EMBO Rep (2011) advance online publication. doi:; DOI: 10.1038/embor.2011.28
The recognition of apoptotic cells by phagocytes is a complex, yet highly orchestrated event. Many receptors have been identified that recognize phosphatidylserine (PS; Fig 1)—which is exposed on early apoptotic cells—leading to downstream signalling and apoptotic cell engulfment. In a paper published this month in EMBO reports, the receptor for advanced glycation end-products (RAGE) is described as a new PS receptor on alveolar macrophages that participates in the clearance of apoptotic cells (He et al, 2011).
…[RAGE] is described as a new phosphatidylserine receptor on alveolar macrophages that participates in the clearance of apoptotic cells
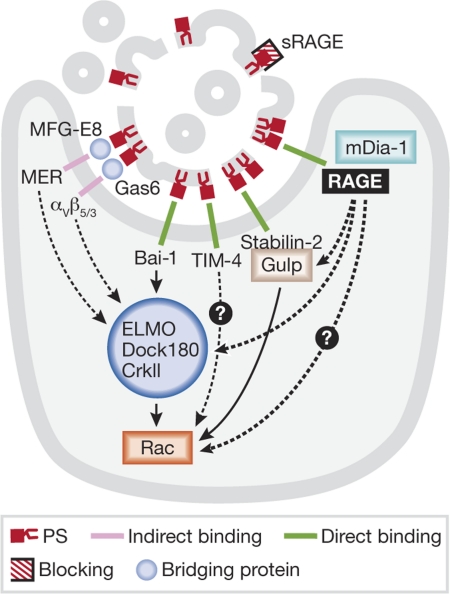
Figure 1. Phosphatidylserine-dependent apoptotic cell recognition.
Schematic of the known PS receptors and downstream signalling to Rac. Dashed lines indicate unknown signalling mechanisms. PS, phosphatidylserine; RAGE, receptor for advanced glycation end-products; sRAGE, soluble RAGE.
More than 200 billion cells undergo apoptosis every day in a human body, yet few apoptotic cells are detected in healthy tissue (Ravichandran, 2010). Apoptotic cells are generated during development, as part of normal homeostatic turnover and in disease states. The efficient clearance of apoptotic cells is crucial to prevent them from becoming secondarily necrotic, thereby limiting the immune response to apoptotic cell-derived self-antigens (Green et al, 2009). Disruptions to the clearance of apoptotic cells are linked to several diseases including atherosclerosis, chronic inflammation and autoimmunity (Elliott & Ravichandran, 2010).
More than 200 billion cells undergo apoptosis every day in a human body, yet few apoptotic cells are detected in healthy tissue
Apoptotic cell engulfment can be divided into several steps. The first is the release of ‘find-me’ signals—such as triphosphate nucleotides (ATP and UTP), sphingosine-1-phosphate (S1P), lysophosphatidylcholine (LPC) and the chemokine CX3CL1—by apoptotic cells (Ravichandran, 2010). Then, phagocytes sense the find-me signals and migrate toward the apoptotic cell. When they are in close proximity, recognition is mediated by the interaction between engulfment receptors on phagocytes and ligands, known as ‘eat-me’ signals, that are expressed on the dying cells (Ravichandran, 2010). The best-studied eat-me signal is PS, which is flipped from the inner leaflet to the outer leaflet of the plasma membrane during early apoptosis. Many receptors have been linked to the recognition of the exposed PS on apoptotic cells, and they are discussed below. The recognition of an apoptotic cell results in a downstream signalling cascade that leads to cytoskeletal rearrangement of the phagocytic membrane and subsequent engulfment of the apoptotic cell. Once the corpse is internalized, the phagocyte must process and digest the cellular contents.
The exposure of PS on the outer leaflet of the membrane is the most-characteristic marker of an apoptotic cell. Phagocytes can recognize PS directly through receptors such as Bai1, TIM-4 and stabilin 2, or through soluble bridging molecules that bind to both PS and specific phagocyte receptors. For example, bridging molecules MFG-E8 and Gas6 interact with αVβ3/5 and MER on the phagocytic membrane, respectively. Other eat-me signals and the molecules that bind to them have been characterized: thrombospondin is recognized by the vitronectin receptor, calreticulin by LRP1, oxidized LDL by scavenger receptors, ICAM3 might bind to CD14 and altered sugars bind to lectins (Lauber et al, 2004). Not all receptors need to be engaged for engulfment to occur, and different cell types have different receptor-expression levels.
In a paper published this month in EMBO reports, the Yamamoto team identify RAGE as a new type of PS receptor on macrophages (He et al, 2011). There are two functional forms of RAGE, an abundant full-length transmembrane form that can initiate signalling through its intracellular tail, and a soluble isoform (sRAGE) that acts as a decoy receptor. RAGE is characteristically regarded as a pro-inflammatory receptor and has a variety of ligands, including advanced glycation end-products (AGEs) and many other damage-associated molecular patterns (DAMPs; Sims et al, 2010). One ligand in particular—high-mobility group protein B1 (HMGB1)—is released by cells undergoing necrosis and has been shown to bind to RAGE and induce inflammation (Sims et al, 2010). Therefore, RAGE might function during pro-inflammatory conditions and—as proposed by He and colleagues—during the anti-inflammatory process of apoptotic cell clearance. RAGE is mainly expressed in the lungs, but levels of it quickly increase at sites of inflammation, mostly on inflammatory and epithelial cells. Given the multitude of RAGE ligands and its inducible expression levels, RAGE is implicated in a variety of inflammation-related pathological states such as neurological and pulmonary disorders, vascular disease, cancer and diabetes (Sims et al, 2010).
He and colleagues suggest that RAGE is a PS receptor during apoptotic cell engulfment in alveolar macrophages (He et al, 2011). Furthermore, sRAGE—which can bind to PS and apoptotic thymocytes—acts as a decoy and inhibits RAGE recognition of PS. By using PS liposomes as an artificial apoptotic target, the authors find RAGE in areas of the membrane in which a pseudopod forms to engulf a PS liposome. Additionally, sRAGE can compete with transmembrane RAGE to block the recognition of PS by the phagocyte and subsequently decrease the engulfment of apoptotic cells. Under homeostatic conditions, alveolar macrophages isolated from RAGE-deficient mice have defects in phagocytosis of apoptotic thymocytes. In a model of lung injury induced by lipopolysaccharide administration, RAGE-deficient mice accumulate neutrophils in the alveolar space and RAGE-deficient macrophages have defects in neutrophil engulfment. Previous works have implicated RAGE expression and/or upregulation in inflammatory conditions. In fact, genetic deletion of RAGE in mice can result in attenuated atherosclerosis, resistance to septic shock and reduced diabetic kidney disease (Ramasamy et al, 2010). Apoptotic cell clearance is generally an immunologically silent process and, therefore, if RAGE significantly contributes to engulfment, RAGE-deficient mice would be expected to have defects in cell clearance, leading to enhanced inflammation and disease. However, this does not seem to be the case. Thus, future studies should examine cell-type specific deletions of RAGE to clarify its apparently contradictory role in cell clearance and inflammation in these diseases.
Given that several modes of PS recognition have been identified (Ravichandran, 2010), there must be some redundancy. The way in which RAGE contributes to this scenario remains to be investigated. Analysis of the expression levels of each PS receptor on different cell types will also help to define their relative importance in individual cells. As RAGE is highly expressed in the lung, it would be interesting to analyse its contribution to apoptotic cell engulfment in this tissue, in comparison with the other PS receptors. Furthermore, RAGE is induced by inflammation, suggesting that it is probably important during disease states to facilitate engulfment and reduce inflammation in the microenvironment.
Another interesting question that remains is how RAGE signals to the phagocyte for engulfment. RAGE signalling results in pro-inflammatory cytokine production through activation of NF-κB (Yan et al, 1994), which seems to be different from the production of anti-inflammatory cytokines—such as IL-10 and TGFβ—by phagocytes during cell engulfment. However, as several RAGE ligands exist, the way in which they bind to RAGE could result in differential signalling. RAGE has also been shown to interact with mouse Dia1, leading to downstream activation of Rac1 and Cdc42, and cell migration (Hudson et al, 2008). Now, He and colleagues suggest that RAGE signals to Rac1 through Dia1 in the context of apoptotic cell clearance, as RAGE-deficient macrophages have decreased Rac1 activity in response to PS-liposome engulfment. Two evolutionarily conserved Rac-dependent pathways have been identified to mediate corpse internalization. Engagement of some engulfment receptors such as Bai1, results in Rac activation through the ELMO–Dock180–CrkII complex. ELMO and Dock180 mediate the exchange of GDP to GTP on Rac, whereas CrkII has been proposed to function as an adaptor protein. Another pathway involves signalling from the engulfment receptor LRP1 or stabilin 2, leading to Rac activation through the engulfment adaptor protein (GULP). Additional work is necessary to determine whether RAGE–mDia1 signalling constitutes a third intracellular signalling pathway for cell engulfment.
Another interesting question that remains is how RAGE signals to the phagocyte for engulfment
The study from the Yamamoto team identifies RAGE as a new PS-recognition molecule implicated in apoptotic cell-clearance in the lung. As each new receptor is identified, we are reminded of the redundancy and cell-type-specific expression of PS receptors. Defects in apoptotic cell-clearance lead to a variety of inflammatory diseases, including cardiovascular and autoimmune diseases. This study could also open an interesting therapeutic avenue; if sRAGE blocks the recognition of PS by RAGE and other PS receptors, it might be beneficial as a therapy by enhancing cell clearance and decreasing the severity of cell-clearance-associated diseases.
References
- Elliott MR, Ravichandran KS (2010) J Cell Biol 189: 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR et al. (2009) Nat Rev Immunol 9: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M et al. (2011) EMBO Rep [Epub 11 Mar] doi:; DOI: 10.1038/embor.2011.28 [DOI] [Google Scholar]
- Hudson BI et al. (2008) J Biol Chem 283: 34457–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber K et al. (2004) Mol Cell 14: 277–287 [DOI] [PubMed] [Google Scholar]
- Ramasamy R, Yan SF, Schmidt AM (2010) Amino Acids doi:; DOI: 10.1007/s00726-010-0773-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS (2010) J Exp Med 207: 1807–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims GP et al. (2010) Annu Rev Immunol 28: 367–388 [DOI] [PubMed] [Google Scholar]
- Yan SD et al. (1994) J Biol Chem 269: 9889–9897 [PubMed] [Google Scholar]