as seen in those generated by p53 inactivation.
VP16 is a known potent transcriptional transactivator. The fact that fusion of VP16 to reprogramming factors led to more iPSC colonies shows that transcriptional activation is crucial for reprogramming. VP16 fusions would target the strong activation to the promoter of the endogenous pluripotency genes, leading to their fast reactivation. The mechanism at this level is clear. As VP16 has several molecular functions—such as increasing transcription initiation by recruiting transcription factors (Gupta et al, 1996; Shen et al, 1996), remodelling the chromatin by unfolding (Carpenter et al, 2005) and recruiting P300 histone acetyltransferase causing local hyperacetylation (Hall & Struhl, 2002)—we are interested to know which of the VP16 functions is most important in creating an improved reprogramming factor.
We have observed previously that ectopic expression of a Nanog–VP16 fusion in mouse embryonic stem cells triggers their differentiation (Wang et al, 2008). This detrimental effect might have been avoided here due to the rapid silencing of transduced synthetic factors in reprogrammed cells during the acquisition of pluripotency. This process is associated with de novo DNA methylation of retroviral promoters (not shown), probably due to the reactivation of endogenous Dnmt3-family methyltransferases. Nevertheless, the timing and duration of the ectopic expression of synthetic factors remain to be determined for optimal iPSC reprogramming. Moreover, the synthetic reprogramming factors themselves could be further improved, for example by improving transcriptional activation, DNA binding or resistance to ubiquitin-mediated degradation (Xu et al, 2009). The optimization process has now become feasible due to the reduction of reprogramming factors to a single factor, Oct4–VP16.
The robust and consistent iPSC generation that we have demonstrated in several contexts highlights the potential for engineered factors in the preparation of safe human iPSCs for clinical applications. Improved versions of other transcription factors than Oct4, Sox2 and Nanog might also be designed to reprogramme cell fate at heightened efficiencies in systems such as the directed differentiation of stem and precursor cells into functional cell types for regenerative medicine.
Methods
Plasmid construction. Full-length complementary DNAs of murine and human Oct4, Sox2 and Nanog genes were ligated with complementary DNA encoding the VP16 activation domain (amino acids 446–490; from MLGDG to DEYGG) with a region encoding a glycine-rich linker (TSGLGGGSGGGGSGGGGSG, for Oct4 and Sox2) or without the linker (Nanog). Fusion genes were cloned into retroviral vector pMXs (Takahashi & Yamanaka, 2006) and inducible lentiviral vector pLV-TRE-EF1a-GFP (Wu et al, 2009). For episomal vector construction, coding sequences for OCT4–VP16 (X), KLF4, SOX2–VP16 (Y) and NANOG–VP16 (Z) were linked in this order with 2A elements (Okita et al, 2008) and subcloned into pCEP4 (Invitrogen).
Retroviral production and mouse iPSC induction. Retroviral production and infection followed the previously published protocol (Takahashi et al, 2007). Oct4–GFP MEF cells (seeded at 5 × 104 cells in each well in a six-well plate) were incubated with virus-containing supernatants for 12 h. Two days after infection, the medium was changed to mouse embryonic stem cell medium. Eight days after infection, the transduced Oct4–GFP MEF cells were re-plated onto mitomycin-C-treated MEF feeder layers at 5 × 104 cells in each well in a six-well plate. Approximately 7 days after re-plating, the numbers of GFP-positive and alkaline phosphatase-positive colonies were scored. Alkaline phosphatase staining was performed with NBT/BCIP (Roche) according to the instructions of the manufacturer.
Generation of iPSCs from MEFs with episomal vector. For reprogramming with oriP/EBNA1-based episomal vector, 5 μg of episomal plasmid pCEP4–XKYZ was transfected into 1 × 106 MEFs by nucleofection (Amaxa). Transfected MEFs were directly plated on 2 × 10-cm feeder-seeded dishes with no drug selection. On day 2 post-transfection, the culture medium was replaced with an optimized culture medium (Chen et al, 2010). The culture medium was changed every two days with no drug selection. GFP-positive colonies with morphology similar to embryonic stem cell were visible on day 18 post-transfection and picked for characterization.
Human iPSC induction. 1 × 105 HFFs seeded in a 6-cm dish were infected with lentiviral supernatants overnight and then cultured in HFF medium supplemented with doxycycline (Sigma) to 1 μg/μl. Three days post infection, transduced HFFs were re-plated onto mitomycin-C-treated MEF feeder layers at a ratio of 1:3 and were changed to human embryonic stem cell medium supplemented with 1 μg/μl doxycycline. Two weeks post-infection, doxycycline was withdrawn. Approximately 3 weeks after infection, iPSC colonies were picked and alkaline phosphatase-positive, human embryonic stem cell-like colonies were scored. The experiments were repeated more than three times.
Methods are provided in detail in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank J. Ellis and C. Walsh for critical reading of the manuscript; R. Bronson for histological analysis of teratomas; X. Ding and L. Xu for human fibroblast cells and help with in vitro human cell differentiation assay; L. Xiao for the human embryonic stem cell line; Y. Jin for human foreskin fibroblast cells; and H. Deng for human transcription factors. This study was supported by grants from the Ministry of Science and Technology China (2006CB943900 and 2009CB941101), the Shanghai Municipal government (08dj1400501) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010301).
Footnotes
The authors declare that they have no conflict of interest.
References
- Carpenter AE, Memedula S, Plutz MJ, Belmont AS (2005) Common effects of acidic activators on large-scale chromatin structure and transcription. Mol Cell Biol 25: 958–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J et al. (2010) Towards an optimized culture medium for the generation of mouse induced pluripotent stem cells. J Biol Chem 285: 31066–31072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Emili A, Pan G, Xiao H, Shales M, Greenblatt J, Ingles CJ (1996) Characterization of the interaction between the acidic activation domain of VP16 and the RNA polymerase II initiation factor TFIIB. Nucleic Acids Res 24: 2324–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Struhl K (2002) The VP16 activation domain interacts with multiple transcriptional components as determined by protein–protein cross-linking in vivo. J Biol Chem 277: 46043–46050 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F et al. (2010) A nonviral minicircle vector for deriving human iPS cells. Nat Methods 7: 197–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D et al. (2009) Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106 [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S (2008) Generation of mouse induced pluripotent stem cells without viral vectors. Science 322: 949–953 [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J (2010) The genetics of induced pluripotency. Reproduction 139: 35–44 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M (1988) GAL4–VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Shen F, Triezenberg SJ, Hensley P, Porter D, Knutson JR (1996) Transcriptional activation domain of the herpesvirus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. J Biol Chem 271: 4827–4837 [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A (2008) Capturing pluripotency. Cell 132: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S (2007) Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2: 3081–3089 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma T, Chi X, Pei D (2008) Aromatic residues in the C-terminal domain 2 are required for Nanog to mediate LIF-independent self-renewal of mouse embryonic stem cells. J Biol Chem 283: 4480–4489 [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R (2008) c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2: 10–12 [DOI] [PubMed] [Google Scholar]
- Wu Z et al. (2009) Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol 1: 46–54 [DOI] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, Jin Y (2009) WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res 19: 561–573 [DOI] [PubMed] [Google Scholar]
- Yamanaka S (2009a) Elite and stochastic models for induced pluripotent stem cell generation. Nature 460: 49–52 [DOI] [PubMed] [Google Scholar]
- Yamanaka S (2009b) A fresh look at iPS cells. Cell 137: 13–17 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y et al. (2008) Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3: 475–479 [DOI] [PubMed] [Google Scholar]
- Zhou H et al. (2009) Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4: 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.