Abstract
The Rsp5 ubiquitin ligase contains a non-covalent binding site for ubiquitin within the amino-terminal lobe (N-lobe) of the HECT domain, and the X-ray crystal structure of the HECT–ubiquitin complex has been determined. Hydrophobic patch residues of ubiquitin (L8, I44, V70) were crucial for interaction with Rsp5, and amino-acid alterations at the Rsp5-binding interface resulted in defects in polyubiquitination. Our results support a model in which the N-lobe-binding site acts to localize and orient the distal end of the ubiquitin chain to promote conjugation of the next ubiquitin molecule.
Keywords: HECT, polyubiquitin chains, Rsp5, ubiquitin
Introduction
HECT (homologous to E6AP carboxyl-terminus) domain ubiquitin ligases (E3s) form a ubiquitin thioester intermediate during catalysis, with ubiquitin being transferred from the E3 to the substrate (Scheffner et al, 1995). HECT domains consist of an approximately 250-amino-acid-elongated amino-terminal lobe (N-lobe) connected by a short flexible linker to an approximately 100 amino-acid globular C-terminal lobe (C-lobe; Huang et al, 1999). The E2-binding site is located on the N-lobe and the active-site cysteine is within the C-lobe. The structure of an E2–ubiquitin thioester complex with the HECT domain of Nedd4l showed the C-lobe of the HECT domain oriented towards the E2–ubiquitin complex and interacting with ubiquitin (Kamadurai et al, 2009), providing insight into how ubiquitin is transferred from the E2 to the E3. Less is known about the mechanism for transferring ubiquitin from the E3–ubiquitin complex to the substrate. One unresolved problem has been how the distal end of the polyubiquitin chain is oriented near to the active site of E3 during a processive polyubiquitination reaction.
Previous studies identified a non-covalent binding site for ubiquitin on the N-lobe of the HECT domain of yeast Rsp5 and human Smurf2; however, these studies came to different conclusions about the function of this binding site. Although the Rsp5 study concluded that the binding site acted to restrict polyubiquitination (French et al, 2009), the Smurf2 study concluded that the binding site facilitated polyubiquitination (Ogunjimi et al, 2005). In this paper, we present the X-ray crystal structure of ubiquitin bound to this site of Rsp5 and show that, consistent with the Smurf2 study, alteration of this binding site results in a defect in polyubiquitination. Furthermore, for some substrates, alteration of the binding site limits the initial substrate ubiquitination event. We propose that the non-covalent ubiquitin-binding site acts to orient the distal end of growing polyubiquitin chains, and might also act as a surface for recruiting and orienting target proteins for the initial ubiquitination event.
Results
Structure of Rsp5 in complex with ubiquitin
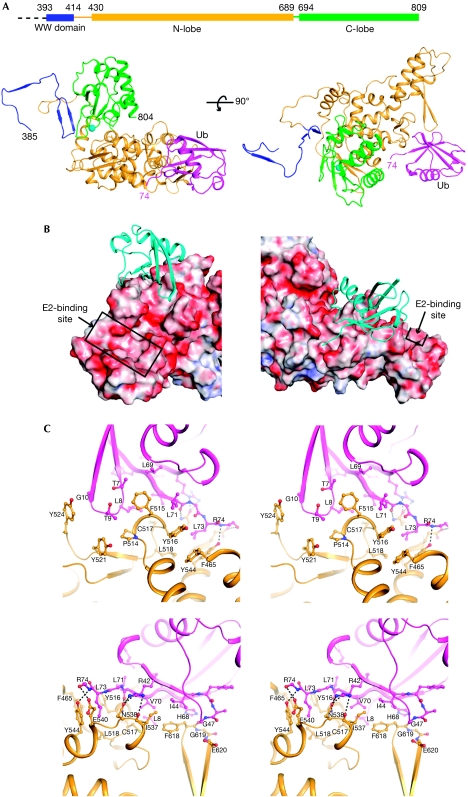
Residues 384–809 of Rsp5, including one WW domain and the C-terminal HECT domain, were co-crystallized with ubiquitin. The crystal structure, determined up to 2.5 Å resolution, contains residues 385–804 of Rsp5 and 1–74 of ubiquitin (Table 1 and Fig 1A). No electron density was observed for residues 493–500 of Rsp5, suggesting that this region is unstructured. The HECT domain folds into the canonical bi-lobed structure, and the position of the C-lobe relative to the N-lobe is similar to that of E6AP (HPV E6 associated protein). A WW domain of an Rsp5/Nedd4 family member has not been previously visualized in relation to the HECT domain. The 15-amino-acid linker (residues 415–429) between the WW and HECT domains had little secondary structure, suggesting flexibility in the relative positioning of these two domains in solution.
Table 1. Data collection and refinement statistics.
|
Data collection
| |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 161.6, 50.3, 79.7 |
| α, β, γ (°) | 90.0, 116.7, 90.0 |
| Wavelength (Å) | 1.03326 |
| Resolution (Å) | 50–2.5 |
| Completeness (%) | 75.2 (33.8)* |
| Rsym | 14.2 (56.0) |
| I/σI† | 10.4 (3.4) |
| Redundancy | 5.3 (2.6) |
|
Refinement
| |
| Resolution (Å) | 50–2.5 |
| No. reflections‡ | 20,360 |
| Rwork/Rfree | 22.0/27.2 |
| No. of atoms | |
| Rsp5 | 3,437 |
| Ubiquitin | 587 |
| Water | 56 |
| B factors (Å2) | |
| Rsp5 | 53.7 |
| Ubiquitin | 102.3 |
| Water | 35.8 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.488 |
| Ramachandran statistics§ (%) | |
| Favoured | 95.63 |
| Outliers | 0.21 |
| *Highest resolution shell is given in parenthesis; | |
| †I/σI indicates the average of the intensity divided by its average standard deviation; | |
| ‡Number of reflections following diffraction anisotropy truncation; | |
| §Ramachandran statistics calculated with MolProbity (http://molprobity.biochem.duke.edu). | |
Figure 1.
Crystal structure of Rsp5384−809 in complex with ubiquitin. (A) Ribbon representation of two orthogonal views showing the WW domain in blue, N-lobe in yellow, C-lobe in green and Ub in pink. The first and last residues of Rsp5 and ubiquitin in the structure are indicated by their residue numbers. The sphere represents the catalytic cysteine (C777). (B) Two views of ubiquitin docking on Rsp5. Ubiquitin is shown as a ribbon representation in cyan. Rsp5 is shown in electrostatic surface potential representation. The E2-binding site is indicated by a box. (C) Stereo diagram of two views at the interface between Rsp5 (yellow) and ubiquitin (pink). Residues making direct contact with the opposite subunit are labelled and shown as a ball-and-stick model (O atoms, red; N atoms, blue). Hydrogen-bonding and salt-bridge interactions are indicated by dashed lines. C-lobe, C-terminal lobe; N-lobe, N-terminal lobe; Ub, ubiquitin.
Previous studies, on the basis of mutagenesis (French et al, 2009) and nuclear magnetic resonance spectroscopy (Ogunjimi et al, 2010), identified residues of Nedd4 family members involved in ubiquitin binding; here, we present the atomic details of the Rsp5–ubiquitin interaction. Ubiquitin sits on the surface of Rsp5 like a tripod (Fig 1B,C). One leg of the tripod is formed by a loop connecting the first and second β-strands of ubiquitin (residues T7 to G10), leading to van der Waals interactions with P514, F515, C517, Y521 and Y524 in Rsp5. The second leg consists of I44 to G47 and H68, which are engaged in van der Waals and electrostatic interactions with I537, F618, G619 and E620. The third leg is near the C-terminus of ubiquitin, including V70, inserting into a surface groove lined by residues F465, F515, Y516, C517, L518, I537, N538, E540, Y544 and F618. Together, the interaction surfaces on ubiquitin encompass the L8-I44-V70 hydrophobic patch that is recognized by a variety of ubiquitin-binding proteins (Dikic et al, 2009). The buried surface area at the N-lobe–ubiquitin interface was approximately 850 Å2 per subunit, suggesting that the interaction was of low affinity (see below). In addition, the R42 of ubiquitin forms three hydrogen bonds with Rsp5 at N538 and I537 (main chain atom). The distance between the Rsp5 active-site cysteine (C777) and the R74 of ubiquitin was 31 Å, suggesting that ubiquitin observed in the crystal does not represent a molecule that could exist in a thioester linkage. The position of ubiquitin is also different from that seen for ubiquitin in the Nedd4L/E2–ubiquitin complex, where ubiquitin made C-lobe contacts (Kamadurai et al, 2009). Binding of ubiquitin to the N-lobe is not predicted to interfere with the binding of E2 to the N-lobe (Fig 1B).
Binding site mutations
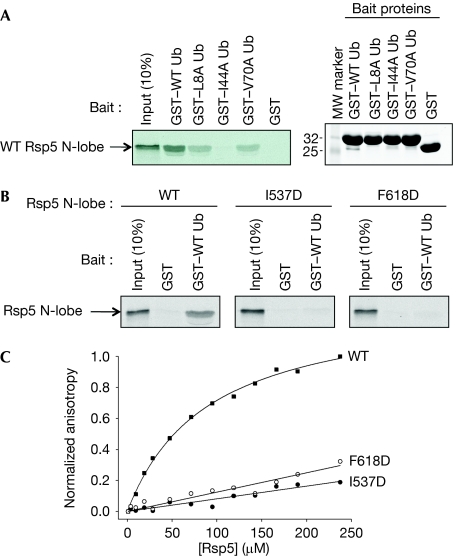
Figure 2A shows a binding assay using glutathione-S-transferase (GST) fusion proteins to ubiquitin or hydrophobic patch mutants of ubiquitin (L8A, I44A, V70A) and 35S-labelled in vitro-translated Rsp5 N-lobe protein. The N-lobe bound to GST–ubiquitin, but not to GST, and the L8A and V70A mutants showed reduced binding. Binding to I44A ubiquitin was undetectable. Several residues of Rsp5 at the interface with ubiquitin were also altered, including Y516, Y521, I537 and F618. Alteration of Y516 and F618 was previously reported to decrease binding of ubiquitin to Rsp5 (French et al, 2009). Y521 is equivalent to residue Y459 of Smurf2, which is implicated in binding of ubiquitin to this E3 (Ogunjimi et al, 2010). The I537D and F618D alterations were assayed for binding to GST–ubiquitin, and both N-lobe mutations reduced binding to ubiquitin (Fig 2B). The dissociation constant (Kd) for the HECT–ubiquitin interaction was determined by fluorescence anisotropy to be approximately 90 μM (Fig 2C), and again both the I537D and F618D mutants nearly completely eliminated ubiquitin binding in this assay.
Figure 2.
Binding of Rsp5 to ubiquitin. (A) Binding of 35S-labelled in vitro-translated Rsp5 N-lobe to GST–ubiquitin or to indicated ubiquitin mutants (left panel). Right panel shows a Coomassie blue-stained gel on input levels of GST–ubiquitin proteins used in the binding assay. (B) Binding of GST–ubiquitin to in vitro-translated Rsp5 N-lobe, wild-type or contact point mutants I537D or F618D. (C) Determination of the dissociation constant of the Rsp5–ubiquitin interaction. The normalized anisotropy of fluorescein-labelled ubiquitin is plotted as a function of Rsp5 concentration. The calculated Kd for wild-type Rsp5 was 90.6±6.4 μM; the binding constant for the two mutants was too low to be determined. GST, glutathione-S-transferase; Kd, dissociation constant; MW, molecular weight; N-lobe, N-terminal lobe; Ub, ubiquitin; WT, wild type.
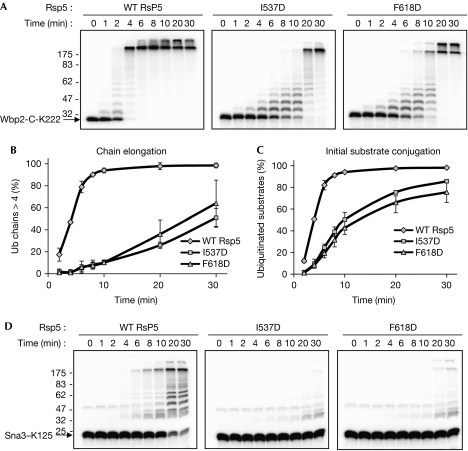
The mutated Rsp5 proteins (Y516A, Y521A, I537D and F618D) were assayed for in vitro ubiquitination of two substrate proteins, both of which contain a single lysine residue. This simplified the reaction products and allowed unambiguous determination of rates of initial substrate ubiquitination and polyubiquitination. Wbp2-C-K222 is a C-terminal fragment of human WW domain binding protein 2 (Wbp2), with K-to-R mutations that leave only K222 intact. Yeast Sna3 is a physiological target of Rsp5, and Sna3–K125 contains K-to-R mutations that leave only K125 intact (Stawiecka-Mirota et al, 2007). Fig 3A shows a time course of ubiquitination of purified Wbp2-C-K222 by Rsp5 and two ubiquitin-binding site mutants, I537D and F618D. Both mutants showed a strong defect in the rates of ubiquitination compared with wild-type Rsp5. The rate of chain elongation was determined as described previously (Kim & Huibregtse, 2009), by quantifying the fraction of the total modified substrate molecules that contained chains of more than four ubiquitin molecules in length (Fig 3B). The rate of initial substrate conjugation was determined by quantifying the amount of unmodified Wbp2-C-K222 remaining at each time point (Fig 3C). These results indicated that the I537D and F618D proteins had defects in both initial substrate conjugation and in polyubiquitination. Y521A had a similar effect to F618A on initial ubiquitination and polyubiquitination, whereas Y516A had only a subtle effect on both phases of the reaction (supplementary Fig S2A online). Importantly, the Rsp5 N-lobe mutations did not affect the rate of ubiquitin thioester formation (supplementary Fig S3 online) or the behaviour of the purified Rsp5 proteins by gel filtration (data not shown), indicating that the mutations did not result in misfolded or unstable proteins.
Figure 3.
Rsp5 proteins defective for ubiquitin binding show a defect in chain elongation and initial substrate conjugation. (A) Time course of ubiquitination of Wbp2-C-K222 by wild-type Rsp5 and mutant forms of Rsp5. (B,C) The results from the three independent experiments conducted as in A were quantified, and the graphs show (B) the percentage of ubiquitinated products that contained more than four ubiquitin molecules, or (C) the percentage of total substrate that was ubiquitinated. (D) As in A, with 32P-labelled Sna3–K125 as the substrate. Ub, ubiquitin; WT, wild type.
Similar results were seen with the Sna3–K125 substrate, although the rate of ubiquitination by wild-type Rsp5 was slower for this substrate. For the I537D and F618D mutants, a small amount of the input substrate had been ubiquitinated, even at late time points (Fig 3D). Polyubiquitination by the mutant forms of Rsp5 with this substrate was too defective to be accurately quantified. In addition, as with Wbp2, the Y521A mutation showed a stronger defect in Sna3 ubiquitination, whereas the effect of the Y516A mutation was less severe (supplementary Fig S2B online).
Rsp5, similarly to many HECT E3 enzymes, will ‘auto-conjugate’ ubiquitin in vitro to one or more of its own lysine residues, and we examined the effect of the N-lobe mutations on HECT domain autoubiquitination. Fig 4A (top panels) shows a time course of autoubiquitination of a HECT domain-containing fragment of Rsp5 (amino acids 420–809) and the corresponding I537D and F618D mutants. Preliminary experiments showed that the only acceptor site in this truncated Rsp5 protein is K432 (the canonical HECT domain beginning at residue 460). A time-dependent increase in polyubiquitin conjugates was observed, with the appearance of products at the top of the gel by 8 min. By contrast, the I537D Rsp5 HECT showed a slower rate of formation of polyubiquitin chains, with most chains never exceeding four ubiquitins in length, even after 20 min. The F618D HECT showed a less-severe defect in chain elongation (see Fig 4B). The rate of initial substrate modification of the wild-type and mutant HECT domains was similar, indicating that the ubiquitin-binding site mutations did not affect conjugation of the first ubiquitin molecule. To confirm this, the assay was performed in the presence of lysine-less ubiquitin (Fig 4A, lower panels), and the rate of monoubiquitination of wild-type Rsp5 and the mutants was similar (Fig 4B, lower panel).
Figure 4.
Rsp5 proteins defective for ubiquitin binding are defective for autoubiquitination. (A) Time course of autoubiquitination of wild-type and mutant forms of the Rsp5 HECT domain, in the presence of either wild-type ubiquitin (top panels) or K(0) ubiquitin (lower panels). (B) The results from the experiments shown in A were quantified and the graph shows the percentage of ubiquitinated products that contained more than four ubiquitin molecules (top panel) or the percentage of substrate that was monoubiquitinated (lower panel). Ub, ubiquitin; WT, wild type.
Finally, although yeast cells expressing the N-lobe ubiquitin-binding site mutants as the sole source of Rsp5 were viable, they showed a strong temperature-sensitive growth defect (supplementary Fig S4A online). On the basis of earlier work showing that chain-type specificity is a function of the C-lobe, we predicted that the short chains formed by the I537D and F618D mutants would still be primarily K63-linked. This was found to be the case, as shown in supplementary Fig S4B online.
Substrate ubiquitination is enhanced by fusion to ubiquitin
We noted that a C-terminal fragment of Sna3 (Sna3C; amino acids 68–133) was a poor substrate for ubiquitination, although it contained the physiological site of ubiquitination (K125) and the crucial PY motif (PPAY; amino acids 106–109; Oestreich et al, 2007). A possible explanation for this is that it is unable to productively engage with the HECT domain. If so, then a translational fusion of ubiquitin to Sna3C might rescue its ubiquitination as a result of the ubiquitin portion of the molecule engaging the N-lobe. Fig 5A shows a schematic of constructs made for testing this. Sna3C was inefficiently ubiquitinated, whereas fusion of ubiquitin to Sna3C (Ub–Sna3C) rescued its ubiquitination (Fig 5B). Ubiquitination of the fusion protein was dependent on both the PY motif within the Sna3 portion of the protein (see Ub–Sna3CPAAA) and hydrophobic patch residues of the ubiquitin portion of the molecule (see UbI44D–Sna3C). Ubiquitination was also dependent on the N-lobe ubiquitin-binding site, as both the I537D and F618D Rsp5 mutants were defective for ubiquitination of Ub–Sna3C (Fig 5C). These results support a model in which the N-lobe ubiquitin-binding domain functions to recruit a substrate-conjugated ubiquitin molecule to promote conjugation of the next ubiquitin molecule in a polyubiquitin chain.
Figure 5.
Ubiquitin–Sna3C is targeted more efficiently than Sna3C. (A) Schematic diagram of wild-type Sna3, Sna3C, ubiquitin–Sna3C and mutant forms of ubiquitin–Sna3C. (B) Time courses of ubiquitination of the indicated substrates by wild-type Rsp5. (C) Time course ubiquitination of ubiquitin–Sna3C by wild-type Rsp5, I537D Rsp5 and F618D Rsp5. Ub, ubiquitin; WT, wild type.
Discussion
The structural and biochemical results presented here indicate that the N-lobe ubiquitin-binding site is crucial for polyubiquitination by Rsp5. We propose that the N-lobe promotes polyubiquitination by localizing the distal end of a ubiquitin chain in the vicinity of the active site for addition of the next ubiquitin molecule. A similar conclusion was reached in a study on Smurf2 (Ogunjimi et al, 2010) and in the accompanying study by Maspero et al (2011), which examined the human Nedd4 HECT E3. These conclusions differ from an earlier study that concluded that the N-lobe ubiquitin-binding site acted to restrict polyubiquitination (French et al, 2009). A distinction in the design of our ubiquitination experiments was that all the substrates examined, including the HECT domain in the autoubiquitination experiments, contained only a single lysine acceptor. This simplified the reaction products and allowed us to unambiguously follow the initial substrate-ubiquitination event and the length and rate of formation of polyubiquitin chains.
Our results suggest that the N-lobe ubiquitin-binding site might also function as a general substrate recruitment site, to promote the initial ubiquitination event. The relative affinities of a target protein for these two regions could influence both the efficiency and type of ubiquitination products. Interestingly, the N-lobe ubiquitin-binding site has so far only been described for the WW domain-containing subfamily of HECT E3s (that is, yeast Rsp5, human Nedd4 and Smurf2). Not surprisingly, the contact points for ubiquitin binding are well conserved among this subfamily, but less well among some other HECT E3s (supplementary Fig S5 online). It is therefore not clear whether the N-lobe ubiquitin-binding domain is a common feature of all HECT–ubiquitin ligases or whether variations in mechanism among HECT E3s might determine the nature of the final ubiquitination products.
Methods
Crystallography. Rsp5384−809 and ubiquitin were expressed as GST fusion proteins in Escherichia coli and purified by means of Superdex 200, 16/60 chromatography (GE Healthcare). GST was removed by TEV protease, and proteins were purified by size-exclusion chromatography and mixed at a 1:2 molar ratio (Rsp5/Ub) in 10 mM Tris–HCl (pH 8.0), 250 mM NaCl and 5 mM dithiothreitol (DTT). Crystals were obtained by the sitting drop vapour diffusion method at 20°C, against a reservoir solution containing 13.5% PEG 4000, 0.2 M MgCl2 and 0.1 M Tris–HCl (pH 9.0; Rsp5 at 0.4 mM, Ub 0.8 mM; protein/precipitant ratio 1:1). Crystals were flash-frozen in reservoir solution supplemented with 10% glycerol. Diffraction data were collected at 100 K on beamline 23-ID at the Advanced Photon Source (GMCA-CAT, Argonne National Laboratory; MARmosaic 300 mm CCD detector) and processed with HKL2000 (Otwinowsi & Minor, 1997). The structure was determined by molecular replacement using Phaser (McCoy et al, 2007) and the N- and C-lobes of WWP1 (Protein Data Bank code: 1ND7) as the search models. ubiquitin (Protein Data Bank code: 1UBQ) was placed manually according to an electron density map calculated using phases from a model containing the HECT domain (supplementary Fig S1 online). Given that diffraction data were severely anisotropic (supplementary Table S1 online), the output-scaled data were imported into a diffraction anisotropy server (Strong et al, 2006) that uses an ellipsoid truncation and scaling at the high-resolution boundary after elimination of weak or missing reflections. The pruned data were then used for structure refinement. The model was improved by manual building in Coot (Emsley & Cowtan, 2004) and refinement using CNS (Brunger et al, 1998) and REFMAC5 (CCP4; Murshudov et al, 1997). Further refinement was carried out in Phenix (CCP4; Afonine et al, 2005) with translation/libration/screw (TLS) parameters generated by the TLSMD server (Painter & Merritt, 2006). The final structure was refined to 2.5 Å with R and Rfree factors of 22.0% and 27.2%. The Protein Data Bank code for this structure is 3OLM.
Biochemical assays. Plasmids for the bacterial expression of Wbp2-C-K222, Sna3–K125, wild-type Rsp5, Rsp5 HECT domain and Ubc4 were described previously (Kim & Huibregtse, 2009). 32P-labelled proteins, generated as described previously (Kim & Huibregtse, 2009), were incubated with 5 μg/ml of Rsp5 in the presence of 10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 0.1 mM DTT, 50 μg/ml of ubiquitin (Boston Biochem), 1.25 μg/ml of yeast E1 (Boston Biochem), 1.25 μg/ml of Ubc4 (40 μl total volume) and 10 mM ATP. All ubiquitination reactions were initiated by the addition of ubiquitin and stopped by the addition of 4 × SDS–polyacrylamide gel electrophoresis (PAGE)-loading buffer containing 0.4 M DTT. Ubiquitinated products were quantified using a Bio-Rad Phosphoimager. GST-ubiquitin proteins were purified on glutathione beads (Novagen). Rsp5 N-lobe proteins were generated by in vitro translation (Promega) in the presence of 35S-methionine and incubated with GST–ubiquitin on beads in buffer (50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 0.1% NP40) for 2 h at 4°C. Total proteins were recovered by boiling the beads in the presence of SDS–PAGE-loading buffer and analysed by SDS–PAGE and autoradiography. For fluorescence polarization assays, ubiquitin was labelled with fluorescein-5-maleimide according to the manufacturer's protocol (Molecular Probes). Fluorescence anisotropy at 25°C was measured using a Beacon 2000 fluorescence polarization instrument (VWR), with the excitation and emission wavelengths of 488 and 535 nm, respectively. Data were averaged from three measurements for wild-type Rsp5 and from two measurements for the mutant Rsp5 proteins. Data were fitted to a single-site binding model using nonlinear regression with SigmaPlot (Eletr et al, 2005).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Photon Source beamline 23-ID for data collection assistance. J.C. is an investigator of the Howard Hughes Medical Institute. This work was supported by a grant to J.M.H. from the National Institutes of Health (CA72943).
Footnotes
The authors declare that they have no conflict of interest.
References
- Afonine PV, Grosse-Kunstleve RW, Adams PD (2005) A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr 61: 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B (2005) E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol 12: 933–934 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- French ME, Kretzmann BR, Hicke L (2009) Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem 284: 12071–12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP–UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science 286: 1321–1326 [DOI] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin–HECT(NEDD4L) complex. Mol Cell 36: 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 29: 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, Polo S (2011) Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep [Epub ahead of print 11 March] doi:10.1038/embor.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ (2007) Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell 18: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL (2005) Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell 19: 297–308 [DOI] [PubMed] [Google Scholar]
- Ogunjimi AA, Wiesner S, Briant DJ, Varelas X, Sicheri F, Forman-Kay J, Wrana JL (2010) The ubiquitin binding region of the Smurf HECT domain facilitates polyubiquitylation and binding of ubiquitylated substrates. J Biol Chem 285: 6308–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowsi Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA (2006) TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr 39: 109–111 [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM (1995) Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373: 81–83 [DOI] [PubMed] [Google Scholar]
- Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P (2007) Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic 8: 1280–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.