Abstract
Several mechanisms have been proposed for the synthesis of substrate-linked ubiquitin chains. HECT ligases directly catalyse protein ubiquitination and have been found to non-covalently interact with ubiquitin. We report crystal structures of the Nedd4 HECT domain, alone and in complex with ubiquitin, which show a new binding mode involving two surfaces on ubiquitin and both subdomains of the HECT N-lobe. The structures suggest a model for HECT-to-substrate ubiquitin transfer, in which the growing chain on the substrate is kept close to the catalytic cysteine to promote processivity. Mutational analysis highlights differences between the processes of substrate polyubiquitination and self-ubiquitination.
Keywords: catalysis, E3 ligase, polyubiquitination, structure, ubiquitin
Introduction
The ubiquitination process is carried out by an enzymatic cascade that includes an activating enzyme (E1), a conjugating enzyme (E2) and a ligase (E3; Dye & Schulman, 2007). The transfer of the ubiquitin moiety from the thioester-linked E3 (in HECT-type ligases) to the acceptor lysine on the substrate is the last step of this process. Subsequent chain elongation requires the modification of specific lysine residues in consecutive ubiquitin moieties. With few exceptions (Petroski & Deshaies, 2005; Jin et al, 2008), little is known about the mechanisms of ubiquitin-chain assembly, although various models have been proposed (Hochstrasser, 2006).
The Nedd4 family of HECT domain E3 ligases is a well-characterized class of enzymes that present a conserved modular organization with an amino-terminal C2 domain that is crucial for membrane localization, between two and four WW domains that recognize substrates and adaptor proteins and a carboxy-terminal catalytic HECT domain. In humans, there are nine members of this family that are implicated in a range of biological processes such as endocytosis, protein transport, viral budding, signalling, cellular growth and proliferation (Rotin & Kumar, 2009). This class of E3 enzymes seems to use a sequential addition mechanism, by which ubiquitin molecules are added one at a time from the catalytic cysteine to the distal lysine of the growing chain (Kim & Huibregtse, 2009). A key question is how E3 enzymes deal with the shifting position of the acceptor site during chain elongation.
Two groups have recently identified a surface implicated in non-covalent ubiquitin binding on the HECT-type E3 ligases Rsp5 and Smurf2 (French et al, 2009; Ogunjimi et al, 2010). This surface was proposed to have a role in regulating polyubiquitination, although opposite mechanisms were suggested by the groups, with the surface being required to either restrict the length of polyubiquitin chains synthesized by the HECT domain (French et al, 2009) or to promote polyubiquitination (Ogunjimi et al, 2010). In this study, we show the crystal structure of the HECT domain of Nedd4 alone and in complex with ubiquitin, and we present molecular insights into the mechanism by which Nedd4 catalyses polyubiquitination.
Results and Discussion
Structure of HECTNedd4 and HECTNedd4:ubiquitin
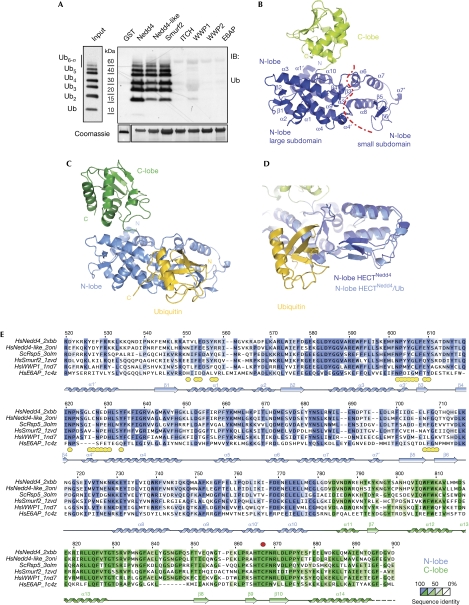
We characterized the interaction of the isolated HECT domain of Nedd4 (HECTNedd4) with ubiquitin in detail. The ubiquitin-binding ability resides in the N-lobe, does not show preference for Lys 63- or Lys 48-polyubiquitin chains and requires the canonical hydrophobic patch on ubiquitin, centred on Ile 44 (supplementary Fig S1 online). We extended this analysis to the other mammalian Nedd4 family members and found that only a subset of these HECT domains binds to ubiquitin, namely Nedd4, Nedd4-like and Smurf2 (Fig 1A).
Figure 1.
Structure of the HECTNedd4 domain in apo form and in complex with ubiquitin. (A) GST pull-down assay with the HECT domains of various Nedd4 family HECT E3 ligases. GST-fusion proteins were incubated for 2 h at 4°C in YY buffer with synthetic Lys 63-polyubiquitin chains and analysed by IB as indicated. Coomassie staining shows comparable loading of GST proteins. Similar results were obtained with linear and Lys 48-polyubiquitin chains (not shown). (B) Overall structure of HECTNedd4 (N-lobe, blue; C-lobe, green). The red dotted line indicates the boundary between the large and small subdomains of the N-lobe. (C) Overall structure of HECTNedd4 in complex with ubiquitin (yellow). The HECT structure is represented in the same orientation as in B; N-lobe, light blue; C-lobe, dark green. (D) Superposition on the large subdomain of the N-lobe of HECTNedd4 and HECTNedd4:ubiquitin. In the complex (light blue), the β5–β6 hairpin of the small subdomain of the N-lobe is closer to the large subdomain, with respect to the isolated HECT (dark blue). (E) Sequence alignment of the HECTNedd4 domain with other crystallized HECT domains. Secondary structure elements are depicted. Dotted line indicates that the residues were not visible in the electron density maps. Yellow circles indicate residues in contact with ubiquitin in the structure of HECTNedd4:ubiquitin (according to PISA; Krissinel & Henrick, 2007). Numbering refers to Nedd4 sequence. GST, glutathione S-transferase; IB, immunoblotting; Ub, ubiquitin.
To understand how ubiquitin binds to the HECT, we determined the crystal structure of the HECTNedd4 in isolation (at 2.5 Å) and in complex with ubiquitin (at 2.7 Å) by molecular replacement (supplementary Table S1 online and supplementary Fig S2 online). In both structures, HECTNedd4 displays the typical HECT fold (Huang et al, 1999; Verdecia et al, 2003; Ogunjimi et al, 2005) composed of two lobes connected by a flexible hinge (Fig 1B,C). The N-lobe, an elongated array of helices and β-hairpins, consists of two moieties, known as the large and small subdomains (Fig 1B). The small subdomain, which hosts the E2-binding site, comprises helices α6–α8 and β-sheets β5–β6 (Huang et al, 1999; Kamadurai et al, 2009). The large subdomain of the N-lobe is present below the C-lobe, an α/β sandwich domain that carries the catalytic cysteine. The orientation of the C-lobe differs in the two HECTNedd4 structures, and both orientations are distinct from those of previously reported HECT domain structures (supplementary Fig S3 online). This highlights the freedom of movement of the C-lobe, which is key for the catalytic function of HECT domains (Verdecia et al, 2003; Kamadurai et al, 2009).
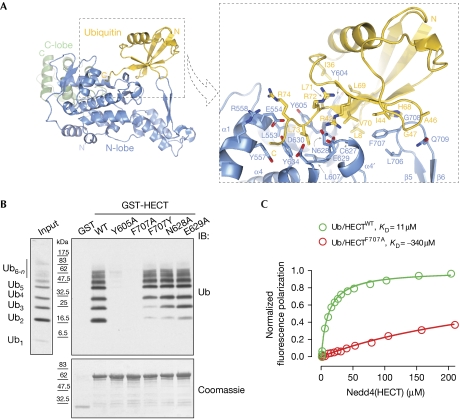
Non-covalent ubiquitin binding to HECTNedd4 conceals a solvent-accessible interaction surface area on ubiquitin of approximately 900 Å2, the largest surface identified so far for ubiquitin-binding domains (supplementary Table S2 online). Ubiquitin makes contact with Glu 554 and neighbouring residues of helix α1; Tyr604, Tyr 605 and Tyr 610 from the region comprising helix α3′ and strand β3; Asn628 and Glu 629 from helix α4′ and the ensuing loop; and Phe707 and neighbouring residues of the β5–β6 beta hairpin. These residues are distributed in both the small and the large subdomains of the N-lobe (Figs 1E,2A). In the absence of ubiquitin, the relative orientation of the small and large subdomains is not fixed and varies for different structures (Huang et al, 1999; Verdecia et al, 2003; Ogunjimi et al, 2005). Ubiquitin binding might therefore be expected to stabilize a specific reciprocal orientation of the N-lobe subdomains. Indeed, superposition of the large subdomains of HECTNedd4 and HECTNedd4:ubiquitin (root mean square deviation of 0.6 Å over 181 Cα) clearly indicates a relative movement of the β5–β6 hairpin of the small subdomain in the HECTNedd4:ubiquitin structure by approximately 5 Å towards the large subdomain of the N-lobe (Fig 1D).
As predicted by the defective behaviour of the I44A ubiquitin mutant (supplementary Fig S1C online), the interaction surface on ubiquitin involves the canonical Ile 44 hydrophobic patch, which also includes Gly 47, Leu 8 and Val 70 (Fig 2A). However, the surface of interaction is not limited to this patch, but extends to a ‘second hydrophobic patch’, including the residues Ile 36/Leu 71/Leu 73, the role of which has recently been discussed in the context of the E2-to-HECT ubiquitin transfer (Kamadurai et al, 2009; Fig 2A; supplementary Fig S4 online). The Asn 628, Tyr 634 and Glu 554 side chains on Nedd4 form hydrogen bonds with the main chain nitrogen atoms of ubiquitin-Leu 73, Arg74 and Gly 75, whereas the ubiquitin-Leu 73 side chain is stacked between Tyr 634 and Tyr 605 of Nedd4.
Figure 2.
HECTNedd4:ubiquitin interaction and mutant validation. (A) Close-up view of HECTNedd4 N-lobe:ubiquitin interaction. (B) GST pull-down assay with the indicated Nedd4 constructs and Lys 63-linked polyubiquitin chains was performed as described in Fig 1A. (C) Fluorescence-polarization assay with the indicated Nedd4 constructs and monomeric ubiquitin was performed. The HECTNedd4:ubiquitin interaction displays a moderate affinity with a KD of 11 μM, F707A mutant displays a thirty times lower affinity. Details are described in the supplementary Methods online and similar results for the Y605A mutant obtained by SPR assay are in supplementary Fig S1 online. IB, immmunoblotting; GST, glutathione S-transferase; SPR, surface plasmon resonance; Ub, ubiquitin; WT, wild type.
We generated Nedd4 mutants that substantiate the functional importance of both interacting patches on ubiquitin. Mutation of Tyr 605 to Ala (Y605A) or Phe 707 to Ala (F707A) almost abolished HECTNedd4 binding to Lys 63 ubiquitin (Fig 2B). Phe 707 to Tyr (F707Y), Asn 628 to Ala (N628A) or Glu 629 to Ala (E629A) mutations had milder effects, preserving the association with higher molecular weight Lys 63 ubiquitin to varying degrees (Fig 2B). We confirmed these results by measuring the interaction between the HECT domains and monomeric and dimeric ubiquitin by fluorescence polarization and surface plasmon resonance (SPR) assay (Fig 2C; supplementary Fig S1D–F online). Both ubiquitin ligands interact with the HECT with rapid kinetics (fast Kon and Koff rate constants, data not shown). Wild-type HECTNedd4 displays a moderate affinity (KD approximately 11 μM) in the range of those reported for ubiquitin-binding domains (supplementary Table S2 online), whereas Y605A and F707A mutations show from 20- to 30-fold decreases in binding (Fig 2C; supplementary Fig S1F online).
Role of ubiquitin binding in Nedd4 activity
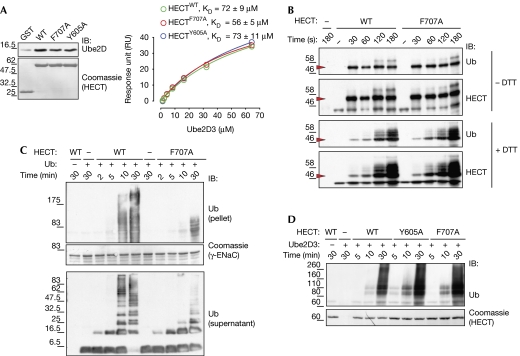
Next, we analysed the catalytic activity of Nedd4 HECT mutants. In principle, the ubiquitin-binding surface might have a role at three stages of the E3 catalysis: binding to the E2, transthiolation process from E2 to E3 or substrate ubiquitination. We tested all of these possibilities using the isolated HECT, which retains the ability to ubiquitinate itself as well as substrates, albeit with reduced efficiency (not shown).
Pull-down and SPR assays showed that mutants have no significant impairment in binding with either the apo or the ubiquitin thioester-linked form of E2 enzyme Ube2D3 (Fig 3A and data not shown). Indeed, the E2 binding is built on an adjacent but non-overlapping surface on the large subdomain of the N-lobe (Huang et al, 1999; Fig 4C). We then tested the importance of the ubiquitin-binding surface in the E2-to-HECT transthiolation process by using a pulse-chase protocol (Fig 3B). Again, no appreciable transthiolation defects were observed for the mutants, supporting the notion that the ubiquitin-binding surface is not involved in the upstream steps of the enzymatic cascade. Of note, the thioester HECT∼ubiquitin bond is unstable and the ubiquitin moiety is immediately transferred to the lysine/s of the HECT, as demonstrated by the appearance of higher molecular-weight proteins that are resistant to dithiothreitol treatment (Fig 3B, lower panels).
Figure 3.
Disruption of HECTNedd4:ubiquitin interaction impairs substrate polyubiquitination. (A) Mutations do not affect E2 binding. Left panel: GST pull-down assay with the indicated HECT mutants and the E2 enzyme Ube2D3. IB was performed as indicated. Coomassie staining shows comparable loading of GST proteins. Right panel: the HECTNedd4:Ube2D3 interaction displays a modest affinity, that is not perturbed by the F707A and Y605A mutations. SPR assay was performed as described in the supplementary methods online. (B) Mutations do not affect the kinetics of the E2-to-HECT transthiolation process. The transfer of ubiquitin was monitored by quenching the reaction at different time points, with the addition of Laemmli buffer with or without the reducing agent (100 mM DTT). Arrow indicates thioesther HECT∼ubiquitin (−DTT) or monoubiquitinated HECT (+DTT) running at the same position. DTT-resistant higher molecular bands represent self-ubiquitinated HECT. Similar results were obtained with Y605A mutant. (C) Mutations impair substrate polyubiquitination. Upper panel: GST-γENaC ubiquitination kinetics with WT HECT and F707A mutant (ubiquitin (pellet)). Middle panel: Coomassie staining showing comparable loading of GST proteins. Lower panel: kinetics of free ubiquitin chain formation (ubiquitin (supernatant)) during the reaction. IB was performed as indicated. (D) Self-ubiquitination kinetics with WT HECT and Y605A and F707A mutants. IB was performed as indicated. Coomassie staining shows comparable loading of HECT proteins. Similar results were obtained with full-length Nedd4 mutants. DTT, dithiothreitol; ENaC, epithelial Na+ channel; GST, glutathione S-transferase; IB, immunoblotting; SPR, surface plasmon resonance; Ub, ubiquitin; WT, wild type.
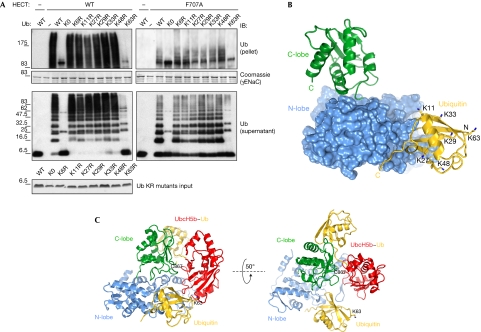
Figure 4.
Ubiquitin binding to the HECTNedd4 domain is compatible with HECTNedd4-like:E2∼ubiquitin complex and does not dictate chain specificity. (A) Substrate ubiquitination assay with the indicated ubiquitin KR mutants was performed. The reaction was quenched after 30 min for the WT HECT and after 60 min for the F707A mutant. Upper panel: GST-γENaC ubiquitination with the indicated constructs. Middle panel: Coomassie staining showing comparable loading of GST proteins. Lower panel: free ubiquitin chain formation during the reaction. IB was performed as indicated. Lower panel: 1 μg of ubiquitin KR mutants were loaded for comparison and visualized by Coomassie staining. (B) Position of ubiquitin lysines in the Nedd4 HECT/ubiquitin complex. HECT N-lobe is shown as surface representation, whereas the C-lobe and ubiquitin are shown as cartoon representations. Ubiquitin lysine side chains are indicated in sticks. Six of the seven ubiquitin lysines are shown, K 6 being in the back. (C) Model of Ubch5B∼Ub:C-lobe complex (Kamadurai et al, 2009) binding to the N-lobe:ubiquitin complex. Details are in the supplementary Methods online. C 867 on the HECT and K 63 on the Ub are shown. ENaC, epithelial Na+ channel; GST, glutathione S-transferase; IB, immunoblotting; KR, lysine-to-arginine mutation; Ub, ubiquitin; WT, wild type.
These results led us to propose that the ubiquitin-binding surface on the HECT might act to bind a ubiquitin moiety that is already conjugated to a substrate, thus promoting polyubiquitination. Indeed, when we assayed F707A and Y605A in an in vitro ubiquitination reaction, we found that mutations in the ubiquitin-binding surface strongly impaired free-chain formation and ubiquitination of all the substrates tested (Fig 3C; supplementary Fig S5 online). The mutant enzymes were efficient in the first cycle of substrate ubiquitination and in ubiquitin dimer formation, using free ubiquitin as a pseudosubstrate (Fig 3C; supplementary Fig S5 online). This was confirmed by using a ubiquitin Lys0 mutant and quantification of the results repeated as fold differences between wild-type HECT and mutants (supplementary Fig S5A online). Interestingly, F707A and Y605A mutations did not affect self-ubiquitination of Nedd4 (Fig 3B, lower panels and Fig 3D), indicating that this in cis reaction is a catalytically distinct process that cannot be used as a surrogate assay for ligase activity on substrates.
Most HECT E3s synthesize polyubiquitin chains with specific linkages (Wang et al, 2006; Kim et al, 2007). To gain insight into the type of chains synthesized by Nedd4, we tested substrate ubiquitination using ubiquitin-bearing individual lysine-to-arginine mutations (KR mutants). We found that Nedd4 has a strong preference for building Lys 63-chains on substrates, a feature retained by the F707A mutant (Fig 4A). Consistent with previous data, however, F707A has defective chain elongation on substrate and shorter free chains, regardless of the type of ubiquitin used (Fig 4A). Therefore, we conclude that the ubiquitin-binding surface on the HECT acts to promote substrate polyubiquitination, but it does not dictate the preference for a specific lysine during elongation. Indeed, recent observations suggest that the C-lobe, rather than the ubiquitin-binding N-lobe, is the crucial determinant of lysine selection during elongation (Kim & Huibregtse, 2009).
CONCLUSIONS
Collectively, our results support the hypothesis that the ubiquitin-binding surface is required for the processivity of the polyubiquitination reaction (Ogunjimi et al, 2010), rather than for limiting chain elongation, as suggested previously (French et al, 2009). How can this occur? It is tempting to envision a model in which the distal ubiquitin on the substrate occupies this surface, promoting retention of the ubiquitinated substrate to the E3, and also keeping the small subdomain of the N-lobe in a conformation that favours processive ubiquitin addition. This could be achieved by either reducing the gap between the catalytic cysteine of the HECT and the C-terminus of ubiquitin linked to the E2 enzyme (Kamadurai et al, 2009) or by facilitating the transfer of a subsequent ubiquitin to the HECT-bound substrate. In support of this model, we found that the non-covalent ubiquitin-binding surface that we mapped remains accessible in the complex of the HECT domain of Nedd4-like with the ubiquitin-loaded E2 (Kamadurai et al, 2009; Fig 4C).
Our data support the notion that Nedd4 adopts a simple sequential-addition model to build a chain on a substrate; after the first ubiquitin is attached, the chain is elongated through Lys 63 linkage, because of the ability of the N-lobe to maintain the growing polyubiquitin chain in close proximity. A similar conclusion about the role of the HECT ubiquitin-binding site in promoting chain elongation was reached in the accompanying study by Kim et al (2011) on Rsp5. Although the position of Lys 63 on bound ubiquitin does not seem to be able to orient the growing chain towards the E2 catalytic cysteine (Fig 4B), the average B factors for the ubiquitin molecules are considerably higher than those of their HECT counterparts (approximately 76 A2 for ubiquitin molecules, approximately 48 A2 for HECT domains; supplementary Table S1 online), suggesting freedom of movement of ubiquitin on its docking site. This could imply that the binding of ubiquitin to the E3 is strong enough to promote polyubiquitination, yet loose enough to allow chain growth, possibly through a slippage mechanism by which the ubiquitin-binding surface specifically binds to the distal ubiquitin at the end of the chain. The moderate affinity and fast kinetic rates of the HECT:ubiquitin interaction fit well with such a mechanism. Future structural studies with Nedd4 in complex with a ubiquitinated substrate might be required to provide a definitive picture of this dynamic process.
The detailed molecular view provided by our structure allows the identification of the crucial residues required for binding (Fig 1E), which can be used to predict the HECT E3 enzymes that are able to bind to ubiquitin. It remains to be established whether the presence of this binding surface might determine the mechanism of chain synthesis adopted by the different HECT ligases to become processive.
At our level of understanding, generalizing the mechanisms that underlie polyubiquitination would be premature, but it is interesting to note from the comparison of the small Nedd4 family of E3, that nature has used a variety of protein architectures to ensure specificity.
Methods
Crystallization and structure determination. Crystals of HECTNedd4 and HECTNedd4:ubiquitin complex were obtained by sitting-drop vapour diffusion at 20°C, using a Honeybee Cartesian robot in 96-well plates. Diffraction-quality crystals were obtained by optimizing the initial conditions in 24-well plates, hanging drops at 20°C. Crystals were all optimized by microseeding. For HECTNedd4, the optimized conditions were 100 mM Na-MES, pH 6.0, 2–4% PEG 400 or PEG 600, 30–60 mM CaCl2 or MgCl2, 5 mM tris(2-carboxyethyl)phosphine, with protein concentration in the 2.5–5 mg/ml range. Crystals were cryoprotected in 100 mM Na-MES, pH 6.0, 4% PEG 400, supplemented with 20% ethylene glycol. The structure was solved with a data set collected at the European Synchrotron Radiation Facility (ESRF) at beamline ID14-2. For the HECTNedd4:ubiquitin complex, initial crystals were obtained in 100 mM Na-HEPES, pH 7–8, 10–20% PEG 2000 MME, 5 mM tris(2-carboxyethyl)phosphine, with proteins purified individually, then mixed in a 1:1 molar ratio and a concentration of approximately 30 mg/ml. To obtain good-quality diffraction and to overcome twinning, the complex was crystallized in the presence of excess ubiquitin (600–900 μM of complex, 1.2 × –2.3 × ubiquitin molar excess), lower concentration of PEG 2000 MME (2–10%), and the crystals were carefully frozen by equilibrating them into cryo-buffer (100 mM Na-HEPES, pH 7.5, 10% PEG 2000 MME) with increasing concentrations of glycerol, reaching a final concentration of 20%. The structure was solved with a data set collected at the ESRF at beamline ID14-1 on a crystal grown in a 1.9 × ubiquitin molar excess.
X-ray diffraction data were processed with HKL2000 (Otwinowski & Minor, 1997) or XDS (Kabsch, 2010). Both structures were solved by molecular replacement with Phaser within the CCP4 suite (CCP4, 1994), using as a search model the HECT domain of Nedd4-like (Protein Data Bank entry 2oni) in the case of HECTNedd4, and HECTNedd4 and a high-resolution structure of ubiquitin (Protein Data Bank entry 1ubi) in the case of HECTNedd4:ubiquitin complex. Initial models were refined with the CNS suite (Brunger, 2007), Refmac (Murshudov et al, 1997), the Phenix suite (Adams et al, 2010) and manual building in Coot (Emsley et al, 2010). For the HECTNedd4:ubiquitin complex, non-crystallographic symmetry (NCS) restraints were used in refinement. In the first cycles of refinement carried out with Refmac, HECT molecules were divided into two NCS groups (the N-lobe and the C-lobe), and ubiquitin molecules were the third NCS group. For further refinement cycles carried out with phenix.refine, five NCS groups were used: ubiquitin molecules and HECT domain residues 522:699, 724:778, 785:828 and 850:891, thus not subjecting HECT domain loops to NCS restraints. Structure representations were generated with Pymol (DeLano Scientific LLC). HECTNedd4 crystallized in spacegroup C2, whereas HECTNedd4:ubiquitin crystallized in spacegroup P21, with two complexes per asymmetrical unit. The two complexes differ slightly in the orientation of the HECTNedd4 C-lobe with respect to the N-lobe, and the relative orientation of HECTNedd4 with respect to ubiquitin, but the interactions discussed here are present in both complexes. Superpositions of pairs of domains of the asymmetrical unit indicate that they are almost identical (root mean square deviations of N-lobes: 0.36 Å over 260 Cα; C-lobes: 0.63 Å over 115 Cα; Ubs: 0.25 Å over 76 Cα).
Ubiquitination assay. Ubiquitination assays were performed with HECT domains produced as glutathione S-transferase (GST) fusion proteins and cleaved with PreScission protease. The E2 enzyme Ube2D3, was produced as a His6-fusion protein and eluted from Ni-NTA Agarose beads (Qiagen). Reaction mixtures contained purified enzymes (20 nM E1, 250 nM of purified His6-tagged Ube2D3, 250 nM HECTNedd4), 300 nM of substrate (γ epithelial Na+ channel and LMP2A were produced as GST fusion proteins and used attached to glutathione beads) and 1 μM of ubiquitin in ubiquitination buffer (25 mM Tris–HCl, pH 7.6, 5 mM MgCl2, 100 mM NaCl, 0.2 μM dithiothreitol, 2 mM ATP). Reactions were incubated at 37°C. At the indicated time point, samples were centrifuged to separate the pellet—containing the ubiquitinated substrates—and the supernatant—containing the enzymes and the soluble ubiquitin chains, if produced. The pellet was washed four times in YY buffer (50 mM Na-HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% triton X-100) before loading on SDS–polyacrylamide gel electrophoresis gel. For self-ubiquitination reaction, the mixtures contained 20 nM E1, 250 nM of purified His6-tagged Ube2D3, 250 nM of GST-HECTNedd4 and 0.5 μM of ubiquitin in ubiquitination buffer. Detection was performed by immunoblotting using specific antibody. Coomassie-stained membrane was used to show loading of GST-fusion protein after immunoblotting.
Reagents and constructs, protein expression and purification, transthiolation assay, pull-down experiments, fluorescence polarization assay and SPR are described in the supplementary Methods online.
Accession codes: Coordinates for HECTNedd4 and the HECTNedd4: ubiquitin complex have been deposited at the Protein Data Bank under accession codes 2xbf and 2xbb, respectively.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank P.R. Romano for critically reading the manuscript, V. Cecatiello for assistance in crystallization, R.A. Steiner for advice, the staff at European Synchrotron Radiation Facility for assistance in data collection and M.P.A. Luna-Vargas for providing reagents. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and the European Community (FP7) to S.P. E.M. is the recipient of an AIRC fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dye BT, Schulman BA (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French ME, Kretzmann BR, Hicke L (2009) Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem 284: 12071–12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286: 1321–1326 [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M (2008) Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin–HECT(NEDD4L) complex. Mol Cell 36: 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 29: 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kim HY, Steffen AM, Oldham ML, Chen J, Huibregtse JM (2011) Structure and function of an HECT domain ubiquitin-binding site. EMBO Rep [Epub ahead of print 11 March] doi:10.1038/embor.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell 19: 297–308 [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Wiesner S, Briant DJ, Varelas X, Sicheri F, Forman-Kay J, Wrana JL (2010) The ubiquitin binding region of the Smurf HECT domain facilitates polyubiquitylation and binding of ubiquitylated substrates. J Biol Chem 285: 6308–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF–Cdc34. Cell 123: 1107–1120 [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409 [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP (2003) Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell 11: 249–259 [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM (2006) Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J 25: 1710–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.